Abstract
Osteosarcoma (OS) is the most frequent primary malignant bone tumor of children and young adults. Although the introduction of combined neoadjuvant chemotherapy has markedly improved survival, the outcome of OS patients with distant metastasis and/or poor response to chemotherapy is still unsatisfactory. Therefore there is a need to develop new therapeutic agents that suppress OS cell proliferation with higher efficacy. The protein kinases are a family of genes that play critical roles in various signaling pathways. Some cancer cells show addiction to constitutive activation of certain signaling pathways for proliferation and survival. To identify new drug targets for OS, we screened a panel of small interfering RNAs (siRNAs) that target 691 genes encoding human protein kinases and related proteins. We found that different constructs of siRNA specifically targeting polo‐like 1 kinase (PLK1) significantly caused mitotic cell cycle arrest and subsequent apoptotic cell death in a variety of OS cell lines. siRNA targeting PLK1 also suppressed the growth of OS xenografts established in immunodeficient mice. Recently, phase I clinical trials of PLK1 chemical inhibitors have been reported. Our results indicate that PLK1 is a promising molecular target for pharmacologic intervention in OS. (Cancer Sci 2009; 100: 2268–2274)
Osteosarcoma (OS) is the most common primary tumor affecting the bones, typically the long tubular bones, of children and adolescents. The prognosis of OS treated by surgery alone was poor, with 5‐year survival rates of <20%,( 1 ) until the introduction of combinational neoadjuvant chemotherapy. The relapse‐free 5‐year survival rate of OS has been greatly improved in the last three decades and can now reach 60–80%.( 2 , 3 ) However, a significant proportion of OS patients respond poorly to chemotherapy and have a high risk of relapse even after curative resection of the primary site. A suitable strategy for management of patients who develop distant metastasis has not been established.( 4 ) Moreover, administration of conventional chemotherapeutic agents is often associated with severe non‐specific toxic effects, and therefore elderly patients or patients whose condition is generally poor may not be able to tolerate the standard chemotherapy.( 5 ) Therefore there is a need to develop a new therapeutic agent with higher efficacy and minimal adverse effects.
An increasing number of therapeutic agents that selectively inhibit molecular components essential for cancer cell growth and survival have begun to be incorporated into oncological practice. However, no specific molecule responsible for the development of OS has been identified. Cytogenetic examinations have revealed histological subtype–specific chromosomal translocation that produce oncogenic fusion genes in other types of sarcomas,( 6 , 7 , 8 ) but not in OS. Genetic aberrations in the retinoblastoma (RB) and p53 pathways have been repeatedly documented in OS,( 9 , 10 ) but these tumor suppressor pathways may not be responsive to current medical and pharmacological technologies. Therefore identification of a new therapeutic target molecule for OS is desirable.
The protein kinases are one of the largest families of human genes and play critical roles in signal transduction, metabolism, gene transcription, cell cycle progression, cell motility, apoptosis, and cell differentiation.( 11 ) Mutations and dysregulation of several protein kinases have been shown to have causal roles in the development of human malignancies, and various protein kinase inhibitors have been successfully introduced for the treatment of cancers.( 12 , 13 ) To identify a molecule that is essential for OS cell proliferation and survival, we performed unbiased genome‐wide functional screening using an small interfering RNA (siRNA) library targeting all known human kinase genes. Here we report the identification of polo‐like 1 kinase (PLK1) as a potential therapeutic target for OS.
Materials and Methods
Cell culture. Human OS cell lines Saos‐2, U‐2, MNNG/HOS, and MG‐63 were purchased from the American Tissue Culture Collection (Rockville, MD, USA). Human OS cell lines HS‐OS‐1, NOS‐1, and HuO‐3N1 were purchased from Riken Bioresource Centre Cell Bank (Tsukuba, Japan). All OS cell lines were cultured as recommended by the suppliers.
siRNA library screening. A pre‐designed Human Protein Kinase siRNA Set (version 2.0), containing 20‐μm pools of four siRNA duplexes for each of 691 genes, was purchased from Qiagen (Valencia, CA, USA). Saos‐2 cells were seeded at 2500 per well into opaque‐walled 96‐well plates in triplicate on the day before transfection to obtain 50–60% confluency. Cells were transfected with 0.5 μL (10 pmol) of siRNA per well using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA, USA). AllStars Negative Control siRNA (Qiagen) was included in each assay plate to serve as a baseline.
Seventy‐two hours after transfection, cell viability was quantified using CellTiter‐Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA). Luminoscence activity was measured with a GloMax 96 Microplate Luminometer (Promega). A 10‐fold serial dilution of standard adenosine triphosphate (ATP) (Promega) (range 10−10 to 10−13 M) was included in each assay plate to generate a standard curve.
PLK1‐targeting and non‐targeting siRNA. Three different constructs targeting PLK1 were designed by Applied Biosystems (Foster City, CA, USA). Silencer Negative Control #1 (NC1) and #2 (NC2) siRNA (Applied Biosystems) were used as negative (non‐targeting) controls.
Real‐time RT‐PCR. The relative expression level of PLK1 mRNA was quantified by real‐time RT‐PCR as described previously.( 14 ) Untreated OS tissue samples were collected from 28 patients who underwent open biopsy or resection at the National Cancer Centre Hospital (Tokyo, Japan). The TaqMan universal PCR master mix and pre‐designed TaqMan Gene Expression probe and primer sets were purchased from Applied Biosystems. Amplification data measured as an increase in reporter fluorescence were collected using the Prism 7000 Sequence Detection system (Applied Biosystems). The use of human materials was reviewed and approved by the ethics committee of the National Cancer Centre (Tokyo, Japan).
Scoring of mitosis. The number of mitotic figures was counted in 10 consecutive high‐power (×400) fields and averaged:( 14 ) score 1 (≤2 per field) and score 2 (>2 per field).
Immunofluorescence microscopy. Cells were grown on cover slips and fixed with 4% paraformaldehyde at room temperature for 30 min. After blocking with 10% normal swine serum (Vector Laboratories, Burlingame, CA, USA) for 60 min at room temperature, the cells were incubated with mouse monoclonal anti‐α‐tubulin antibody (dilution, 1:2000; clone B‐5‐1‐2; Sigma‐Aldrich, St. Louis, MO, USA) and subsequently Alexa 594‐labeled secondary antibody (Invitrogen). Cover slips were mounted in Vectashield Mounting Medium DAPI (Vector Laboratories). Images were taken on a Zeiss Axioplan 2 microscope equipped with a ×100 Plan‐Apochromat objective lens (Carl Zeiss, Gottingen, Germany).( 15 )
Western blot analysis. Proteins were separated by SDS‐PAGE and transferred to polyvinylidene difluoride membranes, as described previously.( 16 ) The membranes were incubated with antibody against either PLK1 (Cell Signaling Technology, Beverly, MA, USA) or β‐actin (AC‐15; Abcam, Cambridge, MA, USA) and subsequently with horseradish peroxidase–conjugated secondary antibody. The protein bands were detected using the LAS‐3000 image analyzer (Fuji Film, Tokyo, Japan).
Flow cytometry. Cells were harvested by trypsinization and centrifuged at 180 g for 5 min at room temperature, and nuclei were stained with a CycleTEST PLUS DNA Reagent Kit (Becton Dickinson, Franklin Lakes, NJ, USA) in accordance with the manufacturer’s instructions. DNA content was measured with the FACSCalibur system (Becton Dickinson). Histograms were generated using ModFit software (Verity, Topsham, ME, USA).
Animal experiments. Ten million Saos‐2 or MNNG/HOS cells suspended in 0.05 mL of PBS were inoculated subcutaneously through 26‐gauge needles into the lower flanks of 7‐week‐old male BALB/c nude (nu/nu) mice (SLC, Tokyo, Japan). When the tumors reached an average volume of 10–20 mm3, PLK1‐targeting or non‐targeting siRNA (30 μm) mixed with 0.5% atelocollagen (Koken, Tokyo, Japan) was injected. Tumor diameters were measured every 2–3 days with a digital caliper, and the tumor volume was calculated according to the formula V = A × B 2/2 (mm3), where A is the largest diameter (mm), and B is the smallest diameter. Animal experiments were reviewed by the institutional ethics committee and performed in compliance with the guidelines for Laboratory Animal Research of the National Cancer Centre Research Institute (Tokyo, Japan).
Results
High‐throughput siRNA‐based screen for kinases required for OS cell viability. To identify protein kinase(s) essential for OS cell proliferation and survival, Saos‐2 cells were transfected with siRNA duplexes (mixtures of four constructs) targeting 691 human kinase and kinase‐related genes on a one‐gene‐per‐well basis. Seventy‐two hours after transfection, the relative abundance of metabolically active cells was determined by examining the production of ATP (Fig. 1a and Table S1). The mean ATP levels in triplicate wells were normalized to those of serially diluted standard ATP and non‐targeting (negative control) siRNA included in the same 96‐well assay plates. We found that siRNA targeting five genes (PLK1, WEE1, EPHA1, CHEK1, and CSNK2B) reduced the ATP production of Saos‐2 cells more than 1.0‐fold relative to the interquartile range (IQR) (whisker, Fig. 1b) from the first 25% quartile (box, Fig. 1b). The five selected genes were then used to challenge six additional cell lines. siRNAs targeting PLK1, followed by WEE1, reduced the ATP production in six out of the seven OS cell lines examined most significantly (48–92.6% inhibition) (Fig. 1c). Transfection with three siRNA constructs targeting different parts of the PLK1 transcripts (namely s‐448, s‐449, and s‐450) at the lowest possible effective concentration resulted in a significant decrease in the expression of PLK1 mRNA (Fig. 1d) and ATP production (52–96.4% inhibition) (Fig. 1e) across all of the seven OS cell lines studied.
Figure 1.
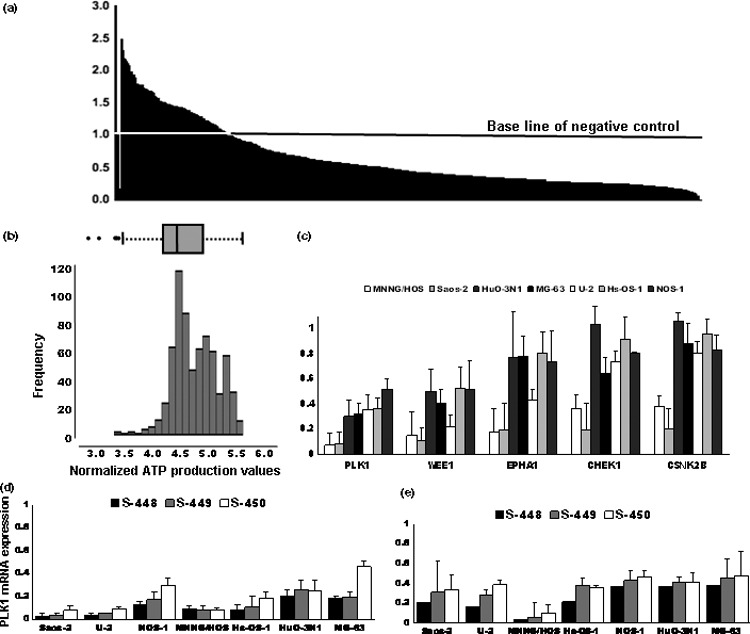
High‐throughput siRNA‐based screen for identifying kinases required for osteosarcoma (OS) cell viability. (a) Relative adenosine triphosphate (ATP) activity of Saos‐2 cells transfected with siRNA targeting 691 kinase and kinase‐related genes. The 691 genes were sorted in order of the normalized values of triplicate means. The value of the negative control siRNA was set as 1. (b) Distribution of the log‐transformed values of ATP production of the 691 genes. Box represents the median value and the 25th to 75th percentile range (IQR). Whiskers indicate the 1.0‐fold range of the IQR from the 25th and 75th percentile values. (c) Effects of siRNA mixtures targeting the five genes (PLK1, WEE1, EPHA1, CHEK1, and CSNK2B) responsible for maximum ATP production in the seven OS cell lines. The value of ATP activity for the negative control siRNA was set as 1. (d,e) Cells were transfected with PLK1‐(s‐448, s‐449, and s‐450) or non‐targeting siRNA. Relative PLK1 mRNA expression (d) and ATP production (e) were determined 48 h after transfection. The values for non‐targeting siRNA were set as 1.
Knockdown of PLK1 induces mitotic spindle aberrations in OS cells. PLK1 is essential for various steps of mitosis progression, including centrosome maturation, spindle formation, and execution of cytokinesis. PLK1‐deficient cells have been reported to be incapable of forming proper bipolar spindles.( 17 , 18 ) Therefore, using immunofluorescence microscopy, we examined the spindle morphology of mitotic OS cells transfected with PLK1‐targeting siRNA (Fig. 2). OS cells transfected with PLK1‐targeting siRNA (s‐448, s‐449, and s‐450) showed disturbance in the organization of microtubules (Fig. 2a, stained red with anti‐α‐tubulin antibody) and chromosomes (Fig. 2a, stained blue with DAPI), indicating failure of proper chromosome segregation. More than 90% of cells showed abnormal/misaligned bipolar (incomplete) and monopolar or multipolar (complete) spindle formation (Fig. 2b), indicating the high sensitivity of OS cells to PLK1 knockdown. No significant morphological abnormality was observed in cells transfected with control siRNA (NC1 and NC2). The knockdown of PLK1 expression was confirmed by Western blotting (Fig. 2c).
Figure 2.
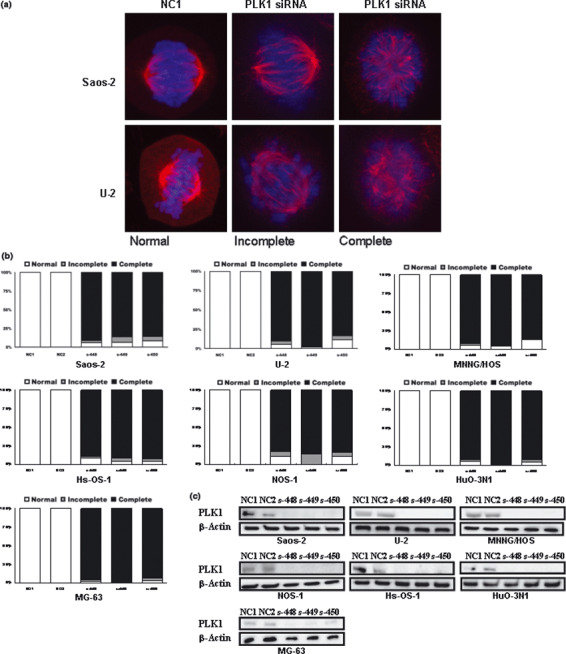
Knockdown of polo‐like 1 kinase (PLK1) induces mitotic spindle aberrations in osteosarcoma (OS) cells. (a,b) Representative appearance (a) and percentages (b) of mitotic cells showing normal (normal), aberrant bipolar (incomplete), and monopolar or multipolar (complete) spindle morphology. Cells were transfected with PLK1 (s‐448, s‐449, and s‐450) or non‐targeting (NC1 and NC2) siRNAs and stained with anti‐α‐tubulin antibody (red) and DAPI (blue) 24 h after transfection. (c) Western blot analysis of PLK1 protein carried out 48 h after transfection. β‐Actin was included as a loading control. Lane 1, negative control #1 siRNA (NC1); lane 2, negative control #2 siRNA (NC2); lanes 3–5, PLK1‐targeting siRNA (s‐448, s‐449, and s‐450, respectively).
Knockdown of PLK1 induces G2/M arrest and apoptosis in OS cells. We next performed cell sorter analysis to examine whether the PLK1‐targeting siRNA caused any abnormalities at particular stages of the cell cycle. As shown in Figure 3, all of the seven OS cell lines examined showed an increase in the proportion of cells in the G2/M phase 24 h after transfection with PLK1‐targeting siRNA (s‐448, s‐449, and s‐450) as compared with two negative control siRNAs (NC1 and NC2), indicating that PLK1‐targeting siRNA induced G2/M arrest. Cells transfected with PLK1‐targeting siRNA induced sub‐G1 DNA peaks, indicative of DNA breakdown and apoptosis.
Figure 3.
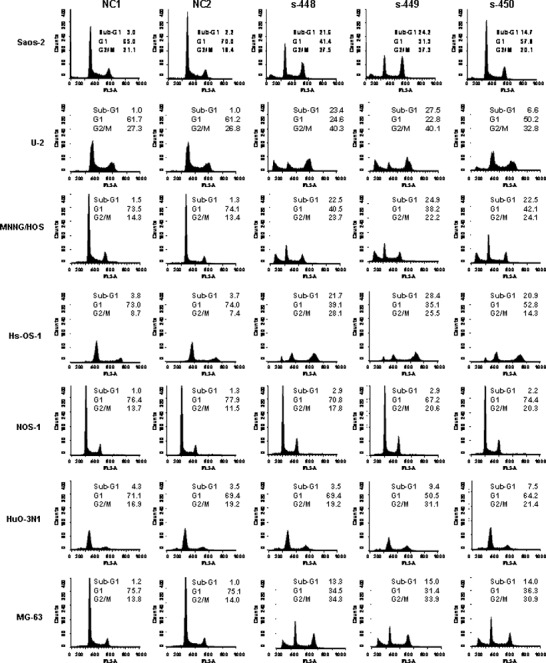
Knockdown of polo‐like 1 kinase (PLK1) induces G2/M arrest and apoptosis in osteosarcoma (OS) cells. Cells were transfected with PLK1‐targeting or non‐targeting siRNAs. Twenty‐four hours after transfection, cells were collected, fixed with 4% paraform aldehyde (PFA), stained with propidium iodide, and subjected to cell sorter analysis. The percentages of sub‐G1, G1, G2/M are indicated.
PLK1 expression in clinical OS. The relative expression level of PLK1 mRNA was determined in fresh‐frozen tissue samples obtained from 28 cases of OS (case no. 1–28) by real‐time PCR (Fig. 4). Although there was a nine‐fold difference, all the cases examined showed a detectable level of expression. The PLK1 expression level showed no significant association with clinicopathological characteristics such as patient age, gender, tumor size, response to chemotherapy, development of metastasis, histological subtype, or frequency of mitosis.( 4 )
Figure 4.
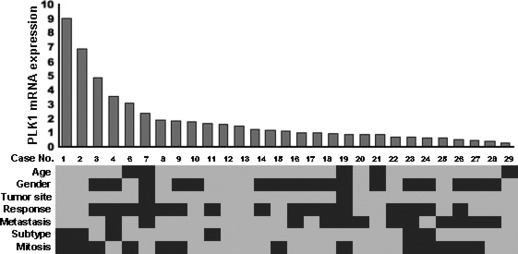
Polo‐like 1 kinase (PLK1) expression in clinical osteosarcoma (OS). Relative expression level of PLK1 mRNA was determined by real‐time PCR, and 28 OS cases were sorted according to the PLK1 expression level. Clinicopathological characteristics of the 29 cases are indicated by gray and black rows as follows: age: gray, <25 years/black, ≥25 years; sex: gray, male/black, female; tumor site: gray, extremity/black, trunk; response to chemotherapy: gray, good [necrosis >90%]/black, poor; metastasis: gray, absent/black, present; histological subtype: gray, osteoblastic/black, other subtypes; mitosis: gray, score 1/black, score 2.
Effects of PLK1 siRNA on OS in vivo. Finally, we examined the effects of PLK1‐targeting siRNA on the growth of established tumors. Saos‐2 or MNNG/HOS cells were injected subcutaneously into immunodeficient mice. One to 2 weeks later, visible tumors (10–20 mm3) developed at the injection sites. siRNA mixed with atelocollagen was injected twice into the tumors with a 1‐week interval. The volume of xenografts was monitored for 21 days after the first siRNA injection (Fig. 5). Tumors injected with PLK1‐targeting siRNA (s‐448, s‐449, and s‐450) showed significant (P < 0.05, Student’s t‐test) growth retardation compared with negative control siRNA (NC1 and NC2), in both Saos‐2 (Fig. 5a) and MNNG/HOS cells (Fig. 5b). Representative photographs showing the appearance of tumors before and after siRNA injection are shown in Figure 5c.
Figure 5.
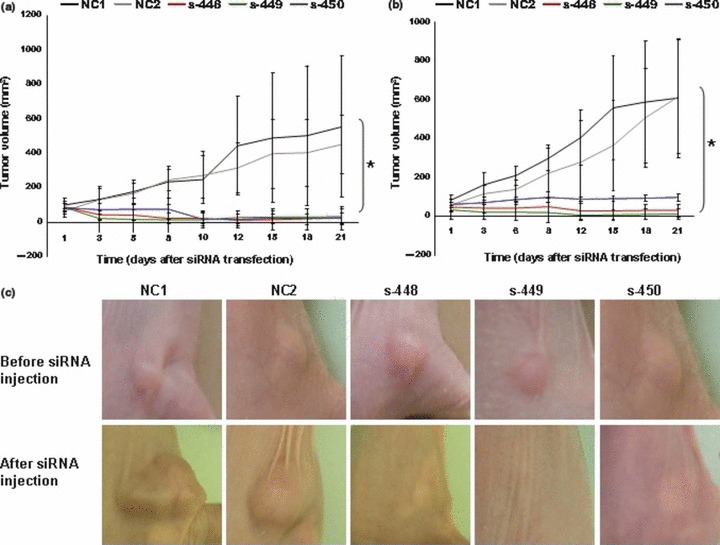
Effects of polo‐like 1 kinase (PLK1) siRNA on osteosarcoma (OS) in vivo. (a,b) Changes in volumes (mean ± SD) of Saos‐2 (a) and MNNG/HOS (b) xenografts after the first injection of non‐targeting (NC1 and NC2) or PLK1‐targeting (s‐448, s‐449, and s‐450) siRNA. Each group comprised five mice. (c) Representative appearance of Saos‐2 xenografts in immunodeficient mice before and 21 days after the first injection of non‐targeting (NC1 and NC2) or PLK1‐targeting (s‐448, s‐449, and s‐450) siRNA.
Discussion
The advent of synthetic interfering RNA (RNAi) libraries has made systemic genome‐wide functional screening possible, and genes that sensitize lung cancer cells to a chemotherapeutic drug and genes required for proliferation and survival of several cancer cell lines have been identified successfully.( 19 , 20 ) To develop therapeutic agents that are effective against a wide spectrum of OS, we screened a siRNA library targeting all 518 protein kinase genes known to be present in the human genome.( 11 ) We initially identified a group of genes (PLK1, WEE1, EPHA1, CHEK1, and CSNK2B), whose loss results in significant reduction of viability of a single OS cell line (Table S1). Further confirmatory experiments revealed that knockdown of PLK1 results in G2/M arrest and apoptosis across all of the seven OS cell lines studied (Fig. 4), confirming that PLK1 is a molecule of interest. OS has been recognized as a single category of disease, but the overall gene expression and clinical behaviors differ markedly among individual cases. We used the ATP production assay and Saos‐2 cells for the initial screen (Table S1). However, the use of other assays and cell lines may have resulted in identification of a different set of genes.
Polo‐like kinases (PLKs), which include PLK1, PLK2, PLK3, and PLK4, belong to a family of serine/threonine kinases that are highly conserved from yeasts to mammals.( 21 , 22 ) The best characterized member of the human PLK family is PLK1. Previous studies have shown that PLK1 is an important regulator of various mitotic events.( 22 ) Generally, PLK1 is expressed in a limited number of normal tissues including thymus, spleen, and testis, but not at a detectable level in other normal tissues.( 21 , 23 , 24 ) Extensive studies have shown that PLK1 is expressed and has prognostic value, in different cancers such as non‐small‐cell lung cancer, head and neck cancer, colorectal cancer, and ovarian cancer.( 25 , 26 , 27 , 28 ) PLK1 is a reliable marker for identifying a high risk of metastasis in melanoma and breast cancer.( 29 , 30 ) Future studies aimed at characterizing the genes responsible for differences in the clinical outcome of OS will be necessary.
Direct application of siRNA to therapeutics is currently not practicable due to the lack of an effective and cell‐specific delivery system. Recently, several small‐molecule PLK1 inhibitors have been characterized. These include the selective PLK1 inhibitor BI 2536, the multi‐targeted inhibitor ON01910, and other ATP competitors such as scytonemin and wortmannin.( 22 ) BI 2536 is a potent ATP‐competing inhibitor with specificity for PLK1 that causes mitotic arrest and apoptosis in a variety of cancer cells.( 18 , 31 ) ON01910 is a non‐ATP‐competitive non‐selective PLK1 inhibitor.( 32 ) Phase I clinical trials of BI 2536 and ON01910 in patients with solid tumors have revealed that both of these compounds have significant activity and favorable pharmacokinetic profiles with manageable adverse effects, even in elderly patients.( 33 , 34 ) Our present results indicate the feasibility of these compounds for the treatment of OS.
Disclosure Statement
T. Yamada received research funding from Astellas Pharma Inc.
Supporting information
Table S1. Inhibition of adenosine triphosphate (ATP) production by siRNA targeting 691 protein kinase genes and related genes.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgments
This work was supported by the Program for Promotion of Fundamental Studies in Health Sciences conducted by the National Institute of Biomedical Innovation of Japan and by the Third‐Term Comprehensive Control Research for Cancer and Research on Biological Markers for New Drug Development conducted by the Ministry of Health, Labor and Welfare of Japan. U. Yamaguchi is an Awardee of a Research Resident Fellowship from the Foundation for Promotion of Cancer Research (Tokyo, Japan). These fund resources played no role in decisions as to the study design or interpretation of the results.
References
- 1. Uribe‐Botero G, Russell WO, Sutow WW, Martin RG. Primary osteosarcoma of bone. Clinicopathologic investigation of 243 cases, with necropsy studies in 54. Am J Clin Pathol 1977; 67: 427–35. [DOI] [PubMed] [Google Scholar]
- 2. Bielack SS, Kempf‐Bielack B, Delling G et al. Prognostic factors in high‐grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 2002; 20 (3): 776–90. [DOI] [PubMed] [Google Scholar]
- 3. Meyers PA, Schwartz CL, Krailo M et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high‐dose methotrexate. J Clin Oncol 2005; 23 (9): 2004–11. [DOI] [PubMed] [Google Scholar]
- 4. Clark JC, Dass CR, Choong PF. A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol 2008; 134: 281–97. [DOI] [PubMed] [Google Scholar]
- 5. Ferrari S, Palmerini E. Adjuvant and neoadjuvant combination chemotherapy for osteogenic sarcoma. Curr Opin Oncol 2007; 19: 341–6. [DOI] [PubMed] [Google Scholar]
- 6. Tomescu O, Barr FG. Chromosomal translocations in sarcomas: prospects for therapy. Trends Mol Med 2001; 7 (12): 554–9. [DOI] [PubMed] [Google Scholar]
- 7. Busam KJ, Fletcher CD. The clinical role of molecular genetics in soft tissue tumor pathology. Cancer Metastasis Rev 1997; 2: 207–27. [DOI] [PubMed] [Google Scholar]
- 8. Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet 2003; 145: 1–30. [PubMed] [Google Scholar]
- 9. Fuchs N, Winkler K. Osteosarcoma. Curr Opin Oncol 1993; 5 (4): 667–71. [DOI] [PubMed] [Google Scholar]
- 10. Toguchida J, Yamaguchi T, Ritchie B et al. Mutation spectrum of the p53 gene in bone and soft tissue sarcomas. Cancer Res 1992; 52 (22): 6194–9. [PubMed] [Google Scholar]
- 11. Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat Methods 2005; 2: 17–25. [DOI] [PubMed] [Google Scholar]
- 12. Dancey J, Sausville EA. Issues and progress with protein kinase inhibitors for cancer treatment. Nat Rev Drug Discov 2003; 2: 296–313. [DOI] [PubMed] [Google Scholar]
- 13. Ghoreschi K, Laurence A, O’Shea JJ. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nat Immunol 2009; 10: 356–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamaguchi U, Nakayama R, Honda K et al. Distinct gene expression‐defined classes of gastrointestinal stromal tumor. J Clin Oncol 2008; 26 (25): 4100–8. [DOI] [PubMed] [Google Scholar]
- 15. Honda K, Yamada T, Hayashida Y et al. Actinin‐4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 2005; 128: 51–62. [DOI] [PubMed] [Google Scholar]
- 16. Shitashige M, Naishiro Y, Idogawa M et al. Involvement of splicing factor‐1 in β‐catenin/T‐cell factor‐4‐mediated gene transactivation and pre‐mRNA splicing. Gastroenterology 2007; 132 (3): 1039–54. [DOI] [PubMed] [Google Scholar]
- 17. Van Vugt MA, Van De Weerdt BC, Vader G et al. Polo‐like kinase‐1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J Biol Chem 2004; 279: 36841–54. [DOI] [PubMed] [Google Scholar]
- 18. Steegmaier M, Hoffmann M, Baum A et al. BI 2536, a potent and selective inhibitor of polo‐like kinase 1, inhibits tumor growth in vivo. Curr Biol 2007; 17: 316–22. [DOI] [PubMed] [Google Scholar]
- 19. Whitehurst AW, Bodemann BO, Cardenas J et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 2007; 446 (7137): 815–9. [DOI] [PubMed] [Google Scholar]
- 20. Schlabach MR, Luo J, Solimini NL et al. Cancer proliferation gene discovery through functional genomics. Science 2008; 319 (5863): 620–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Golsteyn RM, Schultz SJ, Bartek J, Ziemiecki A, Ried T, Nigg EA. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J Cell Sci 1994; 107 (Pt 6): 1509–17. [DOI] [PubMed] [Google Scholar]
- 22. Strebhardt K, Ullrich A. Targeting polo‐like kinase 1 for cancer therapy. Nat Rev Cancer 2006; 6: 321–30. [DOI] [PubMed] [Google Scholar]
- 23. Hamanaka R, Maloid S, Smith MR, O’Connell CD, Longo DL, Ferris DK. Cloning and characterization of human and murine homologues of the Drosophila polo serine‐threonine kinase. Cell Growth Differ 1994; 5: 249–57. [PubMed] [Google Scholar]
- 24. Holtrich U, Wolf G, Brauninger A et al. Induction and down‐regulation of PLK, a human serine/threonine kinase expressed in proliferating cells and tumors. Proc Natl Acad Sci U S A 1994; 91 (5): 1736–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knecht R, Oberhauser C, Strebhardt K. PLK (polo‐like kinase), a new prognostic marker for oropharyngeal carcinomas. Int J Cancer 2000; 89: 535–6. [PubMed] [Google Scholar]
- 26. Takahashi T, Sano B, Nagata T et al. Polo‐like kinase 1 (PLK1) is overexpressed in primary colorectal cancers. Cancer Sci 2003; 94: 148–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weichert W, Denkert C, Schmidt M et al. Polo‐like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br J Cancer 2004; 90 (4): 815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolf G, Elez R, Doermer A et al. Prognostic significance of polo‐like kinase (PLK) expression in non‐small cell lung cancer. Oncogene 1997; 14: 543–9. [DOI] [PubMed] [Google Scholar]
- 29. Ahr A, Karn T, Solbach C et al. Identification of high risk breast‐cancer patients by gene expression profiling. Lancet 2002; 359 (9301): 131–2. [DOI] [PubMed] [Google Scholar]
- 30. Kneisel L, Strebhardt K, Bernd A, Wolter M, Binder A, Kaufmann R. Expression of polo‐like kinase (PLK1) in thin melanomas: a novel marker of metastatic disease. J Cutan Pathol 2002; 29 (6): 354–8. [DOI] [PubMed] [Google Scholar]
- 31. Sumara I, Gimenez‐Abian JF, Gerlich D et al. Roles of polo‐like kinase 1 in the assembly of functional mitotic spindles. Curr Biol 2004; 14: 1712–22. [DOI] [PubMed] [Google Scholar]
- 32. Gumireddy K, Reddy MV, Cosenza SC et al. ON01910, a non‐ATP‐competitive small molecule inhibitor of Plk1, is a potent anticancer agent. Cancer Cell 2005; 7: 275–86. [DOI] [PubMed] [Google Scholar]
- 33. Mross K, Frost A, Steinbild S et al. Phase I dose escalation and pharmacokinetic study of BI 2536, a novel Polo‐like kinase 1 inhibitor, in patients with advanced solid tumors. J Clin Oncol 2008; 26: 5511–7. [DOI] [PubMed] [Google Scholar]
- 34. Jimeno A, Li J, Messersmith WA et al. Phase I study of ON 01910.Na, a novel modulator of the Polo‐like kinase 1 pathway, in adult patients with solid tumors. J Clin Oncol 2008; 26 (34): 5504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Inhibition of adenosine triphosphate (ATP) production by siRNA targeting 691 protein kinase genes and related genes.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


