Abstract
Transcription of human respiratory syncytial virus (RSV) genome RNA exhibited an obligatory need for the host cytoskeletal protein actin. Optimal transcription, however, required the participation of another cellular protein that was characterized as profilin by a number of criteria. The amino acid sequence of the protein, purified on the basis of its transcription-optimizing activity in vitro, exactly matched that of profilin. RSV transcription was inhibited 60 to 80% by antiprofilin antibody or poly-l-proline, molecules that specifically bind profilin. Native profilin, purified from extracts of lung epithelial cells by affinity binding to a poly-l-proline matrix, stimulated the actin-saturated RSV transcription by 2.5- to 3-fold. Recombinant profilin, expressed in bacteria, stimulated viral transcription as effectively as the native protein and was also inhibited by poly-l-proline. Profilin alone, in the absence of actin, did not activate viral transcription. It is estimated that at optimal levels of transcription, every molecule of viral genomic RNA associates with approximately the following number of protein molecules: 30 molecules of L, 120 molecules of phosphoprotein P, and 60 molecules each of actin and profilin. Together, these results demonstrated for the first time a cardinal role for profilin, an actin-modulatory protein, in the transcription of a paramyxovirus RNA genome.
Human respiratory syncytial virus (RSV) is a major pathogen of the lower respiratory tracts of young infants (7). RSV belongs to the Pneumovirus genus within the Paramyxoviridae family. Like other members of this family, RSV has a nonsegmented, negative-strand RNA genome. The RSV genes and genome organization are unique among paramyxoviruses in many respects; the order of genes on the 15,222-kb RSV genomic RNA is 3′-(leader)-NS1-NS2-N-P-M-SH-G-F-M2-L-(trailer)-5′ (14). The RSV nucleocapsid core consists of the viral genomic RNA wrapped with N protein (called the N-RNA template), the phosphoprotein P, the transcription elongation factor M2, and the major subunit of the RNA-dependent RNA polymerase, L (5, 9, 17, 21).
As part of our ongoing investigation of the mechanisms of RSV gene expression, we have embarked on the characterization of the various components of the RSV RNA transcription machinery. Our initial studies showed that the viral nucleocapsid core alone was incapable of transcription in vitro; however, the addition of uninfected cell extract restored transcriptional activity (1). Subsequent fractionation of the cell extract revealed that cellular actin is both necessary and sufficient to reconstitute in vitro transcription (5). While that study constituted the first detailed report of a cytoskeletal protein acting as a bona fide transcription factor for RSV, it remained unknown whether actin-modulatory proteins played any role in the process. In the same study, however, we demonstrated that actin alone did not activate viral transcription to the same degree as the whole-cell lysate did. Thus, it was proposed that at least one other host cell factor was required for optimal viral transcription. Preliminary characterization indicated that this second factor was proteinaceous. In this report, we identify profilin, an actin monomer binding protein that regulates the normal distribution of F-actin structures in vivo, as the second host cell factor required for optimal RSV transcription.
(A preliminary report of this work was presented by E.B. at the 18th Annual Meeting of the American Society for Virology, Amherst, Mass., 10–14 July 1999.)
MATERIALS AND METHODS
Antibodies.
Monoclonal mouse antibody that reacts with all six known actin isoforms was purchased from Roche Molecular Biochemicals (Indianapolis, Ind.). The profilin antibody was raised in rabbits against purified recombinant human profilin-1 (36, 37) and was a generous gift from William Zeile and Frederick Southwick (University of Florida). Secondary antibodies conjugated to horseradish peroxidase were obtained from Sigma (St. Louis, Mo.).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analyses were carried out essentially as described earlier (5), except that the SuperSignal Ultra chemiluminescence procedure (Pierce, Rockford, Ill.) was used for the development of the secondary antibody conjugated to horseradish peroxidase.
The protein concentration was determined by the Bradford assay (3) with bovine serum albumin as a standard.
Purification of HEp-2 cell actin.
To purify HEp-2 cell actin, a cytosolic extract of the cells was fractionated essentially as described previously (5). Briefly, cells from 30 T-150 flasks were harvested, resuspended in 3 ml of buffer A (50 mM Tris-HCl [pH 7.5], 10 mM NaCl, 5 mM β-mercaptoethanol), and lysed by sonication. The lysate was then centrifuged at 120,000 × g for 1 h. The supernatant, designated S120, was loaded on a Sephadex G-200 (Pharmacia, Piscataway, N.J.) column (2 by 150 cm) equilibrated with buffer A. The column was developed with the same buffer, and 0.5-ml fractions were collected. The actin-enriched fractions were identified by SDS-PAGE and immunoblotting (5). These fractions were then pooled, and the actin was further purified by antibody affinity chromatography as previously described (5).
Purification of RBC actin.
Actin was purified from erythrocytes (RBC) essentially as described previously (31). Briefly, 40 ml of human blood was obtained by venipuncture in Vacutainer tubes containing 143 USP units of lithium heparin. Fresh blood was centrifuged at 1,500 × g for 20 min. Packed RBCs were resuspended in 30 ml of lysis buffer (5 mM Na2PO4, 1 mM EDTA [pH 7.6]) and sedimented at 31,000 × g for 15 min at 4°C. This procedure was repeated until the ghosts (empty RBC membranes) became white. To separate the RBC core skeleton from the membrane, the ghosts were incubated for 15 min on ice in 10 ml of Triton buffer (10 mM Na2PO4, 6 M KCl, 1% Triton X-100 [pH 7.6]) and then the core skeleton was sedimented at 35,000 × g for 45 min. The skeleton components were then dissociated by incubation in 2 ml of 2 M Tris-HCl (pH 7.2) at 37°C for 30 min and loaded onto a 160-ml Sepharose 4B gel filtration column which had been equilibrated with 2 M Tris-HCl (pH 7.2). The proteins were eluted with this buffer and collected in 4-ml fractions. Every alternate fraction following the void volume (∼50 ml) was analyzed by SDS-PAGE. The actin-containing fractions were subsequently pooled, dialyzed against 2 mM Tris-HCl (pH 7.5)–10 mM NaCl–5 mM β-mercaptoethanol, and finally concentrated to 500 μg/ml. The purity and identity of the preparation were confirmed by silver stain and immunoblot analyses, and this actin was then used directly in transcription reactions.
The elution profile of both actins from the gel filtration columns corresponded to monomeric actin (i.e., ∼43 kDa). Both preparations were competent for polymerization when incubated in the presence of MgCl2, NaCl, and ATP, as determined by their conversion into a form that failed to enter nondenaturing polyacrylamide gels (15).
Purification of profilin.
To purify native profilin from HEp-2 cells, S120 extracts of HEp-2 cells were fractionated by Sephadex G-200 gel filtration as described for the purification of native actin and the fractions were tested by SDS-PAGE analysis or activity assays as mentioned in Results. Appropriate selected fractions were then pooled and loaded on a 1-ml anion-exchange column (Econo-Q; Pharmacia, Piscataway, N.J.) at a flow rate of 0.5 ml/min. Proteins were eluted with a 0.2 to 0.8 M NaCl gradient in buffer A, and 0.5-ml fractions were collected and tested for profilin.
Recombinant profilin.
Recombinant human platelet profilin-1 was expressed in Escherichia coli and purified essentially as described previously (13). Briefly, profilin cDNA was cloned into the NdeI-EcoRI sites of pMW172 and transformed into E. coli BL21(DE3) to allow isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible protein expression. Expressed profilin was then purified by affinity chromatography with poly-l-proline (PLP)–Sepharose as described below. As shown previously, the recombinant profilin was biochemically and antigenically indistinguishable from native profilin (13).
PLP affinity purification of profilin.
Preparation of PLP-Sepharose was based on the method of Tuderman et al. (35). Briefly, PLP with an Mr of 14,000 (Sigma) was covalently coupled to CNBr-activated Sepharose 4B (Pharmacia) following the directions supplied by the manufacturer. Profilin was purified from HEp-2 cell extract by the PLP affinity method, using published protocols (19), based on the observation that PLP exhibits a strong and specific affinity for profilin (32). Cells from two T-150 flasks were harvested and lysed, and S120 extract, prepared as described above, was applied to a 0.5-ml PLP column. The column was subsequently washed with 1.5 ml of buffer B (20 mM Tris, 150 mM KCl, 0.1 mM dithiothreitol). Actin was removed from the column with 0.5 ml of actin elution buffer (buffer B plus 2 M urea), and 1 ml of wash buffer (buffer B plus 4 M urea) was then applied to remove traces of other proteins. Profilin was eluted with profilin elution buffer (buffer B plus 7 M urea) and subsequently dialyzed against buffer A with several changes. The proper renaturation of profilin by this method had been previously ascertained by nuclear magnetic resonance spectroscopy and actin binding studies (19).
Silver stain.
Silver staining of proteins was carried out as described previously (3, 27). Briefly, following SDS-PAGE, gels were fixed by a 4-h incubation with gentle shaking in 5 gel volumes of ethanol-glacial acetic acid-water (30:10:60), incubated twice for 30 min each in 30% ethanol, and washed three times for 10 min each with distilled water. The gels were then incubated in a 0.1% AgNO3 solution for 30 min and subsequently developed in a solution containing 2.5% sodium carbonate and 0.02% formaldehyde.
Reconstituted RSV transcription.
RSV ribonucleoprotein (RNP) complex was purified from infected cells essentially as previously described (5, 21). The preparation contained N-RNA template, L, and P proteins, as well as trace amounts of cellular actin and the viral M2 protein. The M2 protein was detectable only by immunoblotting with antibodies against bacterially synthesized recombinant M2 protein (V. Bitko and S. Barik, unpublished results).
Reconstitution of a standard viral transcription reaction mixture (20 μl) and measurement of incorporation of [α-32P]UMP by DE81 paper binding were carried out essentially as described earlier (5), except that about 1 μg of N-RNA template was used per reaction. Additionally, 5 U of RNasin (Promega Corp., Madison, Wis.) was included in the reaction mixture. Other additions such as HEp-2 cellular fractions, recombinant proteins, and antibodies were made to the reaction mixture as described herein. The transcription-stimulatory activity of profilin was assayed by using actin-saturated reaction mixtures containing 200 ng of actin per 20-μl reaction mixture. Labeled RNA from a portion of the reaction mixture was analyzed by agarose gel electrophoresis (acidic agarose) and autoradiography as described previously (1).
RESULTS
Determination of optimal actin concentration for RSV transcription.
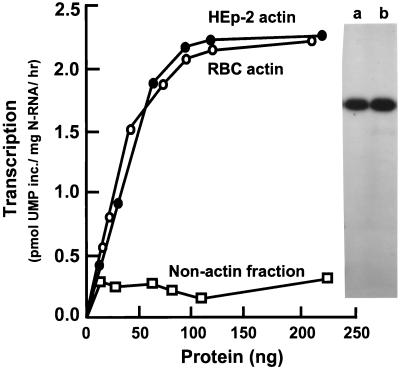
HEp-2 cell actin was purified via antibody affinity chromatography (see Materials and Methods). This actin was then used in increasing amounts in transcription assays in vitro to determine the amount required to reach saturation (Fig. 1). To rule out the possibility that other, nonactin proteins were fortuitously copurified with cellular actin, a second source of actin was sought. As enucleated cells, RBC have relatively few different types of cellular proteins; in fact, there are only three major components of the core cytoskeleton: spectrin, band 4.1, and actin. It is therefore possible to obtain essentially homogeneous (as demonstrated by silver staining) actin from RBC by following established methods of purification (31) such as the one described in Materials and Methods. Actin purified in this manner was capable of activating RSV transcription to the same degree as the affinity-purified actin (Fig. 1), indicating that it was actin alone that contained the transcription-activating property. The high purity of both preparations of actin was confirmed by SDS-PAGE and silver staining (Fig. 1). Because HEp-2 cells are the ex vivo hosts for RSV infection, affinity-purified HEp-2 cellular actin was used for the remainder of the transcription reactions described in this paper.
FIG. 1.
Determination of actin saturation for RSV transcription in vitro. Increasing amounts of either HEp-2 cell actin or RBC actin were used in 20 μl-RSV transcription reactions in vitro, reconstituted as described in Materials and Methods. HEp-2 cell proteins that did not bind to the antiactin column were used as the nonactin fraction and were devoid of activity, as shown. A silver-stained profile of 200 ng of purified RBC (lane a) and HEp-2 (lane b) actin, analyzed by SDS-PAGE, is presented on the right and shows the high purity of both preparations.
Biochemical purification of the transcription-optimizing factor.
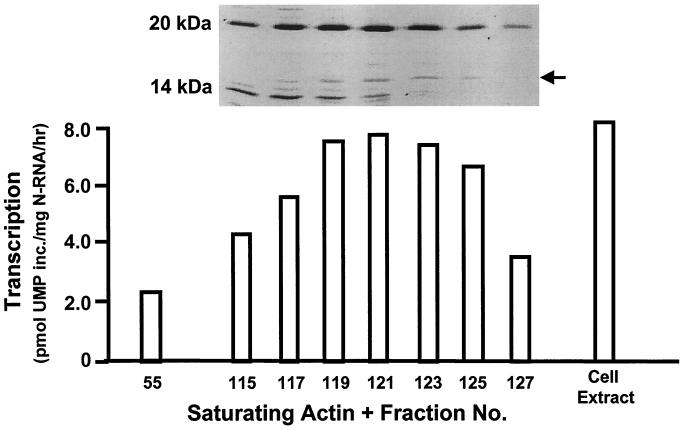
Preliminary characterization of the nonactin host factor required for optimal RSV transcription indicated that it was proteinaceous, unphosphorylated, and devoid of any nucleic acid or lipid component (5). Further characterization of this factor was initiated by gel filtration chromatography of uninfected HEp-2 cell extracts. Alternate fractions from this column were then assayed for RSV transcription optimization activity by adding aliquots of each to actin-saturated in vitro transcription reaction mixtures. As shown in Fig. 2, the activity was localized to low-molecular-mass fractions ranging predominantly from 10 to 25 kDa. When the fractions with the highest activity (fractions 119 to 123) were passed through a PLP column that specifically and strongly binds profilin (19), the resultant preparations showed only “background” transcription activity characteristic of an actin-only reaction (about 3.0 U of transcription). These early results suggested that the transcription-stimulatory factor is profilin, and we will provide further proof of this below.
FIG. 2.
Determination of the molecular size of the RSV transcription-optimizing factor. HEp-2 cell extract was fractionated on a size exclusion column (Sephadex G-200), and alternate fractions (numbers at the bottom) were tested for RSV transcription-optimizing ability in actin-saturated in vitro transcription reactions. SDS-PAGE analysis and silver staining of the relevant fractions are shown at the top. The peak activity, at fraction 121, was comparable to that of the total-cell extract. Fraction 55, far from the activity peak, was used as a negative control. Note the location of the activity in the low-molecular-weight region; the band indicated by the arrow is profilin (see Results).
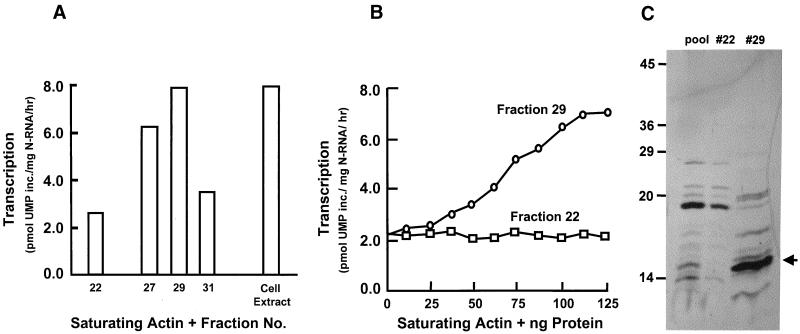
These fractions were subsequently pooled and loaded onto an anion-exchange column, which was then developed with a linear salt gradient. Each of these fractions was then tested as before in an actin-saturated in vitro transcription assay. The results presented in Fig. 3A show that the activity peaked at fraction 29, eluting at 0.3 M NaCl. This fraction proved capable of maximizing transcriptional activity in a dose-dependent manner (Fig. 3B). The silver stain of fraction 29 indicated that the predominant band migrated as a 14.8-kDa polypeptide (Fig. 3C). An aliquot of fraction 29 was then concentrated, subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane (27), and stained with Coomassie blue. The 14.8-kDa polypeptide was excised and subjected to Edman-based microsequencing; the first 15 amino acids at the N terminus (MAGWNAYIDNLMADG) proved to be a perfect match to human profilin-1.
FIG. 3.
Transcription-optimizing activity of anion-exchange column fractions. Fractions from the size exclusion column which contained the peak transcription-optimizing activity were pooled and loaded onto an anion-exchange column (Econo-Q). (A) Peak activity eluted at 0.3 M NaCl, in fraction 29. Fraction 22 was far from the activity peak and was used as a control. (B) Dose-dependent activity of fraction 29. Fraction 22 lacked activity at all concentrations. (C) Silver stain of representative anion-exchange fractions (fractions 22 and 29). Lane “pool” contains pooled size exclusion fractions, loaded onto the anion-exchange column. Molecular masses of standards are indicated in kilodaltons on the left. About 0.4 to 0.8 μg of protein was analyzed in the different lanes. The arrow points to the 14.8-kDa profilin band (see Results).
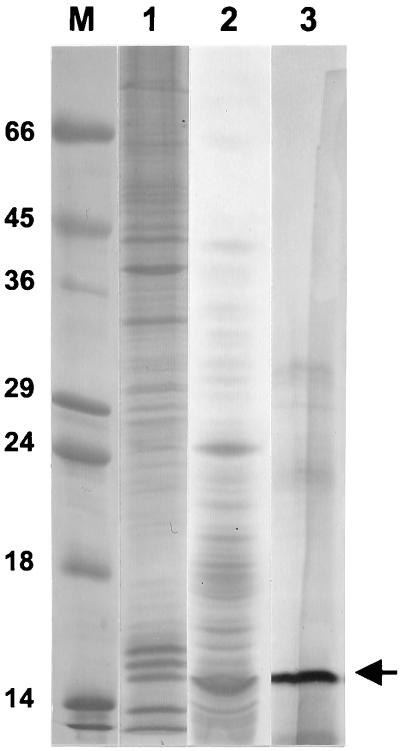
Based on the specific activity calculation of the gel filtration peak, a 57-fold purification had been achieved (Table 1). The polypeptide profile at the major steps of purification is presented in Fig. 4, which clearly demonstrates an enrichment of profilin through purification.
TABLE 1.
Purification of RSV transcription optimizing factor (profilin)
| Purification step | Total amt of protein (μg) | Total activitya | Sp actb | Yield (%)c | Fold purificationd |
|---|---|---|---|---|---|
| Soluble extract (S120) | 16,440 | 27,948 | 1.7 | 100.0 | 1.0 |
| Gel filtration peak | 79 | 2,323 | 29.4 | 8.3 | 17.2 |
| Ion-exchange peak | 21 | 2,085 | 97.0 | 7.4 | 57.0 |
One unit of activity is defined as 1 pmol of UMP incorporated per mg of N-RNA per h, calculated from a 20-μl transcription reaction mixture reconstituted as described in Materials and Methods.
Specific activity is the total activity per microgram of protein.
The yield (protein) of each fraction is expressed as the percentage of that of the soluble extract, taken as 100%.
Fold purification is the ratio of the specific activity of a given fraction to that of the soluble extract, taken as 1.0.
FIG. 4.
Purification of profilin. Approximately 0.4 μg of the following assorted fractions obtained at various stages of purification were analyzed by SDS-PAGE and silver staining: HEp-2 S120 extract (lane 1), gel filtration fraction (pooled fractions 119 to 123 of Fig. 2) (lane 2), and ion-exchange fraction (pooled fractions 27 to 29 of Fig. 3A) (lane 3). Standard protein markers (indicated in kilodaltons) are shown in lane M. The arrow points to the 14.8-kDa profilin band. The specific activities of essentially identical fractions are presented in Table 1.
Profilin as the transcription-optimizing factor.
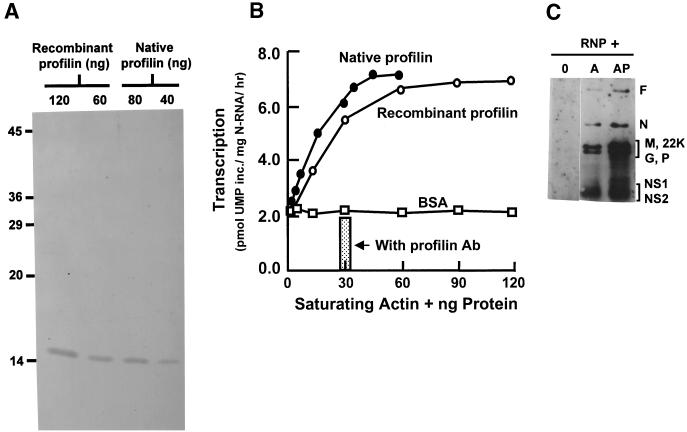
To determine if profilin is in fact the second host cell factor that participates in RSV transcription, both recombinant and native profilin were purified by the PLP affinity method (19) and tested in actin-saturated transcription reactions in vitro. As shown in Fig. 5A and B, the two preparations contained essentially pure profilin of 14.8 kDa and optimized transcription in a dose-dependent manner and with comparable specific activity. Furthermore, the addition of an antiprofilin antibody to the fully reconstituted in vitro transcription reaction mixture negated this transcription-optimizing effect; i.e., it reduced transcription by about 70%, down to the actin-only level.
FIG. 5.
Recombinant and native profilin maximize RSV transcription in a dose-dependent manner. (A) Silver stain of recombinant profilin and of native profilin purified from HEp-2 cells as described in Materials and Methods. (B) The dose-responses of the RSV transcription-optimizing activity of recombinant and native profilin were comparable. Addition of antiprofilin antibody to a reaction mixture with 30 ng of native profilin obliterated this response. Acetylated bovine serum albumin (BSA) did not stimulate transcription. (C) mRNA profile. RSV transcription reactions were carried out in the presence of viral RNP and the following: none (lane 0), actin only (lane A), or both actin and profilin (lane AP). The 32P-labeled RNA were deproteinized and analyzed on acidic agarose gels as described in Materials and Methods. The various gene products are indicated on the right.
Recently, the 22-kDa M2 protein of RSV has been shown to be a transcription antitermination factor that stimulates the ability of the viral transcription complex to generate full-length transcripts (9, 17). The viral phosphoprotein P is important for promoter clearance as well as elongation of the transcription complex (12). It was therefore important to ask whether the increased overall transcription observed with profilin was due to a general increment of all mRNA species or to a preferential transcription of promoter-distal genes resulting from a stimulation of the processivity of the polymerase. Transcripts synthesized in the presence of actin or a combination of actin and profilin were therefore analyzed by gel electrophoresis. The results (Fig. 5C) clearly show an essentially identical mRNA profile synthesized in the absence and presence of profilin, except that in its presence all the mRNAs were transcribed in greater quantities. Profilin therefore stimulates the overall transcription activity of the viral polymerase, the exact mechanism of which will require further study.
Optimal transcriptional activity of 1 μg of N-RNA required an average of about 100 ng of actin and 30 ng of profilin, which is equivalent to about 60 molecules each of actin and profilin per molecule of N-RNA (assuming roughly 1,200 molecules of N protein per N-RNA template). The equimolar amounts of actin and profilin may indicate their joint role in viral transcription.
Packaging of profilin in mature RS virions.
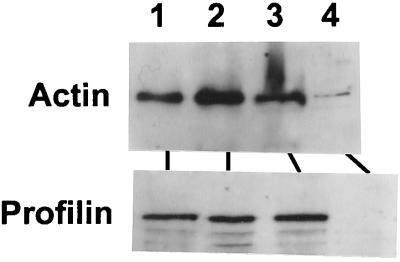
We have previously demonstrated that cellular actin is present in the mature RS virion (5), indicating that it plays an important role in the viral life cycle. Thus, it was of interest to determine if profilin is also packaged with RSV. Because it is formally possible that large aggregates of actin or cytoskeletal material will contaminate RSV preparations, a portion of the purified virus was immunoprecipitated by a polyclonal anti-RSV antibody described under Materials and Methods. The actin and profilin contents of the purified as well as the immunoprecipitated RSV were analyzed by SDS-PAGE and Western blotting. The results (Fig. 6) clearly show that highly purified virus preparations contain profilin as well actin. In contrast, purified viral RNP that we have used for transcription in this study and previously (1) contained only traces of actin and virtually no profilin (Fig. 6), confirming our previous finding (1). This may explain the requirement for actin as well as profilin in transcription by the RNP. Antibodies against various other cytoskeletal proteins were previously shown to not react with the purified virus (5), further indicating that this association may be specific to cellular proteins that play a critical role in the viral life cycle.
FIG. 6.
Profilin content of the RSV virion. The following were subjected to SDS-PAGE and Western blot: purified standard proteins (40 ng; lane 1), purified RSV (30 μg; lane 2), immunoprecipitated RSV (lane 3), and purified RSV RNP (4 μg; lane 4). RSV was immunoprecipitated by standard procedures involving the binding of a goat anti-RSV antibody (generated against complete RSV and reacts with all the structural proteins of the virus) (5) and protein A-Sepharose to 50 μg of RSV, followed by cycles of centrifugation and washing essentially as described previously (5). Western blotting was carried out with either antiactin or antiprofilin antibody as shown. The autoradiogram from chemiluminescence detection is presented.
DISCUSSION
In this communication, we have demonstrated a role for profilin in RSV transcription. While not absolutely required for viral transcription, profilin did induce an approximately fourfold increase in transcription levels in vitro. This activity was seen with both recombinant and purified native profilin and was inhibited by an antibody specific to profilin. Furthermore, the specific packaging of profilin with the mature RS virion implied an important role in the RSV life cycle. Taken together, these data suggest that the nonactin host cell factor responsible for optimizing RSV transcription in vitro is indeed profilin. These results confirm and extend our previous finding that both actin and a nonactin cellular factor are required for optimal RSV transcription (5).
Cytoskeletal proteins play an important role in the transcription of several negative-strand RNA viral genomes. The transcription complex of Newcastle disease virus irreversibly assembles and synthesizes viral mRNA on the cytoskeletal framework (16). Tubulin is utilized by Sendai virus (22), measles virus (23), and vesicular stomatitis virus (22). Actin has proven necessary for both RSV (5) and human parainfluenza virus-3 transcription (11). For human parainfluenza virus-3 transcription, filamentous actin was required. It was shown that polymerization of nucleocapsid-associated actin imparted a helical structure to the viral nucleocapsid, which was thought to result in transcriptional activation (11). In contrast, either filamentous or globular actin was capable of activating RSV transcription (5), implying a different and perhaps more direct mechanism. Recent studies also implicated cellular actin in human immunodeficiency virus type 1 (HIV-1) cell entry and reverse transcriptase activity. The large subunit of the HIV-1 reverse transcriptase was shown to specifically interact with actin (18). Chemical disruption of actin filaments impaired both reverse transcriptase activity and viral entry via cell fusion (4).
Despite these instances of cytoskeletal involvement in viral transcription, there have been no published reports of a role for cytoskeletal modulatory proteins. Thus, the use of both profilin and actin for RSV transcription is an intriguing combination, due to the known role of profilin as an actin-modulatory protein. The role of profilin in actin filament dynamics has been the subject of intense scrutiny in recent years, due to its seemingly contradictory abilities to either promote or inhibit filament formation (20, 28, 30, 33). It now appears that profilin is in fact a temporal and spatial regulator of actin filament assembly that is capable of either promoting or inhibiting actin polymerization, depending on the immediate environment as well as extracellular signals. In vivo, the scenario is further complicated by the presence of a diverse array of actin binding proteins. For example, various proteins, known as capping proteins, can bind to the growing end of an actin filament and prevent the further addition of actin monomers (20, 28, 30, 33).
As discussed above, both monomeric and filamentous actin are capable of activating RSV transcription in vitro, and chemical disruption of actin filaments ex vivo does not inhibit the production of viral proteins (5). Thus, filamentous actin is not required for viral transcription. Therefore, the role of profilin in viral transcription is not likely to be that of promoting actin polymerization. Perhaps the profilin-bound form of actin is better able to interact with the viral polymerase or with the N-RNA template. This mechanism could explain why profilin is not absolutely required for RSV transcription—the interaction between actin and the viral macromolecules occurs in the absence of profilin, but the addition of profilin makes it more favorable. The propensity of profilin to increase the rate of nucleotide exchange on actin (20, 30) could also be relevant to its role in transcription independent of its role in filament promotion. The conformation of ATP-actin may be transcriptionally superior to that of ADP-actin; alternatively, the hydrolysis of ATP that is normally coupled to actin polymerization may be used to promote RSV transcription. Recent crystallographic studies have indeed demonstrated that profilin can alter the dynamics and structure of actin such that the profilin-bound actin can attain and sample a range of structural states (8, 28, 29). Such conformational plasticity of the profilin-actin complex may underlie the mechanism for profilin-induced enhancement of the RNA transcriptional activity of actin.
Although filamentous actin was not required for RSV transcription, it appeared to be critical for viral budding. Treatment of infected cells with cytochalasin D was shown to result in an approximately 104-fold reduction in viral titer (5). Viral budding may be viewed as a highly specialized type of cell motility. Interestingly, a proposed mechanism for cell motility of various types is one in which profilin acts to provide a pool of assembly-competent monomeric actin at the leading edge of the moving cell (6), with the elongating filament providing the motive force. Recently, it has become apparent that various intracellular pathogens have adapted this strategy for their own use. The use of a profilin-actin motility system has been implicated in the cell-cell spread of the bacterial pathogens Listeria monocytogenes (34) and Shigella flexneri (36). Vaccinia virus is known to use an actin-based motility system for cell-cell spread (10), and a role for profilin in the vaccinia virus-induced actin “rocket tails” has been suggested (37). Curiously, vaccinia virus encodes a viral homolog of profilin. However, a mutant virus from which the profilin coding sequence had been deleted still displayed characteristic viral disruption of actin fibers, intracellular viral movement, and release of mature virions (2). At this time, it is not known if cellular profilin can substitute for the viral homolog during vaccinia virus morphogenesis. Other viruses that utilize actin filaments during their morphogenesis include human parainfluenza virus (Q. Yao and R. Compans, Abstr. 17th Annu. Meet. Am. Soc. Virol., abstr. W42-9, p. 135, 1998), frog virus 3 (24), HIV (25), and Black Creek Canal virus (26). These examples of the use of filamentous actin in the cell-cell spread of various pathogens, as well as the data indicating a requirement for filamentous actin in RSV morphogenesis, may suggest that during the postreplicative stages of the RSV life cycle, profilin acts to promote actin polymerization, thereby facilitating viral budding. Because RSV is a highly cell-associated virus, a role for the new outgrowth of actin in the cell-cell spread of the virus is a distinct possibility.
Since the actin binding surface of profilin is well defined (29), it will be possible to selectively mutate the relevant amino acid residues in order to ascertain whether a direct interaction between actin and profilin is required for optimal viral transcription. Further studies are clearly needed to elucidate the exact mechanism of involvement of these proteins in viral transcription, and they will be facilitated by the functional characterization of selected deletion mutants of both recombinant actin and recombinant profilin in our in vitro transcription reactions.
ACKNOWLEDGMENTS
We thank Frederick Southwick and William Zeile (University of Florida, Gainesville, Fla.) for the antiprofilin antibody.
This research was supported in part by Public Health Service grants AI37938 (to S.B.) and GM53807 (to S.C.A.) from the National Institutes of Health.
REFERENCES
- 1.Barik S. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s) J Virol. 1992;66:6813–6818. doi: 10.1128/jvi.66.11.6813-6818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco R, Cole N B, Moss B. Sequence analysis, expression, and deletion of a vaccinia virus gene encoding a homolog of profilin, an eukaryotic actin-binding protein. J Virol. 1991;65:4598–4608. doi: 10.1128/jvi.65.9.4598-4608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcriptase complex involves the cytoskeleton. J Exp Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke E, Dupuy L, Wall C, Barik S. Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology. 1998;252:137–148. doi: 10.1006/viro.1998.9471. [DOI] [PubMed] [Google Scholar]
- 6.Carlier M F. Control of actin dynamics. Curr Opin Cell Biol. 1998;10:45–51. doi: 10.1016/s0955-0674(98)80085-9. [DOI] [PubMed] [Google Scholar]
- 7.Cate T R. Impact of influenza and other community-acquired viruses. Semin Respir Infect. 1998;13:17–23. [PubMed] [Google Scholar]
- 8.Chik J K, Lindberg U, Schutt C E. The structure of an open state of β-actin at 2.65 A resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 9.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 11.De B P, Burdsall A L, Banerjee A K. Role of cellular actin in human parainfluenza virus type 3 genome transcription. J Biol Chem. 1993;268:5703–5710. [PubMed] [Google Scholar]
- 12.Dupuy L C, Dobson S, Bitko V, Barik S. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase: phosphorylation by casein kinase 1 occurs mainly at Ser215 and is without effect. J Virol. 1999;73:8384–8392. doi: 10.1128/jvi.73.10.8384-8392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedorov A A, Pollard T D, Almo S C. Purification, characterization and crystallization of human platelet profilin expressed in Escherichia coli. J Mol Biol. 1994;241:480–482. doi: 10.1006/jmbi.1994.1522. [DOI] [PubMed] [Google Scholar]
- 14.Galinski M S. Annotated nucleotide and protein sequences for selected Paramyxoviridae. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 537–568. [Google Scholar]
- 15.Gao Y, Thomas J O, Chow R L, Lee G H, Cowan N J. A cytoplasmic chaperonin that catalyzes β-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 16.Hamaguchi M, Nishikawa K, Toyoda T, Yoshida Y, Hanaichi Y, Nagai Y. Transcriptive complex of Newcastle disease virus. II. Structural and functional assembly associated with the cytoskeletal framework. Virology. 1985;147:295–308. doi: 10.1016/0042-6822(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 17.Hardy R W, Harmon S B, Wertz G W. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J Virol. 1999;73:170–176. doi: 10.1128/jvi.73.1.170-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hottiger M, Gramatikoff K, Georgiev O, Chapponnier C, Schaffner W, Hubscher U. The large subunit of HIV-1 reverse-transcriptase interacts with beta-actin. Nucleic Acids Res. 1995;23:736–741. doi: 10.1093/nar/23.5.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janmey P A. Polyproline affinity method for purification of platelet profilin and modification with pyrene-maleimide. Methods Enzymol. 1991;196:92–99. doi: 10.1016/0076-6879(91)96011-f. [DOI] [PubMed] [Google Scholar]
- 20.Korenbaum E, Nordberg P, Bjorkegren-Sjogren C, Schutt C E, Lindberg U, Karlsson R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry. 1998;37:9274–9283. doi: 10.1021/bi9803675. [DOI] [PubMed] [Google Scholar]
- 21.Mazumder B, Barik S. Bacterial expression of human respiratory syncytial viral phosphoprotein P and identification of Ser237 as the site of phosphorylation by cellular casein kinase II. Virology. 1994;205:104–111. doi: 10.1006/viro.1994.1623. [DOI] [PubMed] [Google Scholar]
- 22.Moyer S A, Baker S C, Lessard J L. Tubulin: A factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc Natl Acad Sci USA. 1986;83:5406–5409. doi: 10.1073/pnas.83.15.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyer S A, Baker S C, Horikami S. Host cell proteins required for measles virus reproduction. J Gen Virol. 1990;71:775–783. doi: 10.1099/0022-1317-71-4-775. [DOI] [PubMed] [Google Scholar]
- 24.Murti K, Chen M, Goorha R. Interaction of frog virus 3 with the cytomatrix: role of microfilaments in virus release. Virology. 1985;142:317–325. doi: 10.1016/0042-6822(85)90340-x. [DOI] [PubMed] [Google Scholar]
- 25.Pearce-Pratt R, Malamud D, Phillips D. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravkov E, Nichol S, Peters C J, Compans R. Role of actin microfilaments in black creek canal virus morphogenesis. J Virol. 1998;72:2865–2870. doi: 10.1128/jvi.72.4.2865-2870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schluter K, Jockusch B M, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 29.Schutt C E, Myslik J C, Rozycki M D, Goonesekere N C, Lindberg U. The structure of crystalline profilin-β-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 30.Selden L, Kinisian H, Estes J, Gershman L. Impact of profilin on actin-bound nucleotide exchange and actin polymerization dynamics. Biochemistry. 1999;38:2769–2778. doi: 10.1021/bi981543c. [DOI] [PubMed] [Google Scholar]
- 31.Shartava A, Monteiro C, Bencscath F, Schneider K, Chait B, Gussio R, Casoria-Scott L, Shah A, Heuerman C, Goodman S. A posttranslational modification of β-actin contributes to the slow dissociation of the spectrin-protein 4.1-actin complex of irreversibly sickled cells. J Cell Biol. 1995;128:805–818. doi: 10.1083/jcb.128.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka M, Shibata H. Poly(l-proline)-binding proteins from chick embryos are a profilin and a profilactin. Eur J Biochem. 1985;151:291–297. doi: 10.1111/j.1432-1033.1985.tb09099.x. [DOI] [PubMed] [Google Scholar]
- 33.Theriot J A, Mitchison T J. The three faces of profilin. Cell. 1993;75:835–838. doi: 10.1016/0092-8674(93)90527-w. [DOI] [PubMed] [Google Scholar]
- 34.Theriot J, Rosenblatt J, Portnoy D, Goldschmidt-Clermont P, Mitchison T. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell. 1994;76:505–517. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 35.Tuderman L, Kuuti E R, Kivirikko K I. An affinity-column procedure using poly(l-proline) for the purification of prolyl hydroxylase. Purification of the enzyme from chick embryos. Eur J Biochem. 1975;52:9–16. doi: 10.1111/j.1432-1033.1975.tb03967.x. [DOI] [PubMed] [Google Scholar]
- 36.Zeile W L, Purich D L, Southwick F S. Recognition of two classes of oligoproline sequences in profilin-mediated acceleration of actin-based Shigella motility. J Cell Biol. 1996;133:49–59. doi: 10.1083/jcb.133.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeile W L, Condit R C, Lewis J L, Purich D L, Southwick F S. Vaccinia locomotion in host cells: evidence for the universal involvement of actin-based motility sequences ABM-1 and ABM-2. Proc Natl Acad Sci USA. 1998;95:13917–13922. doi: 10.1073/pnas.95.23.13917. [DOI] [PMC free article] [PubMed] [Google Scholar]