Abstract
A hypoxic microenvironment is a characteristic feature of pancreatic cancer, and induces the expressions of various genes involved in malignant behaviors. Insulin‐induced gene 2 (Insig2) has recently been shown to be correlated with cellular invasion in colon cancer. However, there have been no reports regarding its expression in pancreatic cancer. In this study, we evaluated Insig2 mRNA expression and the biological function of Insig2 in pancreatic cancer. We measured Insig2 mRNA expression in cultured pancreatic cancer cell lines and invasive ductal carcinoma (IDC) cells, normal pancreatic epithelial cells, and pancreatic intraepithelial neoplasia cells obtained by laser‐capture microdissection. We also investigated the effects of Insig2‐targeting siRNAs on the cell proliferation and cell invasion of pancreatic cancer cell lines. All pancreatic cancer cell lines expressed Insig2 mRNA. The PANC‐1 and MIA PaCa‐2 pancreatic cancer cell lines showed >2‐fold higher Insig2 mRNA expression levels under hypoxic conditions (1% O2) than under normoxic conditions (21% O2). Cell proliferation was significantly decreased in SUIT‐2 cells and cell invasion was significantly decreased in SUIT‐2, Capan‐2, and CFPAC‐1 cells after transfection of the Insig2‐targeting siRNAs. In analyses of microdissected cells, cells from IDC tissues expressed significantly higher levels of Insig2 mRNA than normal pancreatic cells (P < 0.001) and pancreatic intraepithelial neoplasia cells (P = 0.082). In analyses of IDC cells, the levels of Insig2 mRNA expression were significantly higher in late‐stage patients than in early‐stage patients. The present data suggest that Insig2 is associated with the malignant potential of pancreatic cancer under hypoxic conditions. (Cancer Sci 2011; 102: 1137–1143)
Pancreatic cancer is the fourth leading cause of cancer‐related death in Western Europe and has the lowest survival rate of any solid tumor.( 1 , 2 ) Owing to a lack of early detection methods and the absence of effective biomarkers, pancreatic cancer patients are usually diagnosed at late stages and have a 5‐year survival rate of <5%. The first‐line agent gemcitabine produces some clinical benefits in the advanced setting but yields limited disease control, with <15% of patients being progression‐free at 6 months after diagnosis.( 3 , 4 ) Recently, advances in our understanding of the genetics and epigenetics of pancreatic cancer have revealed that alterations of several tumor‐related genes, including K‐ras, p53, MMP, HGF, and EGFR,( 5 , 6 , 7 , 8 , 9 , 10 ) could underlie the aggressiveness of this neoplasm and its resistance to conventional therapies.( 11 ) There is a great need to understand the biological mechanisms that contribute to pancreatic cancer development and progression. Therefore, the identification of effective markers for pancreatic neoplasms to more effectively detect pancreatic cancer and its precursors may contribute to the discovery of new approaches to treat this fatal disease.
Pancreatic cancer is characterized by intratumoral hypoxia, which is involved in early and aggressive local invasion and metastatic potential. Hypoxia‐inducible factor (HIF)‐1 is the major transcriptional activator of hypoxia‐responsive genes, and intratumoral hypoxia is associated with an increased risk of metastasis.( 12 ) In this study, we carried out microarray analyses using the PDAC cell line under normoxic conditions (21% O2) and hypoxic conditions (1% O2), and our expression profiling identified several genes that were aberrantly expressed in PDAC cells under hypoxic conditions. Some of these genes, namely PFKFB4, ADM, ANKRD37, ENO2, HIG2, ANGPTL4, SPAG4, and UPK1A, have been reported to be differentially expressed in pancreatic cancer or other cancers.( 13 , 14 , 15 )
Insulin‐induced gene 2 (Insig2) was also upregulated in this expression profiling, and was recently reported to be a biomarker for colon cancer.( 16 ) The Insig2 gene encodes closely related endoplasmic reticulum proteins that regulate the proteolytic activation of sterol regulatory element‐binding proteins, comprising transcription factors that activate the synthesis of cholesterol and fatty acids in animal cells.( 17 , 18 ) Insig2 is correlated with cellular invasion in colon cancer and has a univariate negative prognostic capacity to discriminate human colon cancer survivorship. Overexpression of Insig2 appears to suppress chemotherapeutic drug treatment‐induced expression of Bcl2‐associated X protein (Bax) expression and activation in human colorectal cancer cells.( 19 , 20 ) Insig2 was also found to localize to the mitochondria/heavy membrane fraction and to associate with conformationally altered Bax in HeLa cells.( 21 ) Moreover, Insig2 alters the expressions of several additional apoptosis genes located in mitochondria, further supporting its newly described functional role in regulating mitochondria‐mediated apoptosis in human colorectal cancer cells.( 16 ) These data suggest that Insig2 plays an important role in carcinogenesis or cancer progression. However, there are no reports regarding the involvement of Insig2 in pancreatic cancer.
In the present study, we analyzed Insig2 expression in cultured pancreatic cancer cell lines exposed to normoxic and hypoxic conditions and in invasive ductal carcinoma (IDC) cells, normal pancreatic epithelial cells, and pancreatic intraepithelial neoplasia (PanIN) cells obtained by laser‐capture microdissection. We also investigated the biological function of Insig2 by examining the effects of Insig2‐targeting siRNAs on the cell proliferation and cell invasion of pancreatic cancer cell lines. Our data suggest that Insig2 is associated with carcinogenesis and malignant behaviors in pancreatic cancer.
Materials and Methods
Cell lines. The following 15 pancreatic cancer cell lines were used in this study: human pancreatic cancer cell lines SUIT‐2, AsPC‐1, BxPC‐3, PANC‐1, KP‐1N, KP‐2, and KP‐3 (generously donated by Dr. H. Iguchi, National Shikoku Cancer Center, Matsuyama, Japan); MIA PaCa‐2 (Japanese Cancer Resource Bank, Tokyo, Japan); Capan‐1, Capan‐2, CFPAC‐1, H48N, Hs 766T, and SW1990 (American Type Culture Collection, Manassas, VA, USA); and NOR‐P1 (established by Dr. N. Sato in our laboratory).( 22 ) A human pancreatic ductal epithelial (HPDE) cell line (HPDE6‐E6E7 clone6) immortalized by transduction with the E6/E7 genes of human papillomavirus 16 (kindly provided by Dr. Ming‐Sound Tsao, University of Toronto, Toronto, Canada) was also used. The cells were maintained as described previously.( 23 , 24 )
Clinical samples. Invasive ductal carcinoma cells from 29 patients, PanIN cells from nine patients, and normal pancreatic ductal epithelial cells from 32 patients were selectively isolated using a laser microdissection and pressure catapulting system (PALM Microlaser Technologies, Bernried, Germany) in accordance with the manufacturer’s protocols. All the tissues were taken from surgically resected specimens at the Department of Surgery and Oncology, Kyushu University Hospital (Fukuoka, Japan) and its affiliated hospitals from December 2005 to November 2008. All the tumors were staged according to the TNM classification system of the International Union against Cancer.( 25 ) The histologic grading of the tumors and the diagnosis of PanIN lesions were carried out according to the classification system of the World Health Organization.( 26 ) Other pathological variables (lymphatic invasion, vascular invasion, and perineural invasion) were assessed using the classification system of the Japan Pancreas Society.( 27 ) This study was carried out in accordance with the principles embodied in the Declaration of Helsinki. The study was also approved by the Ethics Committee of Kyushu University and carried out according to the Ethical Guidelines for Human Genome/Gene Research enacted by the Japanese Government.( 28 )
Quantitative assessment of Insig2 mRNA levels by one‐step real‐time qRT‐PCR. Total RNA was extracted from pellets of cultured cells using a High Pure RNA Kit (Roche Diagnostics, Mannheim, Germany) with DNase I treatment (Roche Diagnostics) according to the manufacturer’s instructions. We designed specific primers (Insig2: forward, 5′‐TCA CAC TGG CTG CAC TAT CC‐3′ and reverse, 5′‐ACA GTT GCC AAG AAG GCA AT‐3′; 18S rRNA: forward, 5′‐GTA ACC CGT TGA ACC CCA TT‐3′ and reverse, 5′‐CCA TCC AAT CGG TAG TAG CG‐3′), and carried out blast searches to ensure the specificity of each primer. The extracts were analyzed by qRT‐PCR using a QuantiTect SYBR Green RT‐PCR Kit (Qiagen, Tokyo, Japan) and a Chrom4 real‐time PCR Detection System (Bio‐Rad Laboratories, Hercules, CA, USA). Each reaction mixture was initially incubated at 50°C for 30 min to allow reverse transcription, in which first‐strand cDNA was synthesized by priming the total RNA with the same gene‐specific primer (reverse). The PCR was initiated by incubation at 95°C for 15 min to activate the polymerase, followed by 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. Each primer set used in this study produced a single prominent band of the expected size after electrophoresis. Each sample was analyzed twice, and any samples showing more than 10% deviation in the qRT‐PCR values were tested a third time. The level of mRNA expression in each sample was calculated by reference to a standard curve generated using total RNA from the PANC‐1 human pancreatic cancer cell line. The expression of Insig2 mRNA was normalized by the expression of 18S rRNA mRNA. A cut‐off point for Insig2 mRNA expression was selected by searching for the cut‐off point yielding the smallest log‐rank P‐value, and the expression was divided to high and low level groups.
Microdissection‐based quantitative analysis of Insig2 mRNA. Frozen tissue samples were cut into 5‐μm thick sections. One section from each sample was stained with H&E for histologic examination. Similar numbers of cells were isolated from sections of IDC lesions, PanIN lesions, and normal ductal epithelium. More than 500 cells were obtained from each IDC section, whereas 3–10 sections were required to isolate sufficient normal ductal epithelial cells and PanIN cells owing to the lower numbers of these cells per section. After the microdissection, total RNA was extracted from the selected cells and subjected to qRT‐PCR for quantification of Insig2 mRNA expression.( 29 , 30 )
Microarray analysis. We carried out microarray analyses using MIA PaCa‐2 cells cultured under normoxic conditions (21% O2) and hypoxic conditions (1% O2) for 24 h. The qualities of the RNA samples were evaluated using a 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany) as described previously.( 31 ) We used a HumanWG‐6 Expression BeadChip (Illumina, San Diego, CA, USA) for these analyses. The data were analyzed using BeadStudio software version 3.2.3 (Illumina). All of the microarray data were deposited in CIBEX under the accession number CBX147 (http://cibex.nig.ac.jp/index.jsp).
Transfection of Insig2‐targeting siRNAs. Pancreatic cancer cells were transfected with Insig2‐si1 (sense, 5′’‐cuauguucguucuugguuatt‐3′; antisense, 5′‐uaaccaagaacgaacauagtt‐3′) and Insig2‐si2 (sense, 5′‐cucacacuggcugcacuautt‐3′; antisense, 5′‐auagugcagccagugugagtt‐3′) siRNAs (Qiagen) by electroporation using a Nucleofector System (Amaxa Biosystems, KölnCologne, Germany) according to the manufacturer’s instructions. To verify the specificity of the Insig2 knockdown effects, we used negative control siRNAs (Qiagen).
Cell proliferation assay. Cell proliferation was evaluated by measuring the fluorescence intensity of propidium iodide (PI) as described previously.( 32 ) Pancreatic cancer cells were transfected with the Insig2‐targeting siRNAs and negative control siRNAs, then seeded in 90‐mm dishes at 1 × 106 cells/dish. At 24 h after the transfection, viable cells were plated in 24‐well tissue culture plates (Becton Dickinson Labware, Bedford, MA, USA) at 2 × 104 cells/well and cultured for the indicated hours. Cell proliferation was then evaluated by the PI assay. All experiments were carried out in triplicate wells and repeated at least three times.
Invasion assay. Using SUIT‐2, Capan‐2, and CFPAC‐1 cells, the invasion of pancreatic cancer cells was evaluated by the number of cells invading Matrigel‐coated Transwell chambers (Becton Dickinson Labware). Briefly, Transwell inserts with 8‐μm pores were coated with Matrigel (20 μg/well for all cells; Becton Dickinson) and reconstituted with DMEM supplemented with 10% FBS for 2 h before the experiment. Cells (2 × 105 cells/mL) were seeded into the upper chambers in 250 μL DMEM supplemented with 10% FBS. The same medium (750 μL) was placed in the lower wells. Thereafter, all cells were incubated for 4848 h. Cells that had degraded the Matrigel and invaded to the lower surface of the Matrigel‐coated membranes were fixed with 70% ethanol, stained with H&E, and counted in five random fields at ×400 magnification under a light microscope. The results were expressed as the average number of invasive cells per field.
Statistical analysis. Comparison between two groups was done using Student’s t‐test. Survival analyses undertaken using Kaplan–Meier analyses and curves were compared using the log‐rank test. Values are expressed as the mean ± SD. All experiments were repeated twice. Statistical significance was defined as P < 0.05. All statistical analyses were carried out using JMP 8.01 software (SAS Institute, Cary, NC, USA).
Results
mRNA expression profiling of pancreatic cancer cell lines under hypoxic conditions. We used the MIA PaCa‐2 cell line for the microarray analyses, because MIA PaCa‐2 cells showed the greatest morphological changes under hypoxic conditions compared with normoxic conditions in our experiments. We defined a difference as significant when there was a change of >5‐fold between the hypoxic and normoxic conditions and found that 23 genes were significantly upregulated under hypoxic conditions compared with normoxic conditions. The 23 highest ranking genes are shown in Table 1. Among the genes identified, PFKFB4 and ALDOC were significantly upregulated, as previously reported,( 13 , 33 ) and we therefore considered that the conditions used in this experiment were appropriate.
Table 1.
Twenty‐three highest ranking genes that were significantly upregulated under hypoxic conditions compared with normoxic conditions in microarray analyses
| Overexpression rank | Gene symbol | Gene function | Average difference between normoxia vs hypoxia |
|---|---|---|---|
| 1 | PFKFB4 | Resonse to hypoxia via a hypoxia‐induced factor (HIF)‐1‐dependent mechanism | 22.17 |
| 2 | ADM | Relate invasiveness in pancreatic cancer | 16.11 |
| 3 | ANKRD37 | HIF‐1 target gene | 15.81 |
| 4 | PPFIA4 | Implicated in trafficking of LAR subfamily PTPases and AMPA‐type glutamate receptors | 12.68 |
| 5 | FER1L4 | Mus musculus fer‐1‐like 4 | 10.83 |
| 6 | SPAG4 | Association with the germline nuclei | 10.23 |
| 7 | ALDOC | Bind HIF‐1, and functioning as a hypoxia response element | 9.61 |
| 8 | ANG | Member of the angioprotein family | 8.61 |
| 9 | RNASE4 | Member of the RNase A gene superfamily | 8.14 |
| 10 | UPK1A | Urothelium‐specific markers of terminal urothelial cytodifferentiation | 7.99 |
| 11 | ENO2 | Cytosolic glycolytic enzyme | 7.78 |
| 12 | HIG2 | Canonical Wnt signaling, both as target and activator,and relate invasiveness in renal cell carcinoma (RCC) | 7.72 |
| 13 | C20orf46 | Transmembrane protein | 7.62 |
| 14 | INSIG2 | Inhibits Bax‐mediated apoptosis | 6.66 |
| 15 | TLE6 | Transcriptional repressor and are recruited by transcription factors containing an eh1 or WRPW/Y domain | 6.44 |
| 16 | PDK3 | Induced HIF‐1 and leading to inhibition of mitochondrial respiration | 6.27 |
| 17 | CCNG2 | T‐cell cycle progression and activation | 6.01 |
| 18 | BNIP3L | Promotes autophagy and apoptosis | 5.81 |
| 19 | NDRG1 | N‐myc downstream regulated gene | 5.26 |
| 20 | SCAND2 | Zinc finger protein | 5.21 |
| 21 | P4HA1 | HIF‐1 target gene | 5.12 |
| 22 | ANGPTL4 | HIF‐1 target gene | 5.09 |
| 23 | ANKZF1 | Encode zinc finger domain‐containing protein1 | 5.04 |
Among the upregulated genes, we focused on Insig2, because it has been reported to be a novel biomarker for colon cancer( 16 ) and its expression is correlated with the invasiveness of colon cancer.( 16 )
Insig2 mRNA expression levels in cultured pancreatic cancer cell lines. We investigated the levels of Insig2 mRNA expression in cultures of 15 different pancreatic cancer cell lines and HPDE cells. As shown in Figure 1(a), all 15 pancreatic cancer cell lines and the HPDE cells as well as two primary cultures of fibroblasts isolated from pancreatic cancer specimens expressed Insig2 mRNA. Compared with HPDE cells, SUIT‐2 and KP‐2 cells expressed higher levels of Insig2 mRNA, and the other 13 pancreatic cancer cell lines expressed lower levels of Insig2 mRNA.
Figure 1.
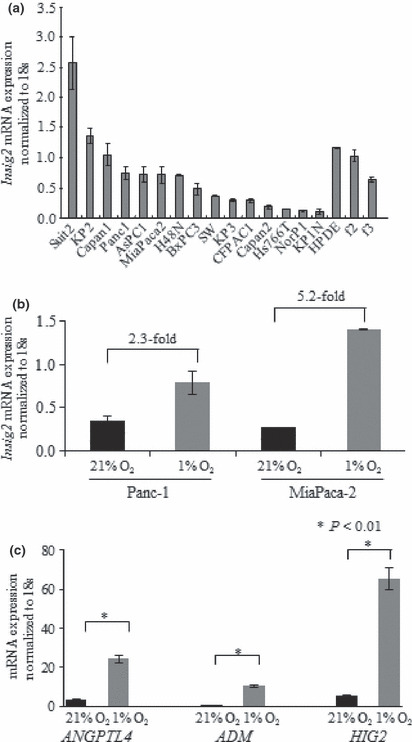
Insig2 mRNA expression levels in cell lines. (a) Insig2 mRNA expression levels in 15 pancreatic cancer cell lines. The expression of Insig2 mRNA was normalized by the expression of 18S rRNA mRNA. All 15 pancreatic cancer cell lines and human pancreatic ductal epithelial (HPDE) cells, as well as two primary cultures of fibroblasts isolated from pancreatic cancer tissues (f2, f3), express Insig2 mRNA. Compared with HPDE cells, SUIT‐2 and KP‐2 cells express higher levels of Insig2 mRNA, whereas the other 13 pancreatic cancer cell lines express lower levels. (b) Comparisons of Insig2 mRNA expression levels under normoxic (21% O2) and hypoxic (1% O2) conditions in two pancreatic cancer cell lines. The Insig2 mRNA expression levels in PANC‐1 and MIA PaCa‐2 cells are 2.3‐fold and 5.2‐fold higher under hypoxic conditions than under normoxic conditions, respectively. (c) Comparisons of the ANGPTL4, ADM, and HIG2 mRNA expression levels under normoxic (21% O2) and hypoxic (1% O2) conditions in the MIA PaCa‐2 cell line. The ANGPTL4, ADM, and HIG2 mRNA expression levels in MIA PaCa‐2 cells are all significantly higher under hypoxic conditions than under normoxic conditions.
Next, we confirmed the effects of hypoxia on Insig2 mRNA expression in the pancreatic cancer cell lines MIA PaCa‐2 and PANC‐1, because these two cell lines have already been shown to induce HIF‐1 protein expression under hypoxic conditions( 34 ) and hypoxia increases the amount of Insig2 in a response mediated by HIF‐1.( 35 ) As shown in Figure 1(b), PANC‐1 and MIA PaCa‐2 cells showed 2.3‐fold and 5.2‐fold higher levels of Insig2 mRNA expression under hypoxic conditions (1% O2) than under normoxic conditions (21% O2), respectively. We also investigated the mRNA expression levels of three other genes, ANGPTL4, ADM, and HIG2, in MIA PaCa‐2 cells under hypoxic conditions to confirm the accuracy of the microarray data, and found that all three mRNAs were expressed at significantly higher levels under hypoxic conditions than under normoxic conditions (Fig. 1c).
Inhibition of Insig2 expression decreases the proliferation of SUIT‐2 pancreatic cancer cells. To investigate the effects of Insig2‐targeting siRNAs, we first measured the levels of Insig2 mRNA expression in pancreatic cancer cells transfected with the Insig2‐targeting or negative control siRNAs using SUIT‐2, Capan‐2, and CFPAC‐1 cells. SUIT‐2, Capan‐2, and CFPAC‐1 cells showed significantly lower levels of Insig2 mRNA at 48 h after transfection with the Insig2‐targeting siRNAs than the control cells transfected with the negative control siRNAs (Fig. 2a, P < 0.01).
Figure 2.
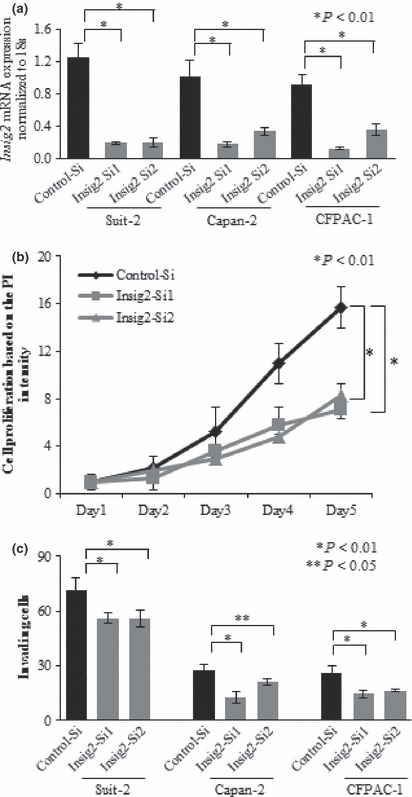
Knockdown of Insig2 expression in pancreatic cancer cell lines by Insig2‐targeting siRNAs and the effects on cell proliferation and invasion. (a) Insig2 expression is significantly suppressed by the Insig2‐targeting siRNAs in SUIT‐2, Capan‐2, and CFPAC‐1 cells. (b) Decreased proliferation is observed in SUIT‐2 cells transfected with the Insig2‐targeting siRNAs compared with the control cells at 120 h after transfection (*P < 0.01, Student’s t‐test). (c) Decreased invasion is observed in SUIT‐2, Capan‐2, and CFPAC‐1 cells transfected seen with the Insig2‐targeting siRNAs compared with the control cells at 48 h after seeding (*P < 0.01, Student’s t‐test). The data are the means ± SD of triplicate measurements.
Next, we carried out PI assays using SUIT‐2 cells, which expressed the highest level of Insig2 mRNA (Fig. 1a) and examined the effects of Insig2 knockdown on cell proliferation. SUIT‐2 cells transfected with the Insig2‐targeting siRNAs showed significantly decreased cell proliferation compared with the control cells at 120 h after transfection (Fig. 2b, P < 0.001). No significant changes in the proliferation of CFPAC‐1 and Capan‐2 cells, which had moderate levels of Insig2 expression, were observed after transfection of the Insig2‐targeting siRNAs (data not shown).
Inhibition of Insig2 decreases the invasion of pancreatic cancer cells. Next, we investigated the effects of Insig2 knockdown on cancer cell invasion, which is an important function for malignant progression and metastasis, using SUIT‐2, Capan‐2, and CFPAC‐1 cells. Capan‐2 and CFPAC‐1 cells, which expressed relatively moderate levels of Insig2, and SUIT‐2 cells, which expressed the highest level of Insig2, showed significantly decreased cell invasion at 48 h after transfection with the Insig2‐targeting siRNAs compared with the control cells (Fig. 2c, P < 0.01 and P < 0.05), Although there were no changes in the proliferation of SUIT‐2, Capan‐2, and CFPAC‐1 cells at 48 h after transfection (Fig. 2b for SUIT‐2 cells; data not shown for Capan‐2 and CFPAC‐1 cells).
Quantitative analyses of Insig2 mRNA expression in microdissected IDC, PanIN, and normal ductal epithelial cells. Among the 32 normal epithelial samples, 15 were normal epithelial samples from pancreatic cancer cases. We obtained the tissues from sites near the tumor stumps, which were pathologically diagnosed as being free of carcinoma cells. The remaining 17 samples were normal epithelial lesions from patients with pancreatic cystic disease, intraductal papillary mucinous neoplasm, pancreatic endocrine tumor and other cancers (cholangiocarcinoma and carcinoma of the ampulla of Vater). Representative images of the dissected cells from pancreatic normal ductal epithelial, PanIN‐1A, and IDC lesions are shown in Figure 3(a). As shown in Figure 3(b), the levels of Insig2 mRNA expression were significantly higher in IDC cells and PanIN cells than in normal ductal epithelial cells (P < 0.001, normal ductal epithelial cells versus IDC cells; P = 0.019, normal ductal epithelial cells versus PanIN cells). Although the median Insig2 mRNA expression level was higher in IDC cells than in PanIN cells, the difference was not significant (P = 0.082).
Figure 3.
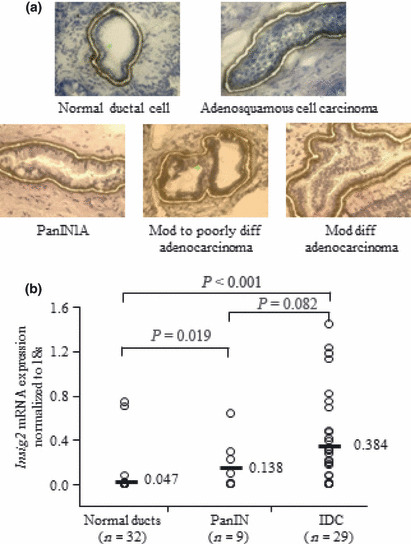
Insig2 mRNA expression levels in microdissected cells. (a) Representative micrographs of cells in pancreatic normal ductal epithelia, pancreatic intraepithelial neoplasia (PanIN) lesions, and invasive ductal carcinoma (IDC) lesions stained with 1% toluidine blue. Diff, differentiated; mod, moderately. (b) Relative levels of Insig2 mRNA expression normalized by the levels of 18S rRNA mRNA expression in microdissected normal pancreatic ductal, PanIN, and IDC cells. The center horizontal lines represent the median values. The levels of Insig2 mRNA expression are significantly higher in IDC cells and PanIN cells than in normal ductal epithelial cells (P < 0.001, normal ductal epithelial cells versus IDC cells; P = 0.019, normal ductal epithelial cells versus PanIN cells). Although the levels of Insig2 mRNA expression in IDC cells tend to be higher than those in PanIN cells, the difference is not significant (P = 0.082).
Relationships between Insig2 mRNA expression and clinicopathological factors. The Insig2 mRNA expression levels were significantly higher in patients with lymph node metastasis than in those without lymph node metastasis (P = 0.003; Fig. 4a). The levels of Insig2 mRNA expression were significantly higher in stages IIB–IV (n = 20) than in stages I–IIA (n = 9) according to the UICC classification (P = 0.0321; Fig. 4b). Regarding recurrence after surgical resection, among 29 pancreatic cancer tissues of microdissected samples, 12 cases showed recurrence and 17 cases remained free from recurrence. Among the 12 cases with recurrence, nine were distant metastatic and peritoneal dissemination cases and three were local recurrence cases. The Insig2 mRNA expression levels were significantly higher in patients with distant metastasis (lung, liver) and peritoneal dissemination than in those with local recurrence (P = 0.0124; Fig. 4c).
Figure 4.
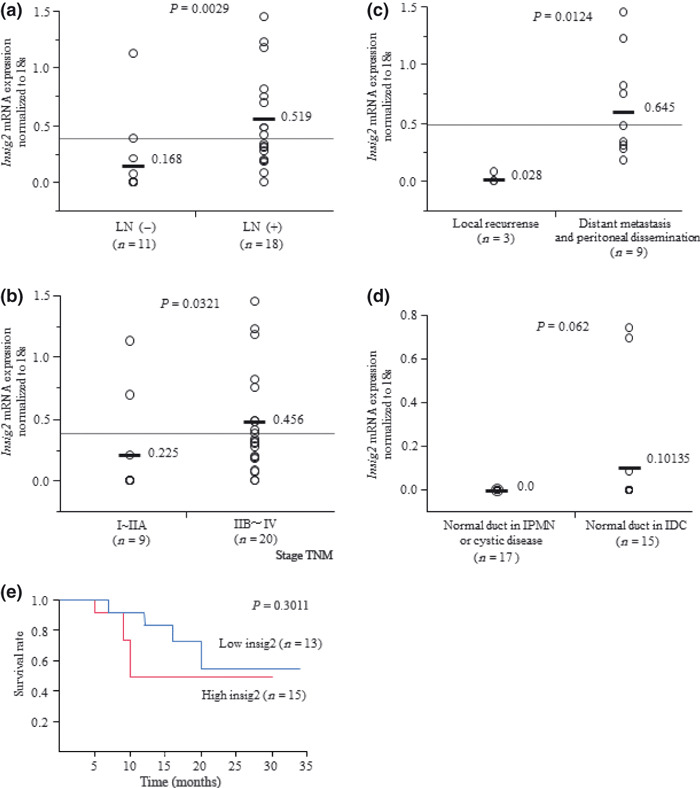
Relationships between Insig2 mRNA expression levels in microdissected cells and clinicopathological factors. (a) The levels of Insig2 mRNA expression are significantly higher in patients with lymph node metastasis (n = 18) than in those without lymph node metastasis (n = 11) (P = 0.003). (b) According to the UICC classification, the levels of Insig2 mRNA expression are significantly higher in stage IIB–IV cases (n = 20) than in stage I–IIA cases (n = 9) (P = 0.0321). (c) The levels of Insig2 mRNA expression are significantly higher in patients with distant metastasis (lung or liver) and peritoneal dissemination (n = 9) than in patients with local recurrence (n = 3) (P = 0.0124). (d) Relative levels of Insig2 mRNA expression in microdissected normal pancreatic ductal cells from 15 cases with IDC and 17 cases with pancreatic cystic disease, intraductal papillary mucinous neoplasm (IPMN), or other cancers. There is a tendency toward higher levels of Insig2 mRNA expression in normal ductal cells of patients with IDC than in those of patients with pancreatic cystic disease, IPMN, or other cancers (P = 0.062). (e) Survival curves based on Insig2 mRNA expression in microdissected tissues.
In the analysis of 32 cases with normal ducts, there was a tendency toward higher levels of Insig2 mRNA expression in the normal ducts of the pancreatic cancer cases than in those of the pancreatic cystic disease, intraductal papillary mucinous neoplasm, and other cancer cases (P = 0.062; Fig. 4d).
In the analysis of the 29 pancreatic cancer cases, one case dropped out from our follow up. Among the remaining 28 cases, there was a tendency for a poor prognosis in the high Insig2 group (n = 15) compared with the low Insig2 group (n = 13) (Fig. 4e). However, the difference was not significant (P = 0.3011), possibly owing to the small number of samples in the present study.
Discussion
In the present study, we found that Insig2 mRNA was expressed in all pancreatic cancer cell lines examined and was correlated with the malignant behaviors of pancreatic cancer in analyses involving microdissection and tissues. This is the first report regarding the involvement of Insig2 in pancreatic cancer under hypoxic conditions.
Insig2 was reported to serve as a novel biomarker for colorectal cancer and to act as a potential tumor promoter with multiple biological functions.( 16 ) In the present study, the Insig2 mRNA expression levels were significantly higher in pancreatic cancer cells than in normal cells in analyses of microdissected cells, although cultured pancreatic cancer cells showed low levels of Insig2 mRNA expression. These findings seem to be inconsistent. However, under hypoxic conditions, which are characteristic features of pancreatic cancer tissues, the levels of Insig2 mRNA expression were significantly upregulated in both Panc1 and MIA PaCa‐2 cells. These findings could support the results of analyses with pancreatic cancer cells isolated from pancreatic cancer tissues, which are possibly under hypoxia.
We also found that Insig2 knockdown decreased the cell proliferation and invasion of SUIT‐2 cells, which expressed a high level of Insig2, suggesting that Insig2 has a functional role in the proliferation and invasion of pancreatic cancer cells with high levels of Insig2. Capan‐2 and CFPAC‐1 cells showed significantly decreased cell invasion after Insig2 knockdown but no changes in their cell proliferation, suggesting that Insig2 may have a functional role in the invasion but not the proliferation of pancreatic cancer cells with moderate levels of Insig2. We also found that the levels of Insig2 mRNA expression were significantly higher in cells from late‐stage tumors than in cells from early‐stage tumors, and were significantly higher in cells from tumors with lymph node metastasis than in cells from tumors without lymph node metastasis. Furthermore, in the analyses of the recurrence patterns after surgery, the levels of Insig2 mRNA expression were significantly higher in cells from tumors with distant metastatic recurrences than in cells from tumors with local recurrences. All these findings suggest that Insig2 may have potential roles in pancreatic cancer progression.
In our analyses of normal duct cells, the normal ducts associated with pancreatic cancer had a tendency to express higher levels of Insig2 mRNA than those associated with non‐pancreatic cancer tissues, such as pancreatic cystic disease, intraductal papillary mucinous neoplasm, endocrine tumors, and other cancers, suggesting that Insig2 was overexpressed in the precancerous lesions of pancreatic cancer, although the cells appeared to be morphologically normal epithelial cells.
In conclusion, our present data showed that Insig2 is overexpressed in pancreatic cancer under hypoxic conditions and is correlated with the malignant behaviors of pancreatic cancer. These data suggest that Insig2 is a possible marker for pancreatic cancer diagnosis, evaluation of malignant behaviors, and prediction of prognosis.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgments
We thank Midori Sato, Emiko Manabe, Miyuki Omori, and Makiko Masuda (Department of Surgery and Oncology, Kyushu University) for their skillful technical assistance, and the Research Support Center, Graduate School of Medical Sciences, Kyushu University for technical support.
References
- 1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin 2009; 59: 225–49. [DOI] [PubMed] [Google Scholar]
- 2. Warshaw AL, Fernandez‐del Castillo C. Pancreatic carcinoma. N Engl J Med 1992; 326: 455–65. [DOI] [PubMed] [Google Scholar]
- 3. Burris HA 3rd, Moore MJ, Andersen J et al. Improvements in survival and clinical benefit with gemcitabine as first‐line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1997; 15: 2403–13. [DOI] [PubMed] [Google Scholar]
- 4. Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet 2004; 363: 1049–57. [DOI] [PubMed] [Google Scholar]
- 5. Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer 2002; 2: 897–909. [DOI] [PubMed] [Google Scholar]
- 6. Jaffee EM, Hruban RH, Canto M, Kern SE. Focus on pancreas cancer. Cancer Cell 2002; 2: 25–8. [DOI] [PubMed] [Google Scholar]
- 7. Di Renzo MF, Poulsom R, Olivero M, Comoglio PM, Lemoine NR. Expression of the Met/hepatocyte growth factor receptor in human pancreatic cancer. Cancer Res 1995; 55: 1129–38. [PubMed] [Google Scholar]
- 8. Bloomston M, Zervos EE, Rosemurgy AS 2nd. Matrix metalloproteinases and their role in pancreatic cancer: a review of preclinical studies and clinical trials. Ann Surg Oncol 2002; 9: 668–74. [DOI] [PubMed] [Google Scholar]
- 9. Jimeno A, Hidalgo M. Molecular biomarkers: their increasing role in the diagnosis, characterization, and therapy guidance in pancreatic cancer. Mol Cancer Ther 2006; 5: 787–96. [DOI] [PubMed] [Google Scholar]
- 10. Sato N, Goggins M. The role of epigenetic alterations in pancreatic cancer. J Hepatobiliary Pancreat Surg 2006; 13: 286–95. [DOI] [PubMed] [Google Scholar]
- 11. MacKenzie MJ. Molecular therapy in pancreatic adenocarcinoma. Lancet Oncol 2004; 5: 541–9. [DOI] [PubMed] [Google Scholar]
- 12. Kizaka‐Kondoh S, Itasaka S, Zeng L et al. Selective killing of hypoxia‐inducible factor‐1‐active cells improves survival in a mouse model of invasive and metastatic pancreatic cancer. Clin Cancer Res 2009; 15: 3433–41. [DOI] [PubMed] [Google Scholar]
- 13. Bobarykina AY, Minchenko DO, Opentanova IL et al. Hypoxic regulation of PFKFB‐3 and PFKFB‐4 gene expression in gastric and pancreatic cancer cell lines and expression of PFKFB genes in gastric cancers. Acta Biochim Pol 2006; 53: 789–99. [PubMed] [Google Scholar]
- 14. Keleg S, Kayed H, Jiang X et al. Adrenomedullin is induced by hypoxia and enhances pancreatic cancer cell invasion. Int J Cancer 2007; 121: 21–32. [DOI] [PubMed] [Google Scholar]
- 15. Sedoris KC, Thomas SD, Miller DM. Hypoxia induces differential translation of enolase/MBP‐1. BMC Cancer 2010; 10: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li CG, Gruidl M, Eschrich S et al. Insig2 is associated with colon tumorigenesis and inhibits Bax‐mediated apoptosis. Int J Cancer 2008; 123: 273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A 1999; 96: 11041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yabe D, Brown MS, Goldstein JL. Insig‐2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element‐binding proteins. Proc Natl Acad Sci U S A 2002; 99: 12753–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang L, Yu J, Park BH, Kinzler KW, Vogelstein B. Role of BAX in the apoptotic response to anticancer agents. Science 2000; 290: 989–92. [DOI] [PubMed] [Google Scholar]
- 20. Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol 2005; 7: 909–15. [DOI] [PubMed] [Google Scholar]
- 21. Schinzel A, Kaufmann T, Schuler M, Martinalbo J, Grubb D, Borner C. Conformational control of Bax localization and apoptotic activity by Pro168. J Cell Biol 2004; 164: 1021–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sato N, Mizumoto K, Beppu K et al. Establishment of a new human pancreatic cancer cell line, NOR‐P1, with high angiogenic activity and metastatic potential. Cancer Lett 2000; 155: 153–61. [DOI] [PubMed] [Google Scholar]
- 23. Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long‐term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol 1996; 148: 1763–70. [PMC free article] [PubMed] [Google Scholar]
- 24. Ohuchida K, Mizumoto K, Murakami M et al. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumor‐stromal interactions. Cancer Res 2004; 64: 3215–22. [DOI] [PubMed] [Google Scholar]
- 25. Sobin LH, Greene FL. TNM classification: clarification of number of regional lymph nodes for pNo. Cancer 2001; 92: 452. [DOI] [PubMed] [Google Scholar]
- 26. Kleihues P, Sobin LH. World Health Organization classification of tumors. Cancer 2000; 88: 2887. [DOI] [PubMed] [Google Scholar]
- 27. Kobari M, Matsuno S. Staging systems for pancreatic cancer: differences between the Japanese and UICC systems. J Hepatobiliary Pancreat Surg 1998; 5: 121–7. [DOI] [PubMed] [Google Scholar]
- 28. Nakata K, Ohuchida K, Nagai E et al. LMO2 is a novel predictive marker for a better prognosis in pancreatic cancer. Neoplasia 2009; 11: 712–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tachikawa T, Irie T. A new molecular biology approach in morphology: basic method and application of laser microdissection. Med Electron Microsc 2004; 37: 82–8. [DOI] [PubMed] [Google Scholar]
- 30. Yu J, Ohuchida K, Nakata K et al. LIM only 4 is overexpressed in late stage pancreas cancer. Mol Cancer 2008; 7: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antonov J, Goldstein DR, Oberli A et al. Reliable gene expression measurements from degraded RNA by quantitative real‐time PCR depend on short amplicons and a proper normalization. Lab Invest 2005; 85: 1040–50. [DOI] [PubMed] [Google Scholar]
- 32. Moriyama T, Ohuchida K, Mizumoto K et al. MicroRNA‐21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther 2009; 8: 1067–74. [DOI] [PubMed] [Google Scholar]
- 33. Jian B, Wang D, Chen D, Voss J, Chaudry I, Raju R. Hypoxia‐induced alteration of mitochondrial genes in cardiomyocytes: role of Bnip3 and Pdk1. Shock 2010; 34: 169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Samulitis BK, Landowski TH, Dorr RT. Inhibition of protein synthesis by imexon reduces HIF‐1alpha expression in normoxic and hypoxic pancreatic cancer cells. Invest New Drugs 2009; 27: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nguyen AD, McDonald JG, Bruick RK, DeBose‐Boyd RA. Hypoxia stimulates degradation of 3‐hydroxy‐3‐methylglutaryl‐coenzyme A reductase through accumulation of lanosterol and hypoxia‐inducible factor‐mediated induction of insigs. J Biol Chem 2007; 282: 27436–46. [DOI] [PubMed] [Google Scholar]


