Abstract
We previously reported that bone morphogenetic protein (BMP)‐4 induces epithelial–mesenchymal transition in a pancreatic cancer cell line. To further investigate the detailed molecular mechanism of BMP action in pancreatic cancer, we carried out comprehensive microarray analysis in Panc‐1 cells. The microarray analysis elucidated novel BMP target genes, and among them, the calcium‐binding protein S100P was identified as an upregulated gene. S100P induction by BMP4 was confirmed by real‐time reverse transcription–polymerase chain reaction and western blot analysis in Panc‐1 and HPDE cells. Short interfering RNA‐based knockdown of S100P expression sufficiently repressed BMP4‐induced cell migration in Panc‐1 cells. Because Panc‐1 and HPDE cells express wild‐type Smad4, we hypothesized that Smad4 might be indispensable for S100P induction by BMP4. S100P induction by BMP4 was not observed in the Smad4‐null cell line BxPC3, and was sufficiently attenuated in short interfering RNA‐based Smad4‐knockdown Panc‐1 cells. Interestingly, detailed promoter analysis revealed that upregulation of S100P by BMP4 was independent of the Smad‐binding element, indicating that an additional unknown downstream factor of the Smad4‐dependent pathway is necessary for this induction. These findings are the first of their kind, and this Smad4‐dependent regulation of S100P by BMP signaling might explain the migratory mechanism of cancer cells, which is still unknown. (Cancer Sci 2009; 100: 103–110)
Abbreviations:
- AcD
actinomycin D
- BMP
bone morphogenetic protein
- BrdU
5‐bromo‐2‐deoxyuridine
- BSA
bovine serum albumin
- CHX
cycloheximide
- DMEM
Dulbecco's modified Eagle's medium
- EMT
epithelial–mesenchymal transition
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde‐3‐phosphate dehydrogenase
- ID
inhibitor of DNA binding and differentiation
- MAPK
mitogen‐activated kinase
- PBS
phosphate‐buffered saline
- PCR
polymerase chain reaction
- RT
reverse transcription
- SBE
Smad binding element
- siRNA
short interfering RNA
- TGF
transforming growth factor.
Pancreatic cancer has high mortality due to its invasive nature, and patients with pancreatic cancer often have distant metastasis by the time of diagnosis. Recent research has uncovered complex regulation of the cellular processes behind cancer invasion and metastasis, but many facets of these processes remain unknown. Among the numerous biological processes involved, EMT is a crucial step for cancer invasion and metastasis.( 1 ) We previously reported the significant contribution of BMP4 to the induction of EMT in the pancreatic cancer cell line Panc‐1.( 2 ) BMP4 belongs to the TGFβ superfamily, and its signal phosphorylates the receptor‐activated Smad proteins Smad1, Smad5, and Smad8.( 3 , 4 , 5 ) Activated Smad protein forms a heteromeric complex with Smad4, then translocates to the nucleus and affects the transcription of target genes.( 6 ) This signaling pathway is referred to as the canonical Smad pathway. In addition to this pathway, the BMP signal is reported to activate p38 MAPK during renal branching morphogenesis.( 7 ) We reported that both the canonical Smad signaling pathway and the p38 MAPK signaling pathway contribute to induction of the homeobox gene MSX2, which is a key molecule for the EMT of pancreatic cancer.( 2 , 8 )
In the present study, we carried out comprehensive examination by using microarray analysis to elucidate possible candidate genes that can contribute to the malignant biological behavior induced by BMP4 in pancreatic cancer cells. As a result, the calcium‐binding protein S100P was found to be upregulated both at the mRNA and protein level by BMP4 stimulation in Panc‐1 cells and the normal human pancreatic duct epithelial cell line HPDE.( 9 ) S100P is a calcium‐binding protein belonging to the S100 protein family, which consists of at least 13 members located as a cluster at 1q21, whereas S100P is located at 4p16.( 10 ) S100P is reported to stimulate cell proliferation and cell survival in NIH3T3 cells via the receptor for advanced glycation end products signaling pathway.( 11 ) S100P is also reported to stimulate cell proliferation and invasion in pancreatic cancer cells.( 12 , 13 ) Furthermore, a recent report suggested that S100P is a marker of pancreatic carcinogenesis.( 14 )
Despite the potent tumor‐promoting effect of S100P, an upstream regulator of its expression has not been well documented. Therefore, we investigated the role of S100P as a target of BMP4 and the possible mechanism of S100P regulation by this cytokine in pancreatic cancer cells. The detailed analysis has clarified that S100P is a novel BMP‐target gene, and Smad4 is indispensable to this process. However, transcriptional regulation of the S100P gene was found to be SBE independent, which indicates that an unknown Smad4‐dependent target molecule is critical in controlling S100P induction in Panc‐1 cells.
Materials and Methods
Cell lines and cell culture. The human pancreatic cancer cell lines BxPC3, Panc‐1, Panc‐1‐derived, and BxPC3‐derived, were maintained in DMEM supplemented with 10% FBS. The normal human pancreatic duct epithelial cell line HPDE cells were provided by Dr M.S. Tsao and maintained as described previously.( 9 ) Cells were incubated in a humidified incubator at 37°C, 5% CO2.
Cytokines and chemical substances. Recombinant human BMP4 was purchased from R. & D. Systems (Minneapolis, MN, USA) and dissolved in PBS supplemented with 0.1% BSA. BMP4 treatment was carried out in DMEM containing 1% serum. CHX was purchased from Wako Pure Chemical Industries (Osaka, Japan) and used at a concentration of 100 µg/mL in the cell culture. AcD was purchased from Sigma Aldrich Japan (Tokyo, Japan) and used at a concentration of 1 µg/mL in the cell culture.
Microarray. CodeLink whole Human Genome Expression Bioarray (Amersham Biosciences, Buckinghamshire, UK), representing approximately 55 000 of the most well‐annotated human genes published in public databases, was used for microarray analysis. The platform uses single‐color detection rather than dual‐color detection, where multiple experimental comparisons are possible without replicating the reference sample. Procedures were carried out according to the manufacturer's protocol and all reagents were provided in the kit.
Gene expression data analysis. Array slides were scanned using an Arrayworx (GE Healthcare Biosciences, Piscatway, NJ, USA), and expression values were measured and manipulated subsequently by CodeLink Expression Analysis v4.0 software (Amersham Biosciences). Each array contained a total of 55 776 spots, of which 54 840 were for human (non‐bacterial) genes. Among the 54 840 gene expression values, low‐intensity spots whose values were less than the detection threshold were all adjusted to the threshold. All good‐quality spots indicated by the ‘G’ flag as well as the low‐intensity spots indicated by the ‘L’ flag were further processed for statistical analysis. Those spots with poor quality were excluded from the analysis. Statistically, 54 470 spots were detected as either ‘G’ or ‘L’ flag in both experiments (Panc‐1 control vs Panc‐1 BMP4), thus most of genes were used subsequently in the following analysis. To normalize data we compared the gene expression values from the two experiments and drew an intensity‐ratio plot (MA‐plot). For each data point, an intensity (A)‐dependent normalization method (Lowess) was applied for adjusting the ratio (M) value.( 15 ) In Lowess, we used the simple adjusting method of log2 M–c(A), where c(A) is the mean ratio of the nearest 10 000 data points surrounding the current data point. After the calibration of M‐values, P‐values for evaluating how significantly those M‐values were apart from the mean were obtained under a cumulative normal distribution model whose variance was calculated with the same 10 000 data points. The extent of each gene expression change was represented by the Z‐score, which was calculated as described previously.( 16 ) Finally, Bon Ferroni correction was applied to each data point and genes showing statistically significant changes in gene expression were obtained. Those genes were analyzed functionally with the annotations and information from the GeneCards homepage (http://www.genecards.org/index.shtml).
RNA isolation and cDNA synthesis. Total cytoplasmic RNA was isolated from cultured cells using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. DNaseI (Qiagen) treatment was carried out to remove any DNA contamination for all of the RNA samples. For real‐time RT‐PCR analyses, cDNA was generated with 1 µg total RNA using the RETROscript kit (Ambion, Austin, TX, USA) according to the manufacturer's protocol.
Real‐time RT‐PCR. For the quantification of S100P, we used real‐time RT‐PCR with LightCycler and LightCycler – FastStart DNA Master SYBR Green I (Roche Diagnostics, Basel, Switzerland). All reactions were carried out according to the manufacturer's protocol. The primer sequences were as follows: ID1 forward 5′‐CGGATCTGAGGGAGAACAAG‐3′ and ID1 reverse 5′‐CTGAGAAGCACCAAACGTGA‐3′; ID2 forward 5′‐CATCTTGGACCTGCAGATCG‐3′ and ID2 reverse 5′‐ATGAACACCGCTTATTCAGC‐3′. The primer sequences for GAPDH, p21, and S100P were described previously.( 14 , 17 , 18 ) The annealing temperatures for these primer sets were 60°C. The specificity of each PCR was confirmed by melting curve analysis. The level of S100P expression in each sample was normalized by the respective GAPDH expression level as described previously.( 19 ) Each experiment was repeated at least twice.
Establishment of Smad4‐expressing BxPC3 cells. To establish the Smad4‐expressing BxPC3 cells, we transfected 2 µg pcDNA3 Flag‐Smad4 vector to BxPC3 cells and selected as described previously.( 2 ) The control cell line B3EV was established as described previously.( 8 )
RNA interference. RNA interference against Smad4 has been described previously.( 2 ) Briefly, the Smad4 siRNA‐expressing vectors were generated by cloning the synthesized and annealed oligonucleotides into the pBAsi‐U6 Neo DNA vector (Takara Bio, Ohtu, Japan) and 1 µg Smad4 siRNA‐expressing vector was transfected to Panc‐1 cells. We previously established the stable Smad4 knockdown cell line 643S4si.( 2 ) In the present study, we established an alternative stable Smad4 knockdown cell line 1446S4si, by targeting GTACTTCATACCATGCCGA, corresponding to the nucleotides of human Smad4 under the control of the human U6 promoter. The control clone Hu6 was generated as described previously.( 2 ) Transient knockdown of S100P was carried out by using HP Validated siRNA against human S100P (Qiagen) at a final concentration of 20 nmol/L. As a negative control, AllStars Negative Control siRNA (Qiagen) was used at the same concentration. The siRNA transfection was carried out using HiPerFect Transfection Reagent (Qiagen) according to the manufacturer's protocol.
Western blotting analysis. Western blotting analysis was done as described previously.( 2 ) The primary antibodies used in the present study were as follows: α‐tubulin (sc‐8035; Santa Cruz Biotechnology, Santa Cruz, CA, USA), S100P (AF2957; R. & D. Systems), and Smad4 (sc‐7966; Santa Cruz Biotechnology). As a secondary antibody, horseradish peroxidase‐conjugated antimouse antibody (Amersham Biosciences) or horseradish peroxidase‐conjugated antigoat antibody (Santa Cruz Biotechnology) was used. Reactive bands were detected using ECL western blotting detection reagents or ECL Plus western blotting detection reagents (Amersham Biosciences). The specific bands were subjected to densitometry analysis using Scion Image Software (Scion Corporation, Frederick, MD, USA).
Cell migration assay. Cell migration was examined using the 8‐µm pore 24‐well BD Falcon Cell Culture Insert (Franklin Lakes, NJ, USA). Cells were transfected with the siRNA indicated and then treated with PBS supplemented with 0.1% BSA or BMP4 at 50 ng/mL for 48 h. After preincubation, 10 000 cells were seeded into the Cell Culture Insert in triplicate, with the bottom chamber containing the same concentration of PBS supplemented with 0.1% BSA or BMP4 for the preincubation. Cells that migrated were counted directly in five random high‐power fields after 12 h incubation.
Cell growth assays. For the cell growth assay, cells were transfected with the indicated siRNA and then seeded 6000 cells per well in 96‐well plates in triplicate. The BrdU assay was carried out after 48 h incubation with or without 50 ng/mL BMP4 using the cell proliferation enzyme‐linked immunosorbent assay BrdU (Roche Diagnostics) according to the manufacturer's protocol.
Reporter assay. The pcDNA3 Flag‐Smad1 and pcDNA3 ALK3QD‐HA vector were provided by Dr T. Imamura. The 5′‐flanking regions of the human S100P gene were PCR amplified from the bacterial artificial chromosome clone RP11‐539L10 and inserted into the pGL4.17 [luc2/Neo] vector (Promega, Madison, WI, USA). The insert DNA sequences were confirmed by direct sequencing. As an internal control, the pRL‐TK vector (Promega) was used. For the luciferase assay, 50 ng/well of the indicated reporter vector, 5 ng/well of the pRL‐TK vector, and 0.5 µg/well of the pcDNA3 Flag‐Smad1 vector plus 0.5 µg/well of the pcDNA3 ALK3QD‐HA vector or 1 µg/well of the pcDNA4/V5‐His vector (empty vector) were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in 12‐well plates. The luciferase assay was done using the Dual‐Luciferase Reporter Assay Kit (Promega). Putative binding elements for the multiple transcription factors were estimated using MatInspector (Genomatix Software, München, Germany).( 20 )
Results
BMP4 treatment upregulates S100P in Panc‐1 and HPDE cells. As a first step, we carried out microarray analysis by comparing BMP4‐treated and vehicle‐treated Panc‐1 cells. As shown in Figure 1a, an intensity‐ratio plot (MA‐plot) showed that typical BMP target genes such as ID1, ID2, ID3, Smad6, and p21 ( 21 , 22 , 23 ) were classified as significantly upregulated by statistical analysis. At the same time, S100P was also classified as a significantly upregulated gene. In contrast, typical TGFβ target genes like plasminogen activator inhibitor‐1( 24 ) were not classified as being significantly changed. The genes that were significantly upregulated or downregulated by BMP4 treatment are summarized in Table 1 and Table 2, respectively. As shown in Figure 1b, real‐time RT‐PCR analysis showed a 20‐fold induction of S100P mRNA in Panc‐1 and HPDE cells after 24 h of BMP4 treatment. The protein‐level induction of S100P was also confirmed in Panc‐1 and HPDE cells, treated with 50 ng/mL BMP4 for 48 h (Fig. 1c). Furthermore, TGFβ treatment at 10 ng/mL did not induce S100P in Panc‐1 cells either at the mRNA or protein level (Fig. 1d).
Figure 1.
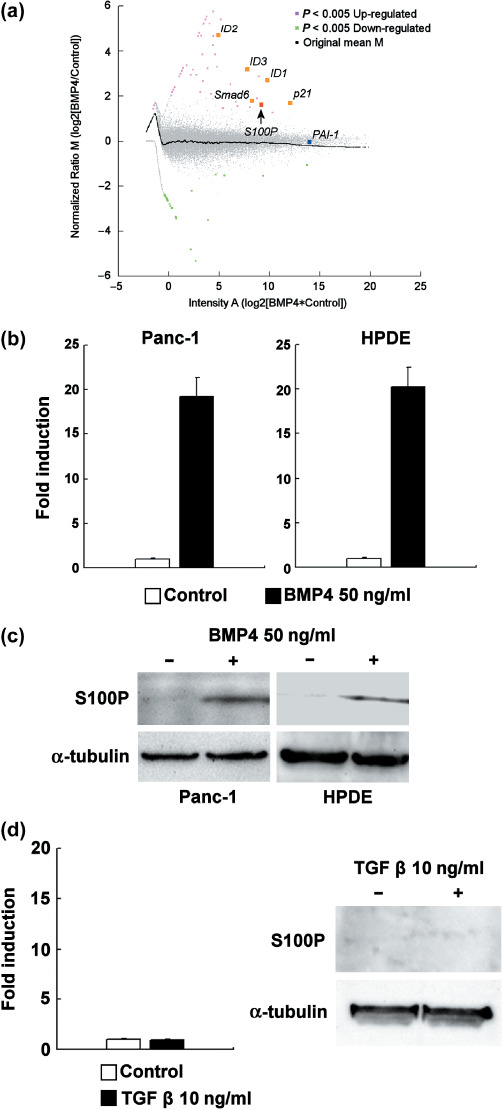
Bone morphogenetic protein (BMP) target gene induction in Panc‐1 cells. (a) The MA‐plot of microarray analysis in Panc‐1 cells. Red spots are significantly upregulated genes and green spots are significantly downregulated genes. Gray spots are unchanged genes. Cells were treated with 50 ng/mL BMP4 or vehicle for 24 h. (b) The real‐time reverse transcription–polymerase chain reaction (RT‐PCR) analysis of S100P induction in BMP4‐treated (50 ng/mL, 24 h) Panc‐1 and HPDE cells (n = 3). The expression level of S100P was normalized by the glyceraldehyde‐3‐phosphate dehydrogenase gene expression level, and the fold of induction was calculated by setting the control‐treated sample as 1. Error bars indicate SE. (c) Western blotting analysis of S100P induction in BMP4‐treated (50 ng/mL, 48 h) Panc‐1 and HPDE cells. α‐Tubulin is displayed as loading control. (d) Real‐time RT‐PCR analysis (n = 3, error bars indicate SE) and western blotting analysis of S100P induction in transforming growth factor (TGF)‐β‐treated (10 ng/mL, 24 h for RT‐PCR, 48 h for western blotting) Panc‐1 cells.
Table 1.
Genes upregulated significantly by bone morphogenetic protein 4 in Panc‐1 cells
| Gene symbol | Gene name | Gene function | Z‐score |
|---|---|---|---|
| LPL | Lipoprotein lipase | Metabolism | 21.5 |
| TAF5 L | TAF5‐like RNA polymerase II | Transcription | 17.6 |
| TNFSF6 | Tumor necrosis factor superfamily, member 6 | Signaling | 17.3 |
| ID1 | Inhibitor of DNA binding 1 | Transcription | 12.2 |
| ZNF221 | Zinc finger protein 221 | Transcription | 11.7 |
| TUB | Tubby homolog | Transcription | 11.4 |
| DUSP2 | Dual specificity phosphatase 2 | Signaling | 10.6 |
| ANP32D | Acidic nuclear phosphoprotein 32 family, member D | Unknown | 9.9 |
| TRPM6 | Transient receptor potential cation channel, subfamily M, member 6 | Signaling | 9.1 |
| TMEM158 | Transmembrane protein 158 | Unknown | 8.9 |
| ID2 | Inhibitor of DNA binding 2 | Transcription | 8.0 |
| ID3 | Inhibitor of DNA binding 3 | Transcription | 7.5 |
| CDKN1A | Cyclin‐dependent kinase inhibitor 1A | Cell cycle | 7.4 |
| GPER | G protein‐coupled estrogen receptor 1 | Signaling | 7.2 |
| SPATA9 | Spermatogenesis associated 9 | Unknown | 7.1 |
| Smad6 | SMAD family member 6 | Signaling | 7.1 |
| SHMT2 | Serine hydroxymethyltransferase 2 (mitochondrial) | Metabolism | 7.0 |
| GATA2 | GATA binding protein 2 | Transcription | 6.9 |
| MAP1D | Methionine aminopeptidase 1D | Metabolism | 6.8 |
| ATOH8 | Atonal homolog 8 (Drosophila) | Transcription | 5.5 |
| PROC | Protein C | Protease | 5.5 |
| LIN7B | Lin‐7 homolog B (C. elegans) | Cell structure | 5.3 |
| S100P | S100 calcium binding protein P | Signaling | 5.3 |
Table 2.
Genes downregulated significantly by bone morphogenetic protein 4 in Panc‐1 cells
| Gene symbol | Gene name | Gene function | Z‐score |
|---|---|---|---|
| KRT15 | Keratin 15 | Cell structure | –10.2 |
| ANAPC2 | Anaphase‐promoting complex subunit 2 | Cell cycle | –8.6 |
| SOX14 | Sex determining region Y‐box 14 | Transcription | –8.1 |
| PDEF | Prostate epithelium‐specific Ets transcription factor | Transcription | –7.9 |
| DDX4 | DEAD/H (Asp‐Glu‐Ala‐Asp/His) box polypeptide 4 | Signaling | –7.3 |
| NOS1 | Neuronal nitric oxide synthase | Metabolism | –7.1 |
| CISH | Cytokine‐inducible SH2‐containing protein | Signaling | –6.9 |
| SLC25A40 | Mitochondrial carrier family protein | Unknown | –6.3 |
| RFC2 | Replication factor C (activator 1) 2 | Cell cycle | –6.3 |
| ISLR | Immunoglobulin superfamily containing leucine‐rich repeat | Cell structure | –6.0 |
| TM4SF1 | Transmembrane 4 superfamily member 1 | Unknown | –6.0 |
| FLEG1 | Upregulated in lung cancer 9 | Unknown | –5.8 |
| BMP4 | Bone morphogenetic protein 4 | Signaling | –5.7 |
| SPC25 | AD024 protein | Cell cycle | –5.3 |
| MCM10 | Minichromosome maintenance deficient 10 | Cell cycle | –5.3 |
Induction of S100P by BMP4 is at the transcription level and requires de novo protein synthesis. As shown in Figure 2a, the time‐course experiment showed a remarkable increase in S100P mRNA after 12 h of BMP4 treatment. To confirm the transcriptional regulation of S100P, we treated the BMP4‐stimulated Panc‐1 cells (50 ng/mL, 24 h) with 1 µg/mL AcD. After the addition of AcD, the S100P mRNA expression level decreased immediately, compared with the control treatment (Fig. 2b). Based on this result, we concluded that BMP4 increases the transcription of S100P. We also evaluated the requirement of de novo protein synthesis for this phenomenon by treating Panc‐1 cells with 100 µg/mL CHX for 1 h before the addition of BMP4. The CHX treatment repressed S100P induction by BMP4 to the basal level, indicating that de novo protein synthesis is indispensable in this biological process (Fig. 2c).
Figure 2.
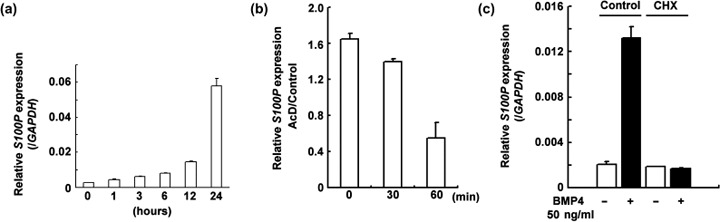
S100P induction by bone morphogenetic protein (BMP)‐4 in Panc‐1 cells. (a) Time‐course experiment of S100P induction in Panc‐1 cells (n = 3, BMP4 50 ng/mL). The expression level of S100P was normalized by the glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) expression level. (b) mRNA decay after actinomycin D (AcD) treatment in Panc‐1 cells treated for 24 h with 50 ng/mL BMP4 (n = 3). The expression level of S100P was normalized by the S100P expression level of control‐treated Panc‐1 cells at the corresponding time point. Error bars indicate SE. (c) S100P induction under the influence of cycloheximide (CHX) (n = 3). Cells were treated with 100 µg/mL CHX or vehicle for 1 h and treated with 50 ng/mL BMP4 or vehicle for 24 h. The expression level of S100P was normalized by the GAPDH expression level. Error bars indicate SE.
Knockdown of S100P attenuates BMP4‐induced cellular migration. As a previous report has shown that overexpression of S100P in Panc‐1 cells results in enhanced cellular migration in the absence of BMP4,( 12 ) we examined the contribution of S100P to the BMP4‐induced EMT in Panc‐1 cells by knocking down S100P. As shown in Figure 3a, transient transfection of siRNA against S100P reduces its gene expression by approximately 60% compared to that of control siRNA‐introduced cells. We then assessed the cellular migration of these cells. The knockdown of S100P attenuates approximately 50% of BMP4‐induced cellular migration (Fig. 3b). As a previous report indicated a stimulatory effect of S100P on cell proliferation,( 11 , 12 ) we also investigated the proliferation of control siRNA‐treated and S100P siRNA‐treated Panc‐1 cells with or without BMP4 treatment for 48 h. The BMP4 treatment reduced BrdU incorporation by 60% compared with the control treatment both in the control siRNA‐treated and S100P siRNA‐treated Panc‐1 cells (Fig. 3c). From this, we concluded that growth inhibition is the dominant effect of BMP4 in Panc‐1 cells.
Figure 3.
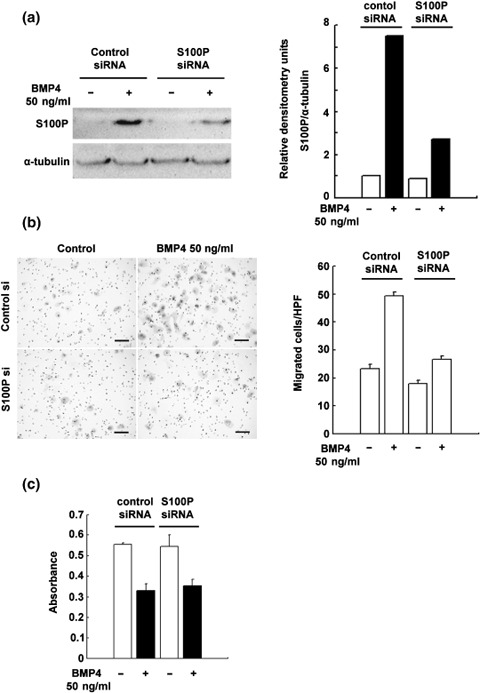
The effect of S100P knockdown by short interfering RNA (siRNA) on bone morphogenetic protein (BMP)‐4‐induced cell migration in Panc‐1 cells. (a) Western blotting analysis of S100P in control siRNA‐ or S100P siRNA‐treated Panc‐1 cells with or without BMP4 treatment (50 ng/mL, 48 h). Right panel represents the densitometry analysis. (b) The transwell migration assay in control siRNA‐ or S100P siRNA‐treated Panc‐1 cells with or without BMP4 treatment (n = 3, 50 ng/mL, 60 h). Error bars indicate SE. Black bar shows 100 µmol/L. (c) The 5‐bromo‐2‐deoxyuridine (BrdU) incorporation assay in control siRNA‐ or S100P siRNA‐treated Panc‐1 cells with or without BMP4 treatment (n = 3, 50 ng/mL, 48 h). Error bars indicate SE.
Smad4 is indispensable for S100P induction by BMP4. Because Panc‐1 and HPDE cells express wild‐type Smad4,( 25 , 26 ) we hypothesized that the induction of S100P by BMP4 might be Smad4 dependent. Thus, we examined whether BMP4 would upregulate S100P in the Smad4‐null pancreatic carcinoma cell line BxPC3.( 27 ) As expected, induction of S100P was not observed in BMP4‐treated BxPC3 cells (Fig. 4a). To further investigate Smad4‐dependent S100P induction by BMP4, we used the previously established stable Smad4 knockdown cell line 643S4si and the control cell line Hu6.( 2 ) In the present study, we also established a new stable Smad4 knockdown cell line, 1446S4si, by targeting a different position of human Smad4. As shown in Figure 4b, in 643S4si and 1446S4si cells, the expression of Smad4 was sufficiently repressed either with or without BMP4 treatment. The induction of S100P by BMP4 in these Smad4‐knockdown cells was decreased to basal levels (Fig. 4c), indicating an essential role for the Smad4‐dependent pathway in this transcriptional regulation. We decided to evaluate whether Smad4 is involved directly in the transcriptional regulation of S100P, and established the Smad4‐expressing cell lines BS4‐11 and BS4‐13 (Fig. 4d). The forced expression of Smad4 in BxPC3 cells could not restore S100P induction by BMP4 at the mRNA level (Fig. 4e), whereas induction of other representative BMP target genes such as ID1, ID2, and p21 could restore it. This suggests that induction of S100P by BMP4 requires an unknown factor that is induced by the BMP4–Smad4 pathway in Panc‐1 and HPDE cells, but not in BS4‐11 or BS4‐13 cells.
Figure 4.
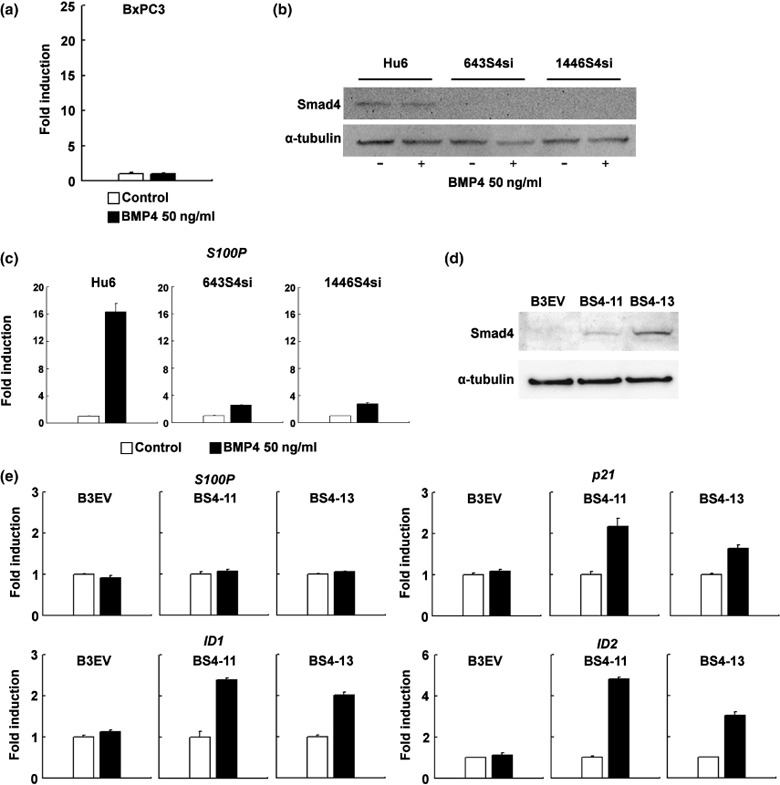
S100P induction by bone morphogenetic protein (BMP)‐4 in Smad4‐null or Smad4‐knockdown cell lines. (a) Real‐time reverse transcription–polymerase chain reaction (RT‐PCR) analysis of S100P induction in the Smad4‐null cell line BxPC3 (n = 3, 50 ng/mL BMP4, 24 h). The expression level of S100P was normalized by the glyceraldehyde‐3‐phosphate dehydrogenase gene expression level, and the fold of induction was calculated by setting the control‐treated sample as 1. Error bars indicate SE. (b) Western blotting analysis of Smad4 expression in Hu6, 643S4si, and 1446S4si cell lines with or without BMP4 treatment (50 ng/mL, 48 h). α‐Tubulin is displayed as a loading control. (c) Real‐time RT‐PCR analysis of S100P induction by BMP4 in Hu6 and Smad4‐knockdown cell lines (n = 3, 50 ng/mL BMP4, 24 h). Error bars indicate SE. (d) Western blotting analysis of Smad4 expression in B3EV, BS4‐11, and BS4‐13 cell lines. α‐Tubulin is displayed as a loading control. (e) Real‐time RT‐PCR analysis of S100P, ID1, ID2, and p21 induction by BMP4 in B3EV, BS4‐11, and BS4‐13 cell lines (n = 3, 50 ng/mL BMP4, 24 h). Error bars indicate SE.
Bone morphogenetic protein signal increases S100P transcription in a SBE‐independent manner. To clarify the mechanism of S100P induction by BMP4, we investigated the detailed transcriptional regulation of S100P by reporter assay in Panc‐1 cells. Figure 5a shows a schema of the S100P gene structure and reporter constructs. The contribution of the canonical Smad4‐dependent signal was evaluated by cotransfecting vectors that express a constitutively active form of the BMP receptor (ALK3QD) and Smad1. As shown in Figure 5b, the reporter activity of pGL4‐448, pGL4‐208, and pGL4‐171 was increased by cotransfection with pcDNA3 Flag‐Smad1 plus pcDNA3 ALK3QD‐HA compared to cotransfection with the empty vector, but this increase was lost with pGL4‐138 and pGL4 without an insert sequence. Because pGL4‐208 and pGL4‐171 do not contain putative SBE, we concluded that this reporter activity increase is regulated by a SBE‐independent mechanism via the promoter region at –171 to –138 bp. Figure 5c summarizes the transcription factors that possibly bind to the promoter region at –171 to –138 bp.
Figure 5.
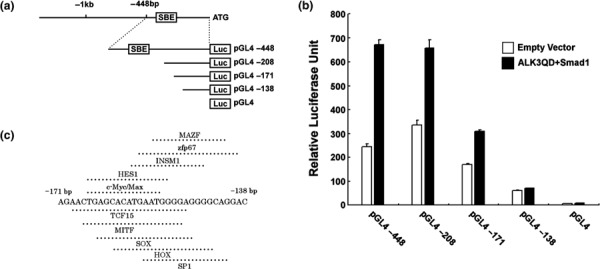
Reporter assay of S100P promoter deletion mutant constructs in Panc‐1 cells. (a) Promoter structure of the human S100P gene and the deletion mutant constructs. (b) Reporter assay of deletion mutant constructs in Panc‐1 cells (n = 3). The firefly luciferase activity of each sample was normalized by renilla luciferase activity. Error bars indicate SE. (c) Transcription factor binding sites in the promoter region at –171 to –138 bp.
Discussion
In the present study, we focused on the downstream target gene of BMP signaling in pancreatic carcinoma cells. We used microarray analysis to find novel BMP‐target genes, and among them, the calcium‐binding protein S100P gene was identified as being significantly upregulated by BMP. This finding is the first of its kind, and sheds light on the regulatory mechanism of this tumor‐promoting molecule.( 12 , 13 ) Altered expression of S100P has been reported in a wide variety of human cancers, including pancreatic cancer,( 14 ) uterine squamous carcinoma, and uterine adenocarcinoma.( 28 ) S100P over expression in breast cancer correlates with poor prognosis, indicating a potent tumor‐promoting effect in vivo.( 29 ) Despite this evidence, the upstream regulator of S100P expression had not been clarified until now. Our present results clearly reveal that BMP4 is the regulator of S100P expression in pancreatic duct epithelial cells. Furthermore, siRNA‐based knockdown of S100P sufficiently reduced the cellular migration of Panc‐1 cells by BMP4, which suggests that S100P is indispensable for BMP4‐induced pancreatic cancer cell migration. This result is in agreement with recent reports, which describe the essential role of S100P during cancer cell migration through the activation of multiple signaling pathways.( 12 , 30 ) The contribution of each signaling pathway activated by S100P during BMP4‐induced migration needs to be clarified by further analysis.
In the current study, we found that BMP4 induces S100P in pancreatic duct epithelial cell lines and that Smad4 is indispensable in this pathway. In addition, the induction of S100P by BMP4 requires de novo protein synthesis and the transcriptional regulation of S100P is independent of SBE, suggesting that an unknown downstream target molecule for Smad4 might play an essential role in this process. Furthermore, forced expression of Smad4 could not restore S100P induction by BMP4 whereas induction of other BMP‐target genes was restored, indicating that a target molecule of the BMP‐Smad4 pathway that can be induced in Panc‐1 and HPDE cells but not in BxPC3 cells is essential to induce S100P. Such a regulatory mechanism has not yet been fully described in pancreatic cancer, but a recent report has suggested the essential role of Smad4 as a mediator of the BMP signaling pathway in neural crest cells for development of the cardiac outflow tract.( 31 ) Conditional inactivation of Smad4 in neural crest cells results in multiple organ‐formation defects accompanied by alterations in the expression of numerous transcription factors. The present study has revealed the involvement of multiple transcription factors in the BMP signaling pathway. Further study is required to identify the downstream target of BMP signaling, which critically regulates the induction of S100P.
Acknowledgments
This work was supported in part by Grants‐in‐Aid #19590745 and #19890015 from the Ministry of Education, Science, Sports, and Culture of Japan. We thank Dr T. Imamura for providing the pcDNA Flag‐Smad4, pcDNA3 Flag‐Smad1, and pcDNA3 ALK3QD‐HA vectors. We thank Dr M.S. Tsao for providing HPDE cells.
References
- 1. Huber MA, Kraut N, Beug H. Molecular requirements for epithelial–mesenchymal transition during tumor progression. Curr Opin Cell Biol 2005; 17: 548–58. [DOI] [PubMed] [Google Scholar]
- 2. Hamada S, Satoh K, Hirota M et al . Bone morphogenetic protein 4 induces epithelial–mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J Cell Physiol 2007; 213: 768–74. [DOI] [PubMed] [Google Scholar]
- 3. Kretzschmar M, Liu F, Hata A, Doody J, Massagué J. The TGF‐β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev 1997; 11: 984–95. [DOI] [PubMed] [Google Scholar]
- 4. Nishimura R, Kato Y, Chen D, Harris SE, Mundy GR, Yoneda T. Smad5 and DPC4 are key molecules in mediating BMP‐2‐induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J Biol Chem 1998; 273: 1872–9. [DOI] [PubMed] [Google Scholar]
- 5. Kawai S, Faucheu C, Gallea S et al . Mouse smad8 phosphorylation downstream of BMP receptors ALK‐2, ALK‐3, and ALK‐6 induces its association with Smad4 and transcriptional activity. Biochem Biophys Res Commun 2000; 271: 682–7. [DOI] [PubMed] [Google Scholar]
- 6. Lagna G, Hata A, Hemmati‐Brivanlou A, Massagué J. Partnership between DPC4 and SMAD proteins in TGF‐β signalling pathways. Nature 1996; 383: 832–6. [DOI] [PubMed] [Google Scholar]
- 7. Hu MC, Wasserman D, Hartwig S, Rosenblum ND. p38MAPK acts in the BMP7‐dependent stimulatory pathway during epithelial cell morphogenesis and is regulated by Smad1. J Biol Chem 2004; 279: 12051–9. [DOI] [PubMed] [Google Scholar]
- 8. Satoh K, Hamada S, Kimura K et al . Up‐regulation of MSX2 enhances the malignant phenotype and is associated with twist 1 expression in human pancreatic cancer cells. Am J Pathol 2008; 172: 926–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long‐term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol 1996; 148: 1763–70. [PMC free article] [PubMed] [Google Scholar]
- 10. Schafer BW, Wicki R, Engelkamp D, Mattei MG, Heizmann CW. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium‐binding protein family. Genomics 1995; 25: 638–43. [DOI] [PubMed] [Google Scholar]
- 11. Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE). J Biol Chem 2004; 279: 5059–65. [DOI] [PubMed] [Google Scholar]
- 12. Arumugam T, Simeone DM, Van Golen K, Logsdon CD. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res 2005; 11: 5356–64. [DOI] [PubMed] [Google Scholar]
- 13. Whiteman HJ, Weeks ME, Dowen SE et al . The role of S100P in the invasion of pancreatic cancer cells is mediated through cytoskeletal changes and regulation of cathepsin D. Cancer Res 2007; 67: 8633–42. [DOI] [PubMed] [Google Scholar]
- 14. Ohuchida K, Mizumoto K, Egami T et al . S100P is an early developmental marker of pancreatic carcinogenesis. Clin Cancer Res 2006; 12: 5411–16. [DOI] [PubMed] [Google Scholar]
- 15. Workman C, Jensen LJ, Jarmer H et al . A new non‐linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 2002: 3: research0048.1–.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn 2003; 5: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki T, Miki Y, Nakata T et al . Steroid sulfatase and estrogen sulfotransferase in normal human tissue and breast carcinoma. J Steroid Biochem Mol Biol 2003; 86: 449–54. [DOI] [PubMed] [Google Scholar]
- 18. Han S, Roman J. COX‐2 inhibitors suppress lung cancer cell growth by inducing p21 via COX‐2 independent signals. Lung Cancer 2006; 51: 283–96. [DOI] [PubMed] [Google Scholar]
- 19. Hamada S, Watanabe K, Hirota M et al . β‐Catenin/TCF/LEF regulate expression of the short form human Cripto‐1. Biochem Biophys Res Commun 2007; 355: 240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cartharius K, Frech K, Grote K et al . MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 2005; 21: 2933–42. [DOI] [PubMed] [Google Scholar]
- 21. Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. ID genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem 1999; 274: 19838–45. [DOI] [PubMed] [Google Scholar]
- 22. Ku M, Howard S, Ni W, Lagna G, Hata A. OAZ regulates bone morphogenetic protein signaling through Smad6 activation. J Biol Chem 2006; 281: 5277–87. [DOI] [PubMed] [Google Scholar]
- 23. Haudenschild DR, Palmer SM, Moseley TA, You Z, Reddi AH. Bone morphogenetic protein (BMP)‐6 signaling and BMP antagonist noggin in prostate cancer. Cancer Res 2004; 64: 8276–84. [DOI] [PubMed] [Google Scholar]
- 24. Liu C, Yao J, De Belle I, Huang RP, Adamson E, Mercola D. The transcription factor EGR‐1 suppresses transformation of human fibrosarcoma HT1080 cells by coordinated induction of transforming growth factor‐β1, fibronectin, and plasminogen activator inhibitor‐1. J Biol Chem 1999; 274: 4400–11. [DOI] [PubMed] [Google Scholar]
- 25. Schutte M, Hruban RH, Hedrick L et al . DPC4 gene in various tumor types. Cancer Res 1996; 56: 2527–30. [PubMed] [Google Scholar]
- 26. Qian J, Niu J, Li M, Chiao PJ, Tsao MS. In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K‐ras oncogenic activation in pancreatic carcinogenesis. Cancer Res 2005; 65: 5045–53. [DOI] [PubMed] [Google Scholar]
- 27. Fink SP, Mikkola D, Willson JK, Markowitz S. TGF‐β‐induced nuclear localization of Smad2 and Smad3 in Smad4 null cancer cell lines. Oncogene 2003; 22: 1317–23. [DOI] [PubMed] [Google Scholar]
- 28. Chao A, Wang TH, Lee YS et al . Molecular characterization of adenocarcinoma and squamous carcinoma of the uterine cervix using microarray analysis of gene expression. Int J Cancer 2006; 119: 91–8. [DOI] [PubMed] [Google Scholar]
- 29. Wang G, Platt‐Higgins A, Carroll J et al . Induction of metastasis by S100P in a rat mammary model and its association with poor survival of breast cancer patients. Cancer Res 2006; 66: 1199–207. [DOI] [PubMed] [Google Scholar]
- 30. Nazmi AR, Muller‐Tidow C, Gerke V. Characterization of the Ca2+ regulated ezrin–S100P interaction and its role in tumor cell migration. J Biol Chem Epub 2008 Aug 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ko SO, Chung IH, Xu X et al . Smad4 is required to regulate the fate of cranial neural crest cells. Dev Biol 2007; 312: 435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]


