Abstract
Elevated amounts of soluble mesothelin‐related proteins (SMRP) have already been reported in sera and pleural effusions from mesothelioma patients, providing a useful diagnostic marker for malignant pleural mesothelioma (MPM). However, the origin of SMRP is not yet understood. Production of SMRP could be related to abnormal splicing events leading to synthesis of a secreted protein (release) or to an enzymatic cleavage from membrane‐bound mesothelin (ectodomain shedding). To test these hypotheses, we used a panel of mesothelioma cells established in culture from pleural effusions of MPM patients. Our in vitro results confirmed specific mesothelin expression and SMRP production in supernatants from epithelioid MPM cell lines, thus providing a relevant cellular model to study soluble mesothelin production mechanisms. The expression of mesothelin‐encoding RNA variants was screened by reverse transcription–polymerase chain reaction experiments. Protease involvement in mesothelin cleavage from the cellular surface was investigated by treatment of MPM cells with GM6001, a broad‐spectrum MMP‐ and ADAM‐family inhibitor. GM6001 treatment significantly impaired SMRP production by MPM cell lines, in favor of an enzymatic‐mediated shedding process. In addition, a splice variant transcript of mesothelin (variant 3) was detected in these MPM cell lines, in accordance with the release of a secreted part of the protein. Our results indicate that both mechanisms could be implicated in soluble mesothelin production by epithelioid mesothelioma cells. (Cancer Sci 2008; 99: 590–594)
- Abbrevations: ADAM
a disintegrin and metalloproteinase
- CEA
carcinoembryonic antigen
- CYFRA
cytokeratin 19 fragment
- ELISA
enzyme‐linked immunosorbent assay
- FCS
fetal calf serum
- GPI
glycosylphosphatidylinositol
- MMP
matrix metalloproteinase
- MPF
megacaryocyte potentiating factor
- MPM
malignant pleural mesothelioma
- PCR
polymerase chain reaction
- PE
phycoerythrin
- RT
reverse transcription
- SMRP
soluble mesothelin‐related protein
Malignant pleural mesothelioma arises from the uncontrolled proliferation of mesothelial cells lining the pleura.( 1 ) Asbestos exposure is the main factor involved in MPM pathogenesis. Management of MPM patients remains difficult( 2 ) because diagnosis is usually established late in disease evolution, making patient prognosis poor (survival rate ranging from 4 to 12 months). Moreover, differential diagnosis between MPM and pleural benign diseases (often induced by asbestos exposure), lung adenocarcinoma, or pleural metastasis of diverse origins remains uncertain. Significant advances in MPM treatment suggest the development of early and reliable diagnostic tests, and thus require a better knowledge of MPM‐specific markers.
Mesothelin has been suggested to significantly improve the panel of MPM‐associated markers.( 3 ) Mesothelin is a cell‐surface GPI‐anchored glycoprotein that has putative functions in cell‐to‐cell adhesion.( 4 ) This differentiation antigen is present at low levels in a restricted set of normal adult tissues, including the mesothelium, but is overexpressed aberrantly by several cancers, such as mesothelioma, and pancreatic and ovarian carcinomas.( 4 , 5 ) The primary product of the human mesothelin gene is a 71‐kDa precursor protein, which is cleaved physiologically by a furine‐like protease to produce a 31‐kDa N‐terminal fragment (N‐ERK/mesothelin or MPF) secreted into the blood( 6 ) and a 40‐kDa C‐terminal fragment (mesothelin) expressed at the cell surface.( 5 ) Additionally, Scholler et al. detected SMRP in culture supernatants of several carcinomas.( 7 ) It has since been reported that the levels of SMRP are more elevated in sera from mesothelioma patients than in patients with other cancers, inflammatory diseases, or in healthy controls.( 8 , 9 , 10 ) Moreover, a raised SMRP concentration in pleural effusions was also demonstrated to be relevant to discriminate MPM from benign pleural lesions and other malignant diseases.( 11 , 12 )
Today, the clinical relevance of clarifying SMRP origin relies potentially on the identification of molecules that could be quantified in association with SMRP in order to establish an earlier and more reliable MPM diagnosis. Another benefit relies on the development of less‐invasive diagnostic tests rather than actual standard diagnostic procedures based on immunohistochemical staining of tumor biopsies. Although several hypotheses have already been proposed, the mechanisms involved in soluble mesothelin production by tumor cells remain largely unknown.( 4 ) To understand these mechanisms, we explored two alternative possibilities: release of an aberrant RNA splicing product (mesothelin variant 3) or enzyme‐mediated shedding of membrane‐bound mesothelin (phospholipases, proteases). These in vitro experiments were conducted on epithelioid mesothelioma cell lines established in our laboratory from pleural effusions of MPM patients.
Materials and Methods
Patients. The MPM patients had received no anticancer therapy before the study. Pleural effusions were collected by thoracocentesis. Diagnosis was established by immunohistochemical and immunocytochemical labelings. All patients gave signed, informed consent.
Cell lines. Mesothelioma (Meso11, ‐13, ‐30, ‐34, ‐36, ‐47, ‐56) and adenocarcinoma (ADK3) cells were derived from pleural effusions. Briefly, pleural effusions were centrifuged at 1200 g for 10 min. Cell pellets were resuspended in RPMI‐1640 medium (Cambrex, Verviers, Belgium) and rinsed twice. Cells were then seeded into flasks at a concentration of 106 cells/mL in complete RPMI medium supplemented with 10% heat inactivated FCS (Eurobio, Les Ulis, France), 1% penicillin–streptomycin, and 2 mM l‐glutamine (Sigma Aldrich, St‐Louis, MO, USA). The following cell lines were used as controls: MCF‐7 (ECACC, Salisbury, UK), OVCAR‐3 and CAPAN‐2 (Dr C. Saï, INSERM U601, Nantes, France); MeT‐5A (Dr E. Dopp, Institut für Hygiene und Arbeitsmedizin, Essen, Germany), (IGR)‐Pub (Dr F. Chouaib, INSERM U753, Villejuif, France), and H1648 (Dr J. Roche, UMR 6558, Poitiers, France). Cells were maintained in complete medium and were mycoplasma free (checked by Hoechst 33258 labeling).
RNA extraction and RT‐PCR analysis. Total RNA was extracted from cell pellets using RNeasy kits (Qiagen) according to the manufacturer's protocol. One µg total RNA was reverse‐transcribed using M‐MLV Reverse Transcriptase (Invitrogen, San Diego, CA, USA). cDNA was used as a template for PCR amplification (35 cycles) using GoTaq polymerase (Promega, Madison, WI, USA). The GenBank accession numbers for mesothelin variants are: 1, NM_005823; 2, NM_013404; and 3, AF180951.
Flow‐cytometric analysis. After a brief trypsinization of subconfluent adherent cellular cultures, cell suspensions were washed with FCS‐supplemented RPMI medium and fixed with 70% methanol at –20°C for 7 min. Fixed cells were incubated with 1/50 mesothelin‐specific monoclonal antibody (clone K1; Zymed, San Francisco, CA, USA) or with purified mouse IgG1 control and then with 1/200 PE‐conjugated rat antimouse IgG1 antibody (clone A85‐1; BD Biosciences, Le Pont de Claix, France) at 4°C.
Soluble protein assays. When cell cultures reached confluence, three rinses were carried out with complete RPMI‐1640 medium that was FCS free. Cells were incubated in this medium for 20 min for the first rinse and 1 h for the last two rinses. Then, 2.5 mL of medium was added into a 25‐cm2 flask for culturing. The culture supernatants were collected 24 h later, centrifuged at 800 g for 10 min, and stored at –80°C until assayed.
In the MMP inhibition experiments, three culture conditions were tested for each mesothelioma cell line: untreated, GM6001 (50 µM), or related GM6001‐negative control (Calbiochem, Nottingham, UK). Cells were incubated overnight and the culture supernatants were collected as described previously.
The SMRP titration was carried out using the MESOMARK immunoassay kit (CIS Bio International, Fujirebio Diagnostics), following the manufacturer's recommendations. Culture supernatants were tested at 1/10 dilution.
Statistical analysis. Wilcoxon rank sum tests and Wilcoxon signed rank tests were carried out using the R program (http://www.r‐project.org).
Results
Cellular mesothelin expression and SMRP production by MPM and control cell lines. In the present study, we investigated cellular and secreted forms of mesothelin expressed by epithelioid MPM cells (Meso11, ‐13, ‐30, ‐34, ‐36, ‐47, ‐56) collected from pleural effusions and compared them with a panel of control lung adenocarcinoma (ADK3, H1648, [IGR]‐Pub), pleural carcinoma metastasis (MCF‐7), other mesothelin‐positive carcinoma (OVCAR‐3, CAPAN‐2), and established mesothelial (MeT‐5A) cell lines. Quantitative cellular mesothelin expression was assessed by flow‐cytometric experiments, using the monoclonal antibody K1. The mesothelin‐specific K1 antibody was generated by mouse immunization with the human ovarian carcinoma OVCAR‐3 cell line,( 13 , 14 ) which was used as a positive control for staining (Fig. 1a). Cellular mesothelin expression was compared between the mesothelioma and control groups (Fig. 1a). Our results clearly demonstrate that control cells displayed similar mesothelin expression levels to MPM cells, as no significant difference was observed between the groups (P > 0.2).
Figure 1.
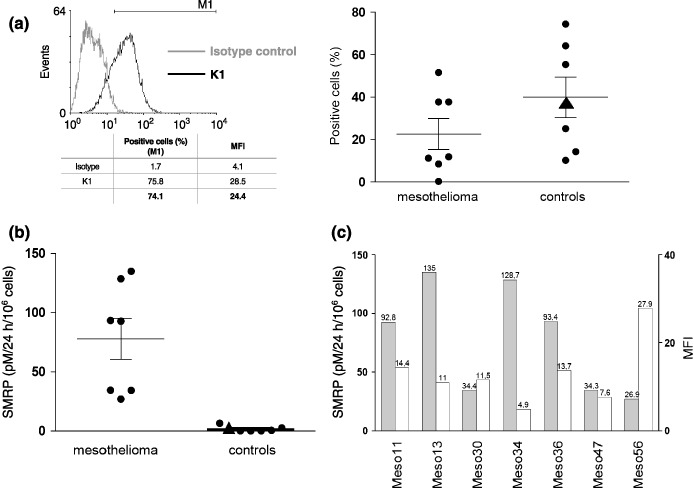
(a) The M1 marker was used to determine the percentage of mesothelin‐positive cells. Mean fluorescence intensity (MFI) was determined by substracting the values of cells labeled with isotype control from those labeled with K1 antibody. (a) Cellular mesothelin expression and (b) soluble mesothelin‐related protein (SMRP) levels from culture supernatants were compared between mesothelioma and control groups (P > 0.2 and P < 0.001, respectively). The MeT‐5A values are indicated by triangle symbols. Mean and SEM are represented for each group. (c) Comparison of cellular mesothelin expression (MFI, white bars) and soluble mesothelin production (SMRP, gray bars) for mesothelioma cell lines.
We next determined the SMRP levels in cell culture supernatants with a double determinant sandwich ELISA (MESOMARK kit), using 4H3 (capture) and OV569 (detection) mesothelin‐specific antibodies.( 7 ) SMRP concentrations were compared between mesothelioma and control cells (Fig. 1b). The SMRP levels in culture supernatants of MPM cells (mean 78 pM/106 cells/24 h) were significantly higher (P < 0.001) than those obtained for adenocarcinoma cells (mean 1.5 pM/106 cells/24 h). A mesothelial cell line, MeT‐5A, was also tested and no SMRP production was detected. In contrast with cellular mesothelin expression, the production of soluble mesothelin in culture supernatants thus appears to be a reliable distinguishing feature of MPM cells. In addition, mesothelin cellular expression (mean fluorescence intensity) was compared with soluble mesothelin production (SMRP) for each mesothelioma cell line tested (Fig. 1c). Interestingly, we observed an inverse correlation between mesothelin expression and SMRP production for Meso13, ‐34, and ‐56 cell lines. This correlation was, however, not clearly established for all MPM cells in the panel.
As mesothelioma tumor cells isolated from pleural effusions of MPM patients retained a major characteristic of the primary tumor (i.e. SMRP production), it provides a relevant in vitro cellular model to examine the physiopathological process for soluble mesothelin production. We therefore used these MPM cells to investigate two previously reported hypotheses: release of an aberrant RNA splicing product (variant 3) or shedding of membrane‐bound mesothelin by enzymatic activity (proteases).
Mesothelin transcript profile of MPM and control cell lines. We used RT‐PCR experiments to study the relative expression of mesothelin‐encoding RNA transcripts by mesothelioma cells. Two different primer pairs were designed to distinguish mesothelin variants 1–2 and 3 (Fig. 2a). We showed that variants 1–2 represent the major mRNA produced by MPM and MeT‐5A cell lines (Fig. 2b). A weak signal corresponding to variant 3 was also detected for most of the MPM cell lines. As confirmed with specific mesothelin variant (V)3 primers, this aberrant splicing variant was solely expressed by mesothelioma cells, and not by mesothelial MeT‐5A cells (Fig. 2b) or the control tumoral cell panel (data not shown).
Figure 2.

(a) The table gives information on mesothelin (MSLN) variant 1–3 and variant 3 primers used in the polymerase chain reaction (PCR) experiments. (b) The relative expression of mesothelin variants was investigated in malignant pleural mesothelioma and MeT‐5A cell lines (black arrow indicates the fragment corresponding to variant 3). β‐Actin was amplified for each cell line to check cDNA integrity. The PCR products were separated by electrophoresis on 2% agarose gels and stained with ethidium bromide.
Implication of proteases in SMRP production by MPM cells. To test whether proteases could be involved in mesothelin shedding by mesothelioma cells, we compared SMRP production in culture supernatants from MPM cells incubated with or without GM6001 (Galardin), a general MMP‐ and ADAM‐family inhibitor (Fig. 3a). SMRP values obtained with the ELISA MESOMARK kit were normalized by cell number, counted for each culture condition. The levels of SMRP were similar between the ‘no treatment’ and ‘GM6001 negative control’ groups (data not shown), demonstrating that the inhibitory doses used are not toxic to MPM cells. A significant inhibition of SMRP production (approximately 50%) was observed in culture supernatants from all MPM cell lines incubated overnight with GM6001 (normalized optical density [OD] means: 0.18 and 0.09 for the ‘no treatment’ and ‘GM6001’ groups, respectively; P = 0.016) (Fig. 3b). This important result thus demonstrates the involvement of zinc‐dependent proteases in soluble mesothelin production and validatee the hypothesis on membrane‐bound mesothelin shedding by an enzymatic activity.
Figure 3.

(a) Soluble mesothelin‐related protein (SMRP) production was quantified in culture supernatants from untreated (white bars) and GM6001‐treated (gray bars) malignant pleural mesothelioma (MPM) and MeT‐5A cell lines. (b) The histogram shows normalized optical density (OD) means obtained for MPM cells incubated with (gray bar) or without (white bar) GM6001. SEM are represented for each group.
Discussion
Mesothelin is a tumor antigen that can be detected by immunohistochemistry staining on mesothelium, carcinoma, and mesothelioma tissues.( 3 , 13 ) Our flow‐cytometric experiments displayed similar mesothelin expression profiles between MPM and control groups (including several cancer and mesothelial cell lines). Apart from the upregulated mesothelin gene expression that has already been described in mesothelioma,( 15 , 16 ) significant overexpression of cellular mesothelin protein was not observed. This observation was not consistent with the feature of a differentiation antigen generally being overexpressed by tumoral cells. We therefore wondered whether this surprising observation could be explained by the capacity of MPM cells to produce soluble mesothelin. Until now, SMRP detection was essentially carried out using body fluids from mesothelioma patients,( 8 , 9 , 10 , 11 , 12 ) and the production of SMRP by mesothelioma cells has never been assessed directly. For that purpose, SMRP concentrations were compared in culture supernatants derived from mesothelioma and control cell lines. Our in vitro results confirm that soluble mesothelin production is associated with MPM‐related tumoral transformation, as previously described by in vivo studies carried out on sera( 8 , 9 , 10 ) and pleural effusions.( 10 , 11 , 12 ) Interestingly, our results also indicate that the production of soluble mesothelin was independent of the cellular or molecular components potentially provided by the tumor environment.( 17 ) Indeed, the mesothelioma cell lines are able to produce SMRP on their own, suggesting that MPM‐associated cellular machinery is sufficient to lead to mesothelin release.
As aberrant splicing phenomena are specific hallmarks of many tumoral cells,( 18 ) the production of soluble mesothelin could be related to abnormal splicing events. Indeed, release of SMRP may result from premature proteosynthesis termination, due to lack of intron splicing between exons 16 and 17, leading to the production of a secreted protein without a membrane‐anchor sequence. This aberrant RNA transcript is already referred as variant 3.( 4 , 7 ) RT‐PCR characterization of the relative expression of mesothelin variants, carried out in ovarian adenocarcinoma cell lines and primary tumors, revealed that mesothelin variant 1 represents the major mRNA detected in these samples.( 19 ) In the same way, we used RT‐PCR experiments with primers specific for variants 1–2 and 3 to distinguish between the different mesothelin‐encoding transcripts expressed by mesothelioma cells. According to our experimental data, it seems that some aberrant splicing events, responsible for the production of a minor RNA transcript (variant 3), can effectively lead to the release of soluble mesothelin by MPM cells. However, detection of mesothelin variant 3 remains to be confirmed in tumor samples from mesothelioma patients. As variant 3 expression by MPM cells appears to be restricted when evaluated by the RT‐PCR method, it may not explain the overall SMRP serum titre detected in mesothelioma patients.( 8 , 9 , 10 , 11 , 12 ) We could not assess the relative importance of aberrant splicing in SMRP production by MPM cells, neither with the MesoMark test that detects both mesothelin variants 1 and 3 without distinguishing between them,( 20 ) nor with western blot experiments because there is currently no commercially available antibody specific for each variant. At present, our experimental results do not exclude the intervention of this mechanism in SMRP production.
Shedding of membrane‐bound mesothelin from the cell surface may be a complementary mechanism. Indeed, sequencing of SMRP isolated from malignant effusions( 20 ) or from tumor cell culture supernatants( 21 ) have identified the extracellular domain of membrane‐bound mesothelin as the biomarker detected previously in published clinical studies, thus strengthening this hypothesis. Mesothelin is a GPI‐linked glycoprotein, and such proteins are frequently shed into the serum. Cleavage of GPI‐anchored proteins from the cell surface can be mediated by phospholipases( 22 ) or various proteases.( 23 ) As demonstrated recently, mesothelin shedding may not require GPI‐specific phospholipase C( 21 ) but could involve protease‐mediated activity.( 24 ) Among the potential candidates, two protease families that share a metalloproteinase domain may be involved: the ADAM and MMP families of proteins. Several studies have already demonstrated that MPM produce numerous MMP, namely MMP‐1, ‐2, ‐3, ‐7, and ‐9.( 25 , 26 ) We also observed that our MPM cell lines produce active MMP, especially MMP‐2 gelatinase activity (Sapede, unpublished data, 2007). Also, several previous studies have reported ADAM expression by human lung cancer (namely ADAM‐8, ‐9, ‐12, ‐15, and ‐28).( 27 ) However, there is still no information on ADAM expression by MPM tumors. The results obtained from the GM6001 inhibition experiments thus validate the hypothesis on membrane‐bound mesothelin shedding by protease activity, and implicate MMP and ADAM in soluble mesothelin production. However, the only partial reduction in SMRP titres observed in the presence of GM6001 strongly suggest that other mechanisms could also be involved: (1) release of an aberrant splicing product (variant 3) might contribute to SMRP production; (2) expression of additional yet‐unidentified molecules (other proteases not targeted by GM6001); or (3) components produced by cells in the stromal environmental (absent in our in vitro model) could also be involved in membrane‐bound mesothelin shedding from the cellular surface. The overall precise mechanisms of mesothelin shedding thus need further investigation. Indeed, identification of the sheddases involved may now rely on screening of candidate proteinases (belonging to the MMP or ADAM families) expressed by human mesothelioma cancer and on the use of more‐specific inhibitors in order to confirm their role in soluble mesothelin production.
Understanding the mechanisms of soluble mesothelin production has some important clinical implications. Indeed, detection of serological or pleural markers could help to establish an early and reliable MPM differential diagnosis,( 8 , 9 , 10 , 11 , 12 ) develop an efficient prognostic test for screening at‐risk asbestos‐exposed individuals,( 8 , 12 ) monitor cancer progression,( 9 ) and assess the relevance of a mesothelin‐targeted therapeutic approach in mesothelioma patients.( 28 ) Identifying new markers (such as soluble members of the MMP and ADAM families) to combine them with the markers already available (SMRP, CA‐125, MPF, osteopontin, CEA, CA‐15.3, CA‐19.9, and CYFRA 21‐1) could be useful for MPM management.( 29 ) For instance, using CA‐125 detection in association with SMRP detection was reported to significantly improve the early diagnosis of ovarian carcinoma.( 30 ) Until now, clinical studies combining SMRP with osteopontin( 31 ) or CA‐125( 32 ) gave disappointing results for the detection of MPM cancer. However, we hope that a combination of MMP or ADAM with SMRP detection in patient sera or pleural effusions could significantly increase the diagnostic and therapeutic efficiency of MPM in the near future.
Acknowledgments
C. S. is a recipient of funding from the Association pour la Recherche contre le Cancer (ARC) and Fondation pour la Recherche Médicale (FRM), and A. G. of la Ligue Nationale Contre le Cancer (LNCC). This work was supported by INSERM and grants from La Fondation Weisbrem‐Benenson and La Ligue Régionale Contre le Cancer (Comités de Vendée, de Pays de Loire et du Morbihan).
MESOMARK enzyme‐linked immunosorbent assay kits were provided, free of charge, by CIS‐Bio International and Fujirebio Diagnostics. CIS‐Bio International and Fujirebio Diagnostics had no role in recruiting patients, carrying out assays, analysing the data, or writing or approving the manuscript.
References
- 1. Robinson BWS, Musk AW, Lake RA. Malignant mesothelioma. Lancet 2005; 366: 397–408. [DOI] [PubMed] [Google Scholar]
- 2. Scherpereel A. Guidelines of the French speaking society for chest medicine for management of malignant pleural mesothelioma. Respir Med 2007; 101: 1265–76. [DOI] [PubMed] [Google Scholar]
- 3. Ordonez NG. Value of mesothelin immunostaining in the diagnosis of mesothelioma. Mod Pathol 2003; 16: 192–7. [DOI] [PubMed] [Google Scholar]
- 4. Hassan R, Bera T, Pastan I. Mesothelin: a new target for immunotherapy. Clin Cancer Res 2004; 10: 3937–42. [DOI] [PubMed] [Google Scholar]
- 5. Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci USA 1996; 93: 136–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kojima T, Oh‐eda M, Hattori K et al . Molecular cloning and expression of megakaryocyte potentiating factor cDNA. J Biol Chem 1995; 270: 21 984–90. [DOI] [PubMed] [Google Scholar]
- 7. Scholler N, Fu N, Yang Y et al . Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci USA 1999; 96: 11 531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robinson BWS, Creaney J, Lake R et al . Mesothelin‐family proteins and diagnosis of mesothelioma. Lancet 2003; 362: 1612–16. [DOI] [PubMed] [Google Scholar]
- 9. Hassan R, Remaley AT, Sampson ML et al . Detection and quantitation of serum mesothelin, a tumor marker for patients with mesothelioma and ovarian cancer. Clin Cancer Res 2006; 12: 447–53. [DOI] [PubMed] [Google Scholar]
- 10. Grigoriu B, Scherpereel A, Devos P et al . Utility of osteopontin and serum mesothelin in malignant pleural mesothelioma diagnosis and prognosis assessment. Clin Cancer Res 2007; 13: 2928–35. [DOI] [PubMed] [Google Scholar]
- 11. Scherpereel A, Grigoriu B, Conti M et al . Soluble mesothelin‐related peptides in the diagnosis of malignant pleural mesothelioma. Am J Respir Crit Care Med 2006; 173: 1155–60. [DOI] [PubMed] [Google Scholar]
- 12. Creaney J, Yeoman D, Naumoff LK et al . Soluble mesothelin in effusions: a useful Tool for the diagnosis of malignant mesothelioma. Thorax 2007; 62: 569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang K, Pastan I, Willingham MC. Isolation and characterization of a monoclonal antibody, K1, reactive with ovarian cancers and normal mesothelium. Int J Cancer 1992; 50: 373–81. [DOI] [PubMed] [Google Scholar]
- 14. Onda M, Willingham M, Nagata S et al . New monoclonal antibodies to mesothelin useful for immunohistochemistry, fluorescence‐activated cell sorting, western blotting, and ELISA. Clin Cancer Res 2005; 11: 5840–6. [DOI] [PubMed] [Google Scholar]
- 15. Frierson HFJ, Moskaluk CA, Powell SM et al . Large‐scale molecular and tissue microarray analysis of mesothelin expression in common human carcinomas. Hum Pathol 2003; 34: 605–9. [DOI] [PubMed] [Google Scholar]
- 16. Wali A, Morin PJ, Hough CD et al . Identification of intelectin overexpression in malignant pleural mesothelioma by serial analysis of gene expression (SAGE). Lung Cancer 2005; 48: 9–29. [DOI] [PubMed] [Google Scholar]
- 17. Zhong J, Gencay MMC, Bubendorf L et al . Erk1/2 and p38 MAP kinase control MMP‐2, MT1‐MMP, and TIMP action and affect cell migration: a comparison between mesothelioma and mesothelial cells. J Cell Physiol 2006; 207: 540–52. [DOI] [PubMed] [Google Scholar]
- 18. Venables JP. Aberrant and alternative splicing in cancer. Cancer Res 2004; 64: 7647–54. [DOI] [PubMed] [Google Scholar]
- 19. Muminova ZE, Strong TV, Shaw DR. Characterization of human mesothelin transcripts in ovarian and pancreatic cancer. BMC Cancer 2004; 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hellstrom I, Raycraft J, Kanan S et al . Mesothelin variant 1 is released from tumor cells as a diagnostic marker. Cancer Epidemiol Biomarkers Prev 2006; 15: 1014–20. [DOI] [PubMed] [Google Scholar]
- 21. Ho M, Onda M, Wang Q, Hassan R, Pastan I, Lively MO. Mesothelin is shed from tumor cells. Cancer Epidemiol Biomarkers Prev 2006; 15: 1751. [DOI] [PubMed] [Google Scholar]
- 22. Lauc G, Heffer‐Lauc M. Shedding and uptake of gangliosides and glycosylphosphatidylinositol‐anchored proteins. Biochim Biophys Acta 2006; 1760: 584–602. [DOI] [PubMed] [Google Scholar]
- 23. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2002; 2: 161–74. [DOI] [PubMed] [Google Scholar]
- 24. Chang K, Pai LH, Batra JK, Pastan I, Willingham MC. Characterization of the antigen (CAK1) recognized by monoclonal antibody K1 present on ovarian cancers and normal mesothelium. Cancer Res 1992; 52: 181–6. [PubMed] [Google Scholar]
- 25. Liu Z, Ivanoff A, Klominek J. Expression and activity of matrix metalloproteases in human malignant mesothelioma cell lines. Int J Cancer 2001; 91: 638–43. [DOI] [PubMed] [Google Scholar]
- 26. Edwards JG, McLaren J, Jones JL. Waller DA, O’Byrne KJ. Matrix metalloproteinases 2 and 9 (gelatinases A and B) expression in malignant mesothelioma and benign pleura. Br J Cancer 2003; 88: 1553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci 2007; 98: 621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Xiang L, Hassan R, Pastan I. Immunotoxin and taxol synergy results from a decrease in shed mesothelin levels in the extracellular space of tumors. PNAS 2007; 104: 17099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scherpereel A, Lee YG. Biomarkers for mesothelioma. Curr Opin Pulm Med 2007; 13: 339–443. [DOI] [PubMed] [Google Scholar]
- 30. McIntosh MW, Drescher C, Karlan B et al . Combining CA 125 and SMR serum markers for diagnosis and early detection of ovarian carcinoma. Gynecol Oncol 2004; 95: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grigoriu BD, Scherpereel A, Devos P et al . Utility of osteopontin and serum mesothelin in malignant pleural mesothelioma diagnosis and prognosis assessment. Clin Cancer Res 2007; 13: 2928–35. [DOI] [PubMed] [Google Scholar]
- 32. Creaney J, Van Bruggen I, Hof M et al . Combined CA125 and mesothelin levels for the diagnosis of malignant mesothelioma. Chest 2007; 132: 1239–46. [DOI] [PubMed] [Google Scholar]


