Abstract
An elevated serum level of NM23‐H1 protein is found in acute myelogenous leukemia (AML), and predicts a poor treatment outcome in AML patients. To investigate the potential pathological link between the elevated serum level of this protein and poor prognosis, we examined the extracellular effects of recombinant NM23‐H1 protein on the in vitro growth and survival of primary cultured AML cells at concentrations equivalent to the levels found in the serum of AML patients. Extracellular NM23‐H1 protein promoted the in vitro growth and survival of AML cells and this activity was associated with the cytokine production and activation of the MAPK and signal transducers and activators of transcription signaling pathways. Inhibitors specific to MAPK signaling pathways inhibited the growth‐ and survival‐promoting activity of NM23‐H1. These findings indicate the novel biological action of extracellular NM23‐H1 and its association with poor prognosis, and suggest an important role for extracellular NM23‐H1 in the malignant progression of leukemia and a potential therapeutic target for these malignancies. (Cancer Sci 2009; 100: 1885–1894)
The NM23 gene was identified by differential hybridization of a cDNA library with total RNA extracted from slightly and highly metastatic melanoma cell lines.( 1 ) NM23 genes have been identified as a family of genes encoding different isoforms of nucleoside diphosphate kinase (NDPK).( 2 ) NM23 genes play critical roles in cellular proliferation, differentiation, oncogenesis, and tumor metastasis, and the mechanisms of these pleiotropic effects are not well understood.( 3 , 4 ) Eight isotypes of the human NM23 gene (NM23‐H1, NM23‐H2, NM23‐H3/DR‐NM23, NM23‐H4, NM23‐H5, NM23‐H6, NM23‐H7, and NM23‐H8) have been identified,( 5 ) of which only NM23‐H1 and NM23‐H2 have been studied extensively in human cancers. The level of NM23‐H1 expression is inversely correlated with the tumor's metastatic potential in experimental rodent cells and in human tumors, such as breast, ovarian, cervical, and gastric cancer, hepatocellular carcinoma, and melanomas.( 4 ) Exogenous overexpression of NM23‐H1 reduces the metastatic potential of multiple types of cancer cells and suppresses in vitro tumor cell motility and invasion.( 6 ) NM23‐H1 is therefore implicated in the regulation of metastasis in a variety of human cancers and its overexpression predicts a favorable patient prognosis; however, overexpression of the NM23‐H1 gene in other neoplasms, including neuroblastoma and hematological malignancies, is indicative of a poor patient prognosis.( 7 , 8 , 9 , 10 , 11 ) The significance of NM23‐H1 overexpression as a prognostic factor is dependent on tumor cell type, although the mechanism of this discrepancy is unknown.
We previously reported that a non‐differentiating myeloid leukemia cell line produced differentiation‐inhibiting factors( 12 , 13 ) and purified one of these factors as a homologue of mouse NM23‐M2.( 14 ) NM23 genes were overexpressed in acute myelogenous leukemia (AML) cells, and a higher level of NM23 expression was correlated with poor prognosis in AML.( 10 , 15 , 16 ) The amount of intracellular NM23‐H1 protein was inversely correlated with differentiation as decreased protein expression was observed during hematopoietic maturation.( 17 , 18 ) We also determined the plasma level of NM23‐H1 protein by ELISA and assessed the association between this and the clinical outcome of patients with AML.( 19 , 20 ) The plasma concentration of NM23‐H1 was higher in patients with AML than in normal controls, and significantly contributed to their prognosis, suggesting that both cellular and extracellular levels of NM23‐H1 in AML play an important role in leukemia cell differentiation and clinical outcome in AML patients. In addition to AML, the clinical significance of the plasma level of NM23‐H1 protein was also shown in neuroblastoma and various types of malignant lymphoma in our study, which involved a number of different institutions and numerous case studies.( 11 , 20 , 21 )
Extracellular NM23 proteins have been reported in the conditioned medium of some tumor cell lines, in body fluids, and on the cell surface of tumor cell lines.( 14 , 19 , 22 , 23 , 24 ) High concentrations of NM23 protein are found in the serum and body fluid of patients with tumors overexpressing NM23, and it is strongly suggested that serum NM23‐H1 protein is derived from tumor cells.( 20 , 25 ) The mechanisms by which NM23‐H1 protein is secreted into the extracellular environment and affects the outcome of patients are unclear. Very little information is available concerning extracellular expression and function, although many studies have examined the expression of intracellular NM23 proteins; therefore, we focused on extracellular NM23‐H1 protein derived from tumor cells because its clinical significance is greater than that of intracellular overexpression and elevated extracellular expression of NM23 has not been found in normal healthy plasma.( 19 ) In the present study, we investigated the extracellular functions of recombinant NM23 (rNH23) proteins in the survival and growth of AML cells and their association with the poor prognosis of AML patients.
Materials and Methods
Normal and leukemia human peripheral blood mononuclear cells and cell culture. AML cells were provided by the Department of Hematology, Saitama Cancer Center after the ethics committee of Saitama Cancer Center approved the study design. Peripheral blood mononuclear cells (PBMNC) from AML patients were prepared using Ficoll Paque‐Plus centrifugation (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and used as AML samples. AML cells were cryopreserved in liquid nitrogen in a freezing mix of 10% (v/v) dimethyl sulfoxide until use. AML cells were cultured at 1 × 106 cells/mL in RPMI‐1640 (Sigma‐Aldrich, St Louis, MO, USA) supplemented with 10% (v/v) heat‐inactivated fetal bovine serum (HyClone, Logan, UT, USA) at 37°C in a 5% CO2 humidified incubator. Cultures were supplemented with polymyxin B (1.25 µg/mL Polymyxin B Sulphate; Sigma‐Aldrich) to inhibit contaminating endotoxin and lipopolysaccharide (LPS). The polymyxin B dose was determined from separate experiments to neutralize the concentration of 100 ng/mL LPS (Difco, Detroit, MI, USA); this polymyxin B dose was neither toxic nor stimulating for the growth and survival of AML cells (data not shown).
Assay of cell growth and survival. The cells were seeded at 1 × 106 cells/mL in a 24‐well multidish (Becton Dickinson, Franklin Lakes, NJ, USA). After culture with or without rNM23 proteins for the indicated days, viable cells were examined using a modified MTT (Sigma‐Aldrich) assay, in which 100 µL MTT solution (1 mg/mL in PBS) was added to each well and cells were incubated for 4 h. The cells were then centrifuged at 1000 g for 10 min, the precipitates dissolved in 1 mL dimethyl sulfoxide, and their absorption determined at 560 nm.
Recombinant NM23 proteins. NM23‐H1 and NM23‐H2 coding regions were cloned into the pGEX‐2T expression vector with a GST tag (Amersham Pharmacia Biotech) and the vectors were transferred into Escherichia coli (JM109; TaKaRa, Shiga, Japan) according to the manufacturer's instructions. DNA sequencing of both strands confirmed the correct orientation of positive clones. Bacterial cultures for induction were prepared in fresh L‐broth media containing 100 µg/mL ampicillin, and recombinant protein expression was induced with 1 mM isopropyl β‐d‐thiogalactopyranoside (Amersham Pharmacia Biotech). Recombinant GST–NM23 fusion protein was prepared with the PediPack GST Purification Module (Amersham Pharmacia Biotech) according to the manufacturer's instructions. GST–NM23 fusion proteins were treated with thrombin, and rNM23 proteins were eluted from the GST column and purified to homogeneity. Contaminating thrombin in rNM23 fractions was removed using Benzamidine Sepharose 6B (Amersham Pharmacia Biotech). The purity and quality of each preparation was assessed using both Coomassie‐stained SDS‐PAGE gels and western blotting using anti‐NM23‐H1, anti‐NM23‐H2, and anti‐NM23‐H1/H2 (data not shown). Using site‐directed mutagenesis of cDNA encoding NM23‐H1, we created an enzymatically inactive mutant NM23‐H1His protein, in which His118 was replaced by Phe.( 26 ) Mutant NM23‐H1His was produced and purified by the same procedure as for wild‐type NM23 proteins described above. All rNM23 fractions were tested for LPS content by the quantitative chromogenic Limulus amebocyte lysate assay (A Cambrex Company, BioWhittaker, Walkersville, MD, USA); preparations contained 20 pg or less of endotoxin/µg rNM23.
Cytokine determination. We analyzed NM23‐regulated cytokines in conditioned medium (CM) of AML PBMNC treated with or without rNM23‐H1 using a human cytokine antibody array system (RayBiotech, Norcross, GA, USA), which simultaneously detects the expression of various cytokines, in accordance with the manufacturer's instructions. The human Inflammation Antibody Array III spotted 40 different cytokine antibodies. To quantify cytokine levels, conventional sandwich ELISA was carried out according to the manufacturer's instructions. The Quantikine Immunoassay (R&D Systems, Minneapolis, MN, USA) for human GM‐CSF, IL‐1β, IL‐8, TNFα, IL‐6, and IP‐10 was used.
Western blot analysis. Cells were packed after washing with cold PBS, and lysed at a concentration of 2 × 107 cells/mL in sample buffer, β‐ME Sample Treatment for Tris SDS (Cosmo Bio, Tokyo, Japan). Whole cell lysates were developed on SDS‐PAGE (10–20% gradient gels). Proteins were transferred electrophoretically from the gel to an Immobilon‐P membrane (Millipore, Bedford, MA, USA) and immunoblotted with anti‐p38 MAPK (Cell Signaling, Beverly, MA, USA), anti‐phospho‐p38 MAPK (Thr180/Tyr182), anti‐p44/42 MAPK, anti‐phospho‐p44/42 MAPK (Thr202/Tyr204), anti‐signal transducers and activators of transcription (STAT) 1, anti‐phospho‐STAT1 (Tyr701, Ser727), anti‐STAT3, anti‐phospho‐STAT3 (Ser727, Tyr705), anti‐STAT‐5, and anti‐phospho‐STAT5 (Tyr694). Unbound antibody was removed by washing with Tris‐buffered saline (pH 7.2) containing 0.5% Tween 20. The membrane was then incubated with the secondary HRP‐conjugated antibody. After extensive washing, protein bands were detected with the Immune‐Star HRP Substrate kit (Bio‐Rad, Hercules, CA, USA), and detected using a Fuji Lumino Image Analyzer LAS‐1000system (Fuji Film, Tokyo, Japan). Blots were stripped using the Western Blot Recycling kit (Alpha Diagnostic, San Antonio, TX, USA) and probed with an antibody against α‐tubulin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) to confirm equal loading.
Results
Effect of extracellular NM23 protein on the in vitro growth and survival of primary cultured AML cells. We examined the effect of rNM23‐H1 protein on primary cultured AML cells obtained from 14 patients (Fig. 1a). These cells were incubated with or without rNM23‐H1 protein at a concentration of 100 ng/mL. The effects on cell growth and survival were measured by MTT assay. The rNM23‐H1 protein promoted the growth and survival of primary cultured AML cells in 11 cases (Fig. 1a,b), although sensitivity to extracellular NM23‐H1 depended on the case. Blast percentages in PBMNC from AML are shown in Figure 1(a). We found no association between the percentage of blasts in PBMNC and rNM23 responsibility in AML cells (Fig. 1a). Although the optimum concentration and time of treatment showed sample variation, extracellular rNM23‐H1 protein overall promoted the growth and survival of primary cultured AML cells at similar concentrations in AML patients (Fig. 1b). Representative data are shown in Figure 1(c) (case 9 on day 3) and Figure 1(d) (case 4 on day 6). AML cells (1 × 106 cells/mL) were incubated with or without rNM23‐H1 (Fig. 1c,d, left) and rNM23‐H2 (Fig. 1c,d, right) proteins in the presence or absence of polymixin B to eliminate any potential effect of endotoxin or LPS. Both rNM23‐H1 and rNM23‐H2 promoted the growth and survival of AML cells; therefore, this activity of NM23 is independent of H1/H2 isotypes. Although LPS also promoted the growth and survival of AML cells, this activity was completely inhibited by polymixin B (Fig. 1e; case 2 on day 5). The growth‐ and survival‐promoting activity of rNM23‐H1 was observed even in the presence of polymyxin B at concentrations neutralizing contaminating LPS (Fig. 1e); therefore, this activity was not due to endotoxin or LPS contamination. In fact, the endotoxin levels of rNM23 fractions used were less than 20 pg/mL (data not shown). Mutant NM23‐H1His protein, which does not have NDPK activity,( 26 ) also promoted the growth and survival of AML cells in the presence or absence of polymixin B (Fig. 1f). These results indicate that the activity of NM23 is independent of its NDPK activity.
Figure 1.
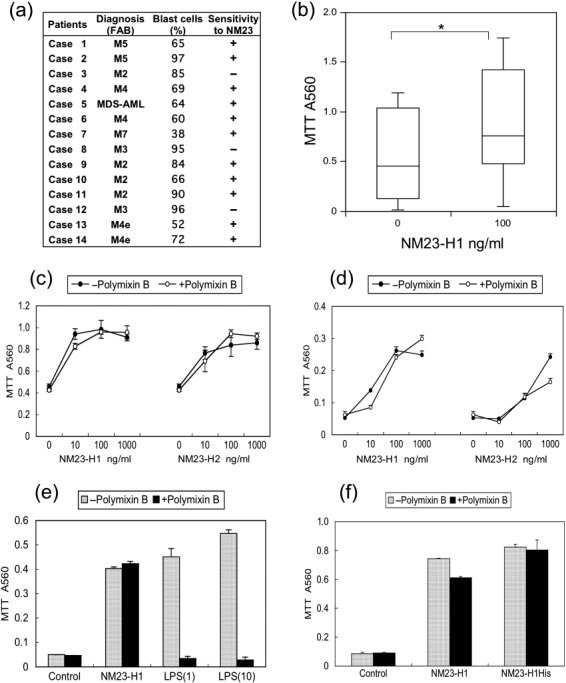
Growth‐ and survival‐promoting activity of extracellular recombinant (r) NM23 proteins on primary cultured acute myelogenous leukemia (AML) cells. (a) We used 14 cases of AML. +, NM23‐responsive cases; –, unresponsive cases. (b) AML cells (1 × 106 cells/mL, n = 14) were incubated with or without 100 ng/mL rNM23‐H1 protein. The effects on cell growth and survival were measured by MTT assay. Box plots: upper and lower lines indicate 10th and 90th percentiles, boxes indicate 25th and 75th percentiles. The line through each box indicates the median. Asterisk represents significant differences (P < 0.05) using Student's t‐test. (c–d) Representative growth‐ and survival‐promoting activity of rNM23‐H1 and rNM23‐H2 proteins in the presence or absence of polymixin B to eliminate any potential effect of LPS. (c) Case 9 on day 3; (d) case 4 on day 6. (e) Effect of LPS inhibitor, polymixin B (1.25 µg/mL), on growth‐ and survival‐promoting activity of extracellular rNM23‐H1 (100 ng/mL) or LPS (1 and 10 ng/mL) (case 2 on day 5). (f) Growth‐ and survival‐promoting activity of mutant NM23‐H1His protein (100 ng/mL), which does not have nucleoside diphosphate kinase (NDPK) activity (case 2 on day 5). Error bars, SD. FAB, French–American–British classification; MDS, myelodysplastic syndrome.
Protein expression and secretion of various cytokines in CM of AML cells treated with rNM23 protein. Four representative cases are shown in Figure 2(a). rNM23‐H1 promoted the growth and survival of AML cells (cases 1, 2, and 4) in a dose‐ and time‐dependent manner, although the pattern was case dependent (Fig. 2a, left panels). It is known that the growth and survival of AML cells is regulated by various cytokines; therefore, we examined cytokine levels in the CM of AML cells treated with or without rNM23‐H1, using cytokine antibody array (Fig. 2b) and ELISA (Fig. 2c). We could detect various cytokines in 48‐h CM of AML cells (cases 1, 2, and 4) whose growth and survival were promoted by rNM23‐H1 using human inflammation antibody array (Fig. 2a, right panels), although the pattern of cytokine expression was not consistent among the cases. These cytokines included IL‐1β, IL‐6, IL‐8, I‐309, IL‐10, GM‐CSF and RANTES (Fig. 2a, right panel). We next quantified representative cytokine levels (TNFα, IL‐6, IL‐1β, GM‐CSF, IL‐8, and IP‐10) by sandwich ELISA (Fig. 2c). The bars in each graph show rNM23‐induced cytokine levels in AML in cases 1, 2, 3, and 4, respectively; for example, TNFα (2‐h CM), IL‐6 (2‐h CM), IL‐1β (48‐h CM), GM‐CSF (48‐h CM), and IL‐8 (48‐h CM) were induced during the CM of AML cells (cases 1, 2, and 4) treated with rNM23‐H1, whereas none of these cytokines, except IL‐8, was detected in the CM of NM23‐unresponsive case 3 (Fig. 2a,c). These results suggest that the cytokine‐inducing activity of rNM23‐H1 may be associated with its growth‐ and survival‐promoting activity in AML cells. Although the patterns of cytokine induction differed among cases, cytokines known as growth factors for AML cells, such as GM‐CSF and IL‐1β, were induced in CM of NM23‐sensitive cases but not NM23‐unresponsive case 3 (Fig. 2a,c). Moreover, the induced cytokine concentrations reached physiologically effective levels. To investigate the correlation between cytokine‐inducing activity and the growth‐ and survival‐promoting activity of rNM23‐H1, we next tried to inhibit the cytokines using specific antibodies (anti‐TNFα, anti‐IL‐1β, anti‐IL‐6 antibodies). These antibodies alone (Fig. 3a) or in various combinations (Fig. 3b) partially inhibited rNM23‐H1‐induced growth and survival of AML cells (case 2 on day 6). Anti‐GM‐CSF antibody also inhibited both GM‐CSF‐induced and rNM23‐induced growth and survival of AML cells (Fig. 3c; case 1 on day 6). These results suggest that the growth‐ and survival‐promoting activity of this protein may be partly mediated by the induction of these cytokines.
Figure 2.
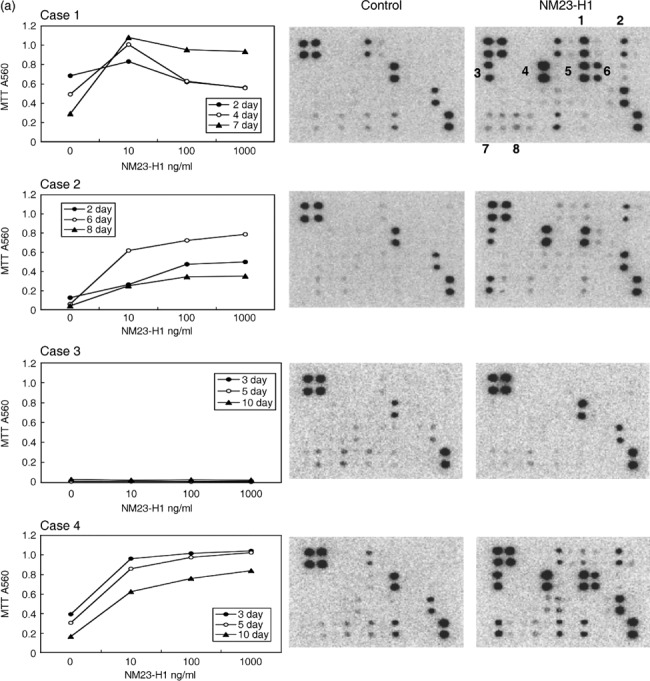
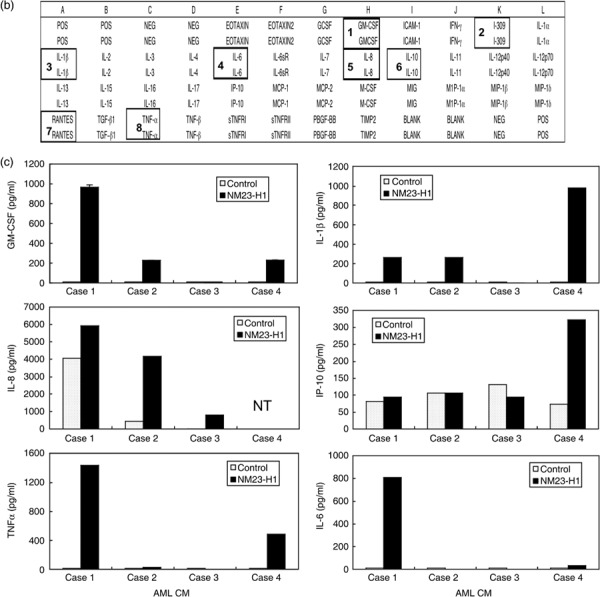
Cytokine‐inducing activity of extracellular recombinant (r) NM23‐H1 protein on primary cultured acute myelogenous leukemia (AML) cells. (a) Growth‐ and survival‐promoting activity (dose response) of rNM23‐H1 protein of four representative cases on days 2–10 (left panels), and the cytokines detected in conditioned medium (CM) of AML cells treated with or without rNM23‐H1 (100 ng/mL) for 48 h, using human inflammation antibody array (right panels), refer to (b). 1, GM‐CSF; 2, I‐309; 3, IL‐1β; 4, IL‐6; 5, IL‐8; 6, IL‐10; 7, RANTES; 8, TNFα. (c) Various cytokine levels in CM (2 h for TNFα, IL‐6 and 48 h for IL‐1β, GM‐CSF, IL‐8, IP‐10) of AML cells treated with or without rNM23‐H1 (100 ng/mL), were measured by sandwich ELISA. Error bars, SD.
Figure 3.
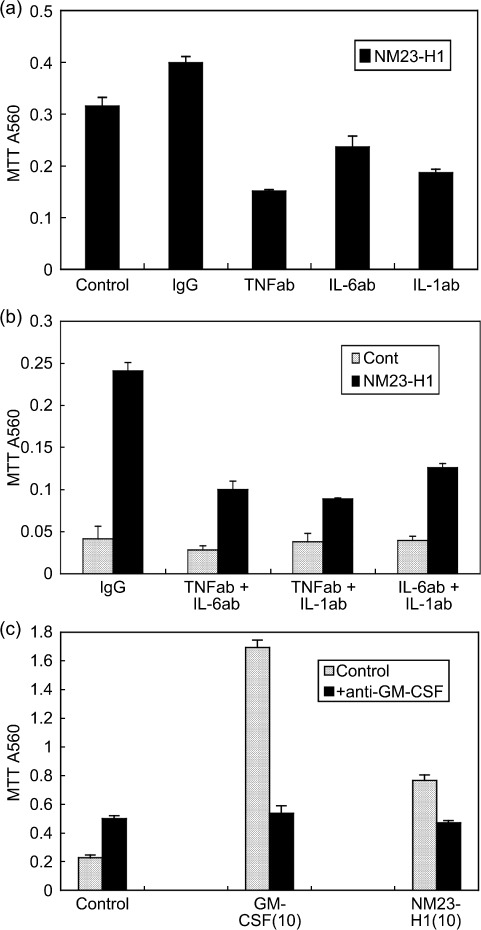
Inhibitory effects of cytokine antibodies on the growth‐ and survival‐promoting activity of NM23‐H1. (a) Growth‐ and survival‐promoting activity of NM23‐H1 in the presence or absence of cytokine antibodies (TNFα antibody, IL‐6 antibody, IL‐1β antibody). IgG, control immunoglobulin. (b) Inhibitory effects induced by the combination of two cytokine antibodies (case 2 on day 6). (c) Anti‐GM‐CSF antibody inhibited the growth‐ and survival‐promoting activity of recombinant (r) NM23‐H1 and rGM‐CSF (case 1 on day 6). Error bars, SD.
Signal transduction pathways induced by extracellular NM23‐H1 protein in primary AML cells. We next investigated the NM23‐induced signal transduction pathways relating to cell proliferation, survival, and cytokine production, namely MAPK (p38, Erk1/2, JNK), STAT, AKT–PI3K, and NFκB in AML cells. Of these pathways, extracellular rNM23‐H1 activated MAPK (p38, Erk1/2) and STAT (Fig. 4a,b), but not others (data not shown). First, we examined MAPK functions, because they are known to have critical roles in the regulation of cell growth and control of cellular response to cytokines. In AML cases 2 and 9, for which we could obtain sufficient cells for western blot analysis, rNM23‐H1 activated p38 MAPK and Erk1/2 MAPK (Fig. 4a). JNK was also activated in case 9, but was not detected in case 2 under the same experimental conditions (data not shown). Next, we examined STAT signaling, because they are known as key proteins acting as signal messengers and transcription factors that participate in normal cellular responses to cytokines. Furthermore, the constitutive activation of STAT3 has been reported to be associated with a wide variety of human malignancies. rNM23‐H1 increased the total amount of STAT1 protein and the phosphorylation of Tyr and Ser in case 9 (Fig. 4b). STAT3 was phosphorylated on Tyr705 in the absence of rNM23‐H1, but Ser727 of STAT3 was also phosphorylated in the presence of rNM23‐H1. Ser phosphorylation of STAT3 has been reported to be required for maximal transcriptional activity.( 27 ) STAT5 was also activated by rNM23‐H1 (Fig. 4b) and similar effects were shown in case 2 (data not shown).
Figure 4.
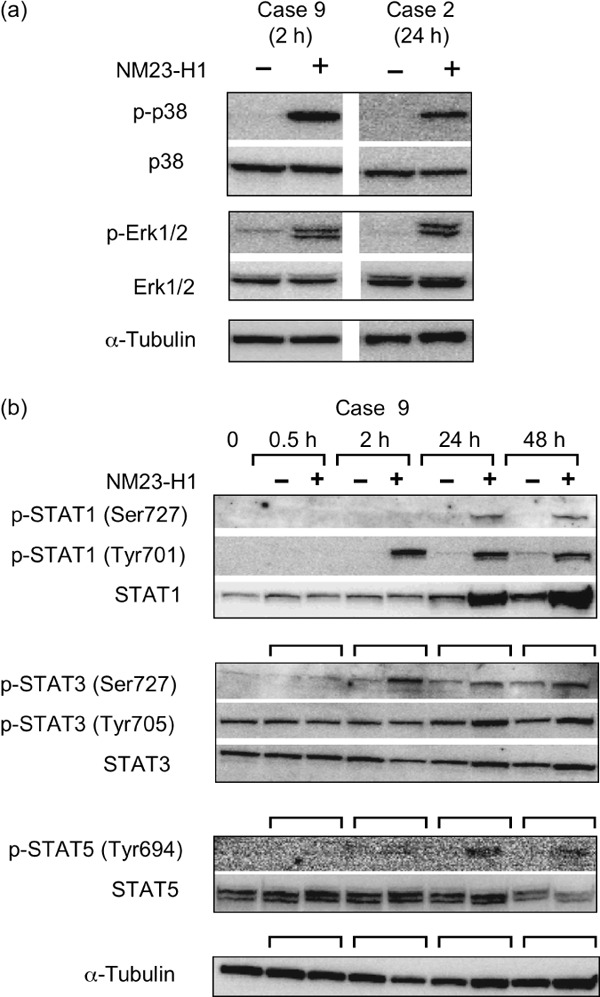
Signal transduction pathways induced by extracellular recombinant (r) NM23‐H1 protein in primary acute myelogenous leukemia (AML) cells. (a) AML cells (case 2 and 9) were treated with extracellular rNM23‐H1 protein (100 ng/mL) for the indicated hours. Activation (phosphorylation) of MAPK (p38 and Erk1/2) was investigated by SDS‐PAGE and immunoblotting. (b) Activation (phosphorylation of Tyr and Ser) of signal transducers and activators of transcription (STAT) 1, STAT3, and STAT5 in AML cells (case 9) by treatment with extracellular rNM23‐H1 protein.
The individual signaling pathways induced by rNM23‐H1 (100 ng/mL) protein were blocked in AML cells (case 2) using specific pharmacological inhibitors, SB202190 and SKF86002 for p38 MAPK, PD98059 for ERK (also known as MEK), and curcumin for STAT3. Viable cells were determined by MTT assay. These inhibitors suppressed the rNM23‐induced growth and survival promotion of AML cells (Fig. 5). These findings indicate that activity is associated with the activation of these signaling pathways in AML cells. To check the direct toxicity of these inhibitors on AML cells, MTT values under conditions without extracellular rNM23 are shown in separate figures with an expanded MTT scale (Fig. 5, right panel). Although direct toxicity was observed in PD98059, other inhibitors did not have any direct toxicity against AML cells. Taken together, these observations suggest that extracellular NM23‐H1 may play an important role in the malignant progression of leukemia, and the inhibitors of extracellular NM23‐H1 protein or inhibitors of extracellular actions of these proteins should be evaluated for their potential treatment as an approach to these malignancies.
Figure 5.
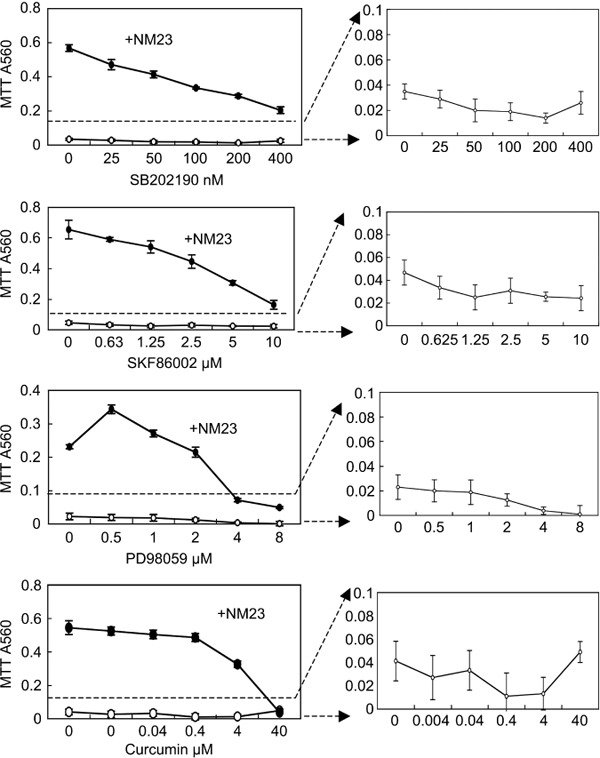
Effects of inhibitors of MAPK and signal transducers and activators of transcription (STAT) 3 on the recombinant (r) NM23‐H1‐promoted growth and survival of acute myelogenous leukemia (AML) cells. AML cells (case 2) were cultured with or without rNM23‐H1 protein (100 ng/mL) and various inhibitors for 5 days. Viable cells were determined by MTT assay. The values are the means ± SD for four independent samples. SB202190 and SKF86002, p38 MAPK inhibitor; PD98059, MEK inhibitor; Curcumin, STAT3 inhibitor, but not specific to STAT3. Right panels have an expanded MTT scale to show the direct toxicity of these inhibitors on AML cells under conditions without rNM23‐H1.
Discussion
In this experiment, we revealed that extracellular NM23‐H1 protein promoted the in vitro growth and survival of AML cells at concentrations equivalent to the levels found in AML patients (Fig. 1). These findings indicate a novel extracellular function of NM23‐H1 and its potential link with poor prognosis. How extracellular NM23‐H1 protein promotes the growth and survival of AML cells remains unclear. This activity was associated with cytokine production and MAPK and STAT signaling activation (2, 3, 4), but not with the NDPK enzyme activity of this molecule (Fig. 1f). MAPK/STAT activation by conventional growth factors takes only a few minutes; however, activation of these signaling pathways by NM23‐H1 required a longer time than various mitogens (Fig. 4), which might indicate indirect activation rather than direct activation by NM23 molecules.
The sensitivity of AML cells to extracellular NM23‐H1 protein was different among cases. We examined NM23‐H1 concentrations of the culture supernatant (48 h) of AML cells in cases 1–4 using ELISA, as it is interesting to know whether these concentrations are different between NM23‐responsive and NM23‐unresponsive AML cells. The results were as follows: case 1, 11 ng/mL; case 2, 9 ng/mL; case 3, 38 ng/mL; and case 4, 13 ng/mL. Of these four cases, the NM23‐unresponsive case 3 showed higher NM23‐H1 levels than the other NM23‐responsive cells; therefore, NM23‐unresponsive cells might secrete sufficient NM23‐H1 themselves to achieve a saturable concentration. However, secreted NM23‐H1 could not support the growth and survival of case 3 cells during culture (Fig. 1a). NM23‐H1 in the culture supernatant of case 3 might be due to leakage from AML cells that could not survive under the culture conditions.
The sensitivity of AML cells to this protein is also completely different from that of established AML cell lines. Extracellular NM23‐H1 protein could not affect the growth and cytokine production of established AML cell lines.( 28 ) It is interesting to examine receptor molecules for extracellular NM23‐H1 on AML cells and the expression difference between primary and established AML line cells.
Willems et al. previously reported that NDPK same as NM23 was present in normal human plasma and that NDPK activity correlated with hemoglobin levels, indicating the presence of NM23 in plasma as a consequence of red blood cell lysis.( 18 ) Moreover, they reported a modulating effect of extracellular NM23 proteins on the terminal stages (CD34 + CD38 + progenitor cells) of normal hematopoietic differentiation.( 29 ) More erythroid burst‐forming units and fewer macrophage colonies were observed in cultures containing any of the NM23 isoforms examined, even the enzymatically inactive H118N mutant of NM23‐H1. This suggested that fairly high concentrations of NM23 constitutively present in plasma and serum could have a physiological role in supporting erythropoiesis and inhibiting excessive macrophage formation. It is interesting that extracellular NM23 serves as an alarm molecule to signal red blood cell lysis and as a supporting molecule in normal erythropoiesis. We also reported that extracellular NM23 could inhibit the erythroid differentiation of human leukemia cell lines (K562, HEL, KU812) without any effect on proliferation,( 30 ) and that serum NM23‐H1 levels in AML‐M6 (acute erythroleukemia classified by the French–American–British [FAB] classification) was especially high and significantly higher than in other FAB subtypes of AML.( 20 ) It would be interesting to examine whether elevated levels of extracellular NM23 in AML‐M6 also function as proliferation‐supporting molecules of erythroleukemia cells as in normal erythropoiesis. In contrast to erythropoiesis, extracellular NM23‐H1 seems to inhibit the growth and survival of normal monocyte lineage cells;( 29 ) however, it could promote the growth and survival of primary cultured acute monocytic leukemia (AML‐M5 classified by FAB classification) cells. Serum NM23‐H1 levels in AML‐M5 were higher than in other FAB subtypes of AML, except AML‐M6.( 20 ) Taken together, these results suggest that an elevated serum level of NM23‐H1 protein in AML affects the biological properties of normal hematopoietic cells and leukemia cells of specific lineages and differentiation stages, resulting in poor prognosis.
We next used extracellular NM23‐H1 molecules and their functions as potential therapeutic targets of these malignancies, as our results suggest an important role of extracellular NM23‐H1 in the malignant progression of leukemia. As shown in Figure 5, inhibitors of MAPK/STAT3 activity suppressed the NM23‐induced growth and survival of AML cells. Various inhibitors are now under development, as MAPK and STAT3 signaling activations and tumor‐induced inflammatory conditions are widely observed in malignant progression.( 31 , 32 , 33 ) These agents might have potential to improve treatment for AML patients with a poor treatment outcome predicted by measuring plasma levels of NM23.
NM23 has no secretion signal peptide but is nonetheless detected in CM of some tumor cell lines and in body fluids.( 14 , 19 , 22 , 23 , 24 ) The mechanisms by which NM23‐H1 protein is secreted into the extracellular environment are unclear. Recently, Keller et al. showed a new pathway for the secretion of many inflammatory response proteins without signal sequences.( 34 ) This unconventional secretion required the catalytic activity of caspase‐1 and could rapidly release a wide variety of proteins involved in trigger detoxification, tissue repair, and cell survival. It would be interesting to examine whether this new secretion pathway secretes NM23 protein; however, unlike secretion, NM23 protein might be released by dying tumor cells overexpressing NM23. Recently, Apetoh et al. identified the Toll‐like receptor (TLR) 4 ligand, high‐mobility‐group box 1 (HMGB1) alarmin protein from dying tumor cells.( 35 ) This indicated that the molecule from tumor cells elicits an immune response involving the induction of inflammatory cytokines in a TLR4‐dependent fashion. Reportedly, a number of endogenous proteins bind and stimulate TLR4: heat‐shock protein 60, heat shock protein 70, oxidized low density lipoprotein (LDL), surfactant protein A, hyaluronan breakdown product, fibronectin, and β‐defensin‐2.( 36 ) The mechanism by which extracellular NM23‐H1 protein induces various inflammatory cytokines in AML cells (Fig. 2) is unknown. The next challenge will be to determine whether extracellular NM23 binds and stimulates TLR4, as does HMGB1.
In conclusion, our results show that the overexpression of extracellular NM23‐H1 derived from tumor cells generates a supportive microenvironment convenient for their growth and survival through cytokine production of primary AML cells (Fig. 6); therefore, the reduction of extracellular NM23‐H1 protein concentration and inhibitors of the extracellular actions of these proteins should be evaluated for their therapeutic potential to combat these malignancies.
Figure 6.
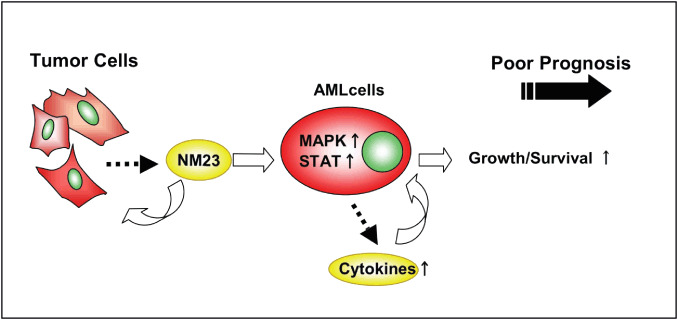
Extracellular function of NM23‐H1 protein derived from tumor cells on primary cultured acute myelogenous leukemia (AML) cells. Overexpression of the NM23 gene found in many hematological malignancies predicts a poor treatment outcome, and the elevated serum level of NM23‐H1 protein is also a poor prognostic factor in patients with these diseases. Extracellular NM23‐H1 protein promotes the growth and survival of primary AML cells mediated by MAPK activation, signal transducers and activators of transcription (STAT) activation and cytokine release. Taken together, these observations suggest that extracellular NM23‐H1 may play an important role in the malignant progression of leukemia, and that inhibitors of these protein or inhibitors of the extracellular functions of these proteins should be evaluated for their treatment potential.
Disclosure Statement
The authors declare no competing financial interests.
Abbreviations
| GM‐CSF | granulocyte macrophage colony stimulating factor |
| IL | interleukin |
| IP | interferon gamma inducible protein 10 |
| MDS | myelodysplastic syndrome |
| NF | nuclear factor |
| PI3K | phosphatidylinositol 3‐kinase |
| RANTES | regulated upon activation, normal T expressed and secreted |
| TNF | Tumor necrosis factor |
Acknowledgments
This work was supported by a Grant‐in‐Aid for Scientific Research (C) (18591091) from Japan Society for the Promotion of Science (J.O.‐K.), a Grant‐in‐Aid for Third Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor, and Welfare of Japan (J.O.‐K. and Y.K.), and a grant from the Kawano Memorial Foundation for the Promotion of Pediatrics of Japan (J.O.‐K. and Y.K.).
References
- 1. Steeg PS, Bevilacqua G, Kopper L et al . Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 1988; 80: 200–4. [DOI] [PubMed] [Google Scholar]
- 2. Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. The human nm23/nucleoside diphosphate kinases. J Bioenerg Biomembr 2000; 32: 247–58. [DOI] [PubMed] [Google Scholar]
- 3. Lascu I, Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J Bioenerg Biomembr 2000; 32: 237–46. [DOI] [PubMed] [Google Scholar]
- 4. MacDonald NJ, De La Rosa A, Steeg PS. The potential roles of nm23 in cancer metastasis and cellular differentiation. Eur J Cancer 1995; 31A: 1096–100. [DOI] [PubMed] [Google Scholar]
- 5. De La Rosa A, Williams RL, Steeg PS. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. Bioessays 1995; 17: 53–62. [DOI] [PubMed] [Google Scholar]
- 6. Horak CE, Lee JH, Elkahloun AG et al . Nm23‐H1 suppresses tumor cell motility by down‐regulating the lysophosphatidic acid receptor EDG2 . Cancer Res 2007; 67: 7238–46. [DOI] [PubMed] [Google Scholar]
- 7. Lacombe ML, Sastre‐Garau X, Lascu I et al . Overexpression of nucleoside diphosphate kinase (Nm23) in solid tumors. Eur J Cancer 1991; 27: 1302–7. [DOI] [PubMed] [Google Scholar]
- 8. Hailat N, Keim DR, Melhem RF et al . High levels of p19/nm23 protein in neuroblastoma are associated with advanced stage disease and with N‐myc gene amplification. J Clin Invest 1991; 88: 341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leone A, Seeger RC, Hong CM et al . Evidence for nm23 RNA overexpression, DNA amplification and mutation in aggressive childhood neuroblastoma. Oncogene 1993; 8: 855–65. [PubMed] [Google Scholar]
- 10. Yokoyama A, Okabe‐Kado J, Wakimoto N et al . Evaluation by multivariate analysis of the differentiation inhibitory factor nm23 as a prognostic factor in acute myelogenous leukemia and application to other hematological malignancies. Blood 1998; 91: 1845–51. [PubMed] [Google Scholar]
- 11. Niitsu N, Okabe‐Kado J, Okamoto M et al . Serum nm23‐H1 protein as a prognostic factor in aggressive non‐Hodgkin's lymphoma. Blood 2001; 97: 1202–10. [DOI] [PubMed] [Google Scholar]
- 12. Okabe‐Kado J, Hayashi M, Honma Y, Hozumi M. Characterization of differentiation inhibitory activity from nondifferentiating mouse myeloid leukemia cells. Cancer Res 1985; 45: 4848–52. [PubMed] [Google Scholar]
- 13. Okabe‐Kado J, Kasukabe T, Honma Y, Hozumi M. Purification of a factor inhibiting differentiation from conditioned medium of nondifferentiating mouse myeloid leukemia cells. J Biol Chem 1988; 263: 10 994–9. [PubMed] [Google Scholar]
- 14. Okabe‐Kado J, Kasukabe T, Honma Y, Hayashi M, Henzel WJ, Hozumi M. Identity of differentiation inhibiting factor for mouse myeloid leukemia cells with Nm23 protein involved in tumor metastasis. Biochem Biophys Res Commun 1992; 182: 987–94. [DOI] [PubMed] [Google Scholar]
- 15. Yokoyama A, Okabe‐Kado J, Sakashita A et al . Differentiation inhibitory factor nm23 as a new prognostic factor in acute monocytic leukemia. Blood 1996; 88: 3555–61. [PubMed] [Google Scholar]
- 16. Wakimoto N, Yokoyama A, Okabe‐Kado J, Nagata N, Motoyoshi K, Honma Y. Combined analysis of differentiation inhibitory factor nm23‐H1 and nm23‐H2 as prognostic factors in acute myeloid leukaemia. Br J Cancer 1998; 77: 2298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamashiro S, Urano T, Shiku H, Furukawa K. Alteration of nm23 gene expression during the induced differentiation of human leukemia cell lines. Oncogene 1994; 9: 2461–8. [PubMed] [Google Scholar]
- 18. Willems R, Van Bockstaele DR, Landon F et al . Decrease in nucleoside diphosphate kinase expression during hematopoietic maturation. J Biol Chem 1998; 273: 13 663–8. [DOI] [PubMed] [Google Scholar]
- 19. Niitsu N, Okabe‐Kado J, Kasukabe T, Yamamoto‐Yamaguchi Y, Umeda M, Honma Y. Prognostic implications of the differentiation inhibitory factor nm23‐H1 protein in the plasma of aggressive non‐Hodgkin's lymphoma. Blood 1999; 94: 541–50. [PubMed] [Google Scholar]
- 20. Niitsu N, Okabe‐Kado J, Nakayama M et al . Plasma levels of the differentiation inhibitory factor nm23‐H1 protein and their clinical implications in acute myelogenous leukemia. Blood 2000; 96: 1080–6. [PubMed] [Google Scholar]
- 21. Okabe‐Kado J, Kasukabe T, Honma Y, Hanada R, Nakagawara A, Kaneko Y. Clinical significance of serum NM23‐H1 protein in neuroblastoma. Cancer Sci 2005; 96: 653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Urano T, Furukawa K, Shiku H. Expression of nm23/NDP kinase proteins on the cell surface. Oncogene 1993; 8: 1371–6. [PubMed] [Google Scholar]
- 23. Anzinger J, Malmquist NA, Gould J, Buxton ILO. Secretion of nucleoside diphosphate kinase (Nm23‐H2) by cells from human breast, colon, pancreas and lung tumors. Proc West Pharmacol Soc 2001; 44: 61–3. [PubMed] [Google Scholar]
- 24. Rumjahn SM, Javed MA, Wong N, Law WE, Buxton ILO. Purinergic regulation of angiogenesis by human breast carcinoma‐secreted nucleoside diphosphate kinase. Br J Cancer 2007; 97: 1372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okabe‐Kado J. Serum nm23‐H1 protein as a prognostic factor in hematological malignancies. Leuk Lymphoma 2002; 43: 859–67. [DOI] [PubMed] [Google Scholar]
- 26. MacDonald NJ, De La Rosa A, Benedict MA, Freije JMP, Krutsch H, Steeg PS. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J Biol Chem 1993; 268: 25 780–9. [PubMed] [Google Scholar]
- 27. Wen Z, Zhong Z, Darnell JE Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995; 82: 241–50. [DOI] [PubMed] [Google Scholar]
- 28. Okabe‐Kado J, Kasukabe T. Physiological and pathological relevance of extracellular NM23/NDP kinases. J Bioenerg Biomemb 2003; 35: 89–93. [DOI] [PubMed] [Google Scholar]
- 29. Willems R, Slegers H, Rodrigus I et al . Extracellular nucleoside diphosphate kinase NM23/NDPK modulates normal hematopoietic differentiation. Exp Hematol 2002; 30: 640–8. [DOI] [PubMed] [Google Scholar]
- 30. Okabe‐Kado J, Kasukabr T, Baba H, Urano T, Shiku H, Honma Y. Inhibitory action of nm23 proteins on induction of erythroid differentiation of human leukemia cells. Biochim Biophys Acta 1995; 1267: 101–6. [DOI] [PubMed] [Google Scholar]
- 31. Lee JC, Kumar S, Griswold DE, Underwood DC, Votta BJ, Adams JL. Inhibition of p38 MAP kinase as a therapeutic strategy. Immunopharmacology 2000; 47: 185–201. [DOI] [PubMed] [Google Scholar]
- 32. Bharti AB, Donato N, Aggarwal BB. Curucumin (Diferuloylmethane) inhibits constitutive and IL‐6‐inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol 2003; 171: 3863–71. [DOI] [PubMed] [Google Scholar]
- 33. Karin M. Inflammation and cancer: the long reach of Ras. Nature Med 2005; 11: 20–1. [DOI] [PubMed] [Google Scholar]
- 34. Keller M, Rüegg A, Werner S, Beer H‐D. Active caspase‐1 is a regulator of unconventional protein secretion. Cell 2008; 132: 818–31. [DOI] [PubMed] [Google Scholar]
- 35. Apetoh L, Ghiringhelli F, Tesniere A et al . Toll‐like receptor 4‐dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med 2007; 13: 1050–9. [DOI] [PubMed] [Google Scholar]
- 36. Seong PY, Matzinger P. Hydrophobicity: an ancient damage‐associated molecular pattern that initiates innate immune responses. Nat Rev Immunol 2004; 4: 469–78. [DOI] [PubMed] [Google Scholar]


