Abstract
Runt‐related transcription factor‐3 (RUNX3), being a tumor suppressor gene in gastric cancer, plays an important role in inhibiting cellular growth by participating in the transforming growth factor‐β‐dependent apoptosis. The aim of this study was to determine the expression of RUNX3 in normal salivary glands and adenoid cystic carcinomas (ACCs), comparing the results with clinicopathological factors and patient survival. The quantitative reverse transcription–polymerase chain reaction (RT‐PCR) analysis and Western blot analysis revealed the expression of RUNX3 both in normal salivary glands and ACCs. Nuclear and cytoplasmic immunoreactivities against RUNX3 in ductal luminal cells and acinous cells, but immunonegative in myoepithelial cells, were detected in normal salivary glands. In ACC, the RUNX3 immunostaining was shown in the cytoplasm of tumor cells; however, no nuclear location of RUNX3 was found. Lower RUNX3 expression showed significant correlation to distant metastasis and histological growth pattern (P = 0.009 and P = 0.025, respectively). On univariate analysis, low level of RUNX3 immunolabeling (P = 0.012), stage T4 (P = 0.017), lymph node involvement (P = 0.007), and distant metastasis (P < 0.001) were significantly associated with decreased overall survival. Multivariate analysis showed only distant metastasis had an independent prognostic effect on overall survival (P = 0.043). Our results demonstrate the expression of RUNX3 in normal salivary glands and salivary ACCs. The low level of RUNX3 protein in salivary ACCs might play a pivotal role in tumor progression and have prognostic values in ACCs. (Cancer Sci 2008; 99: 1334–1340)
Abbreviations:
- ACCs
adenoid cystic carcinomas
- GAPDH
glyceraldehyde‐3‐phosphate dehydrogease
- LI
labeling indices
- RUNX3
runt‐related transcription factor‐3
- TGF‐β
transforming growth factor‐β
- WHO
World Health Organization
Salivary ACC comprises approximately 10% of all epithelial salivary tumors and most frequently involves the parotid, submandibular, and minor salivary glands. Typically ACC presents persistent slow growth, high rates of recurrence, and distant metastasis resulting in poor patient survival.( 1 , 2 , 3 ) The 5‐year survival rate is approximately 35% but the long‐term survival is poor, and 80% to 90% of patients die of the disease in 10–15 years.( 1 ) Various other parameters, including positive surgical margins, clinical stage, histopathological patterns, and perineural invasion, have been reported as relevant prognostic factors in patients with ACC.( 4 , 5 ) The histopathological patterns of ACC can be categorized into three subtypes: cribriform, tubular, and solid. Most studies suggest that a solid growth pattern ACC is correlated with a poor prognosis compared to those with a cribriform or tubular pattern.( 6 , 7 ) In addition to these clinicopathological factors, several investigators have explored the possible relevance of the proliferation or apoptosis‐associated proteins in human salivary ACC, such as proliferation cell nuclear antigen,( 8 ) Ki‐67,( 8 , 9 ) bcl‐2 oncoprotein,( 9 , 10 ) the human homolog of murine double minute 2 oncoprotein,( 10 ) and p53 tumor suppressor protein.( 8 , 10 , 11 ) However, the precise mechanism responsible for its carcinogenesis has not been fully clarified. Therefore, it is important to detect the molecular mechanism involved in the development of salivary ACC.
The human RUNX3 was originally cloned as AML2 and localized on human chromosome 1p36.1.( 12 ) It is widely accepted that RUNX3 as a candidate tumor suppressor gene acts as a master regulator of gene expression in major physiological and pathological processes.( 13 , 14 , 15 ) Because RUNX3 cooperates with SMADs and is a downstream target of the transforming growth factor‐β (TGF‐β)–induced apoptosis pathway,( 16 , 17 , 18 , 19 ) expression of RUNX3 might provide a better prognostic indicator for a variety of malignant tumors.( 20 , 21 , 22 ) Moreover, the lack of RUNX3 expression is probably associated with peritoneal metastasis of gastric cancer.( 23 ) Although there have been no reports on its expression in salivary ACCs, which presents typically distant metastasis resulting in a fatal outcome, RUNX3 expression might be conceivably associated with the tumorigenesis and progression of salivary ACC.
In the present study, we investigated the expression of RUNX3 by quantitative real‐time reverse transcription–polymerase chain reaction (RT‐PCR), Western blot analysis, and immunohistochemistry analysis in normal salivary glands and ACCs. Furthermore, we compared the relationships between immunohistochemical scores and histological or clinicopathological profiles to examine the putative predictors of outcome, including the pathological role of RUNX3 protein expression.
Materials and Methods
Tissue samples. Nine normal adult salivary glands and seven adenoid cystic carcinomas frozen tissues were obtained in 2007, and 73 formalin‐fixed, paraffin‐embedded salivary ACC specimens were selected from the files from 1996 to 2006 in the Department of Oral and Maxillofacial Surgery, Hospital of Stomatology, College of Medicine, Zhejiang University and from the Department of Head and Neck Surgery, Zhejiang Cancer Hospital, China. All cases were histologically reviewed using hematoxylin–eosin staining to confirm the diagnosis according to WHO classification.( 1 ) The perineural invasion and histological patterns of the tumor growth were also studied (Table 1).
Table 1.
Relationship between runt‐related transcription factor‐3 (RUNX3) expression and clinicopathological parameters in 73 adenoid cystic carcinomas
| Variables | Number of cases | RUNX3 expression † | P‐values** | ||
|---|---|---|---|---|---|
| Low | Intermediate | High | |||
| Cases | 73 | 18 | 37 | 18 | |
| Age at diagnosis | |||||
| <46 years | 36 | 6 | 20 | 10 | |
| ≥46 years | 37 | 12 | 17 | 8 | 0.293 |
| Gender | |||||
| Female | 43 | 9 | 19 | 15 | |
| Male | 30 | 9 | 18 | 3 | 0.052 |
| Tumor site | |||||
| Major salivary gland | 29 | 10 | 14 | 5 | |
| Minor salivary gland | 44 | 8 | 23 | 13 | 0.222 |
| T‐phage | |||||
| T1‐3 | 31 | 4 | 16 | 11 | |
| T4 | 42 | 14 | 21 | 7 | 0.061 |
| Histotypes | |||||
| Cribriform | 32 | 3 | 17 | 12 | |
| Tubular | 16 | 4 | 9 | 3 | |
| Solid | 25 | 11 | 11 | 3 | 0.025* |
| Distant metastasis | |||||
| + | 7 | 5 | 2 | 0 | |
| – | 66 | 13 | 35 | 18 | 0.009* |
| Perineural invasion | |||||
| + | 41 | 11 | 19 | 11 | |
| – | 32 | 7 | 18 | 7 | 0.703 |
| Lymph node involvement | |||||
| N+ | 8 | 4 | 3 | 1 | |
| N0 | 65 | 14 | 34 | 17 | 0.203 |
Statistically significant;
χ2‐test;
tertile cut‐off: low ≤ 6.94%, 6.94% < Intermediate < 14.91%, High ≥ 14.91%.
Quantitative real‐time RT‐PCR. Quantitative real‐time RT‐PCR was performed on an ABI Prism 7700 instrument (Applied Biosystems, Foster City, CA, USA) after isolation of RNA from normal salivary glands and ACCs by using Trizol (Invitrogen, Carlsbad, CA, USA) and a reverse‐transcribed total RNA with PrimeScipt RT reagent Kit (Takara Bio, Otsu, Japan) according to the manufacturer's instructions. RUNX3 mRNA levels were determined by quantitative RT‐PCR by using RUNX3 specific primers: 5′‐CACTGGCGCTGCAACAAGA‐3′ (sense primer) and 5′‐CACGAAGCGAAGGTCGTTGA‐3′ (antisense primer). GAPDH mRNA levels were also amplified in the same PCR reactions as an endogenous control by using the following primers: 5′‐GAAGGTGAAGGTCGGAGTC‐3′ (sense primer) and 5′‐GAAGATGGTGATGGGATTTC‐3′ (antisense primer). A BioEasy SYBR Green I PCR Kit (Bioer Technology, Hangzhou city, China) was used in this study. The dissociation of SYBR Green‐labeled cDNA was done after completion of real‐time PCR by heating the products for 15 s at 95°C and 20 s at 60°C and increasing the temperature slowly up to 95°C over a 20 min interval. The PCR amplification cycles consisted of an initial denaturation at 95°C for 3 min; 40 cycles of denaturation at 95°C for 30 s, annealing at 63°C for 30 s, and extension at 72°C for 30 s; and a final extension for 10 min at 72°C. The comparative cycle threshold (Ct) method (2Δ−Ct) was established for the relative quantification of RUNX3 expression.( 24 ) The dissociation curve for each amplification was analyzed to confirm that there were no non‐specific PCR products.
Western blot analysis. Fifty micrograms of protein was separated by electrophoresis on a sodium dodecyl sulfate‐polyacrylamide gel (12% gel). Then the protein was electrotransferred onto a polyvinylidene difluoride membrane (Amersham Pharmacia Biotech, Piscataway, NJ, USA). After 1 h of incubation in a blocking solution (5% non‐fat milk in phosphate‐buffered saline (PBS)–0.5% Tween 20), the membrane was blotted with the goat antihuman RUNX3 affinity purified polyclonal antibody (0.2 µg/mL; R&D Systems, Minneapolis, MN, USA) and a mouse anti‐GAPDH monoclonal antibody (1:10 000, IgG1 for clone 6C5; Kangchen Bio‐tech, Shanghai, China). The blots were developed with peroxidase‐labeled secondary antibodies. After extensive washing, specific bands were detected using an enhanced chemiluminescence system (Beyotime, Jiangsu, China) and captured on X‐ray film. The Western blot analysis of RUNX3 protein expression was performed in at least three independent experiments.
Immunohistochemical analysis. The immunohistochemical investigations were performed on deparaffined, 5‐µm sections after retrieval using autoclave sterilizer heating in 0.01 M citrate buffer (pH = 6.0). A rabbit polyclonal antihuman RUNX3 primary antibody (1:250; Beijing Biosynthesis Biotechnology, Beijing, China) and a rabbit streptavidin‐peroxidase staining system kit with a peroxidase/diaminobenzidine as the chromogenic substrate (Maixin Bio, Fuzhou, China) were used. After blocking, non‐specific staining sections were incubated overnight with anti‐RUNX3 antibody at 4°C. The sections were lightly counterstained with hematoxylin and mounted. Negative controls in all cases were performed by omitting the primary antibody. Histological samples of RUNX3 nuclear positive in epidermal cells of normal skin tissues were used as a positive control.( 25 ) In addition, tissue specimens of normal salivary glands (three parotid glands, two submandibular glands, two sublingual glands, and two minor salivary glands) were studied.
Evaluation of the RUNX3 immunohistochemical staining. LI of RUNX3 positive cells were counted at 200× with using Image‐Pro Plus (version 5.1; MediaCybernetics, Silver Spring, MD, USA) in each tumor section. This received a supplementary quantification via percentage (%) of cells showing RUNX3 positive staining among at least 1000 tumor cells to be counted.( 20 , 21 ) Continuous variables of RUNX3 LI were divided into three discrete exclusive variables according to the distribution of the samples. The tertile cut‐off values were as follows: LI ≤ 6.94% (low quartile), 6.94% < LI < 14.91%, and LI ≥ 14.91% (high quartile). Thus, RUNX3 immunoreactivity was classified into three groups: the low group (LI ≤ 6.94%), intermediate group (6.94% < LI < 14.91%), and high group (LI ≥ 14.91%).
Statistical analysis. All statistical tests were carried out using SPSS (version 12.0; SPSS, Chicago, IL, USA). The relative quantitative RT‐PCR results were expressed as mean and standard deviation (SD) and tested for statistical significance with Student's two‐tailed t‐test. Significant differences between the expression of RUNX3 and clinicopathological parameters, including age, gender, tumor site, clinical stage, perineural invasion, lymph node involvement, and distant metastasis, were compared by the χ2‐test. Survival analysis was computered by means of the Kaplan–Meier method and significant levels were assessed by means of the log‐rank test. A univariate analysis with the Cox regression model was used to determine identified prognostic factors, and multivariate analysis with the Cox regression model was used to explore combined effects. P‐values < 0.05 were considered to indicate statistical significance.
Results
Down‐regulation of RUNX3 in salivary ACCs analyzed by quantiative RT‐PCR. The relative quantification of RUNX3 mRNA expression in ACCs and normal salivary glands is shown in Figure 1. The RUNX3 mRNA levels (mean ± SD) in seven ACCs and nine normal salivary glands were 0.087 ± 0.139 and 0.192 ± 0.245, respectively. The level of RUNX3 mRNA in ACCs was two‐fold lower than that in normal salivary glands; however, there was no significant difference between the two groups (P = 0.329). This result indicated that RUNX3 mRNA expressed both in normal salivary glands and ACCs, and RUNX3 mRNA was down‐regulated in ACCs compared to the levels in normal salivary glands.
Figure 1.
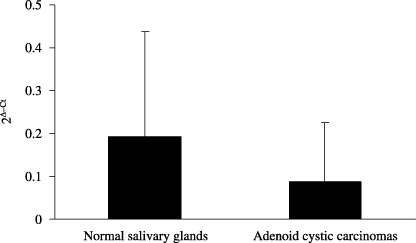
Quantitative reverse transcription–polymerase chain reaction (RT‐PCR) analysis revealed that runt‐related transcription factor‐3 (RUNX3) mRNA levels (mean ± SD) in adenoid cystic carcinomas were down‐regulated compared to the levels in normal salivary glands.
RUNX3 protein expression both in normal and ACC tissues measured by Western blot analysis. Western blot analysis disclosed a RUNX3 protein of 45 kDa both in the human normal salivary glands and ACCs (Fig. 2). There was no significant difference in the level of RUNX3 expression between the ACCs and the normal salivary glands, which is accordant with the results of quantitative RT‐PCR analysis of RUNX3 mRNA levels.
Figure 2.
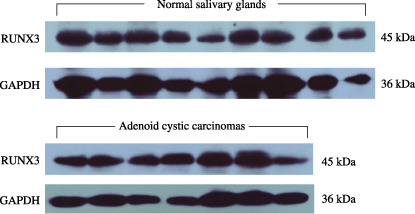
Western blot analysis showed a single band at 45 kDa of runt‐related transcription factor‐3 (RUNX3) protein both in nine normal salivary glands and seven salivary adenoid cystic carcinomas. The expression of glyceraldehyde‐3‐phosphate dehydrogease (GAPDH) served as a quantitative control.
Localization of RUNX3 protein expression by immunohistochemistry. In normal adult salivary glands, RUNX3 immunolocalization was found in all normal adult salivary glands. Expression of RUNX3 protein was detected in the nuclear and cytoplasm of both ductal and serous or mucous acinous cells of the parotid, submandibular, sublingual, and minor salivary glands (Fig. 3a–d). RUNX3 protein was also expressed in a few lymphoid cells (Fig. 3d). However, there was no expression of RUNX3 protein in myoepithelial cells in all normal salivary glands.
Figure 3.
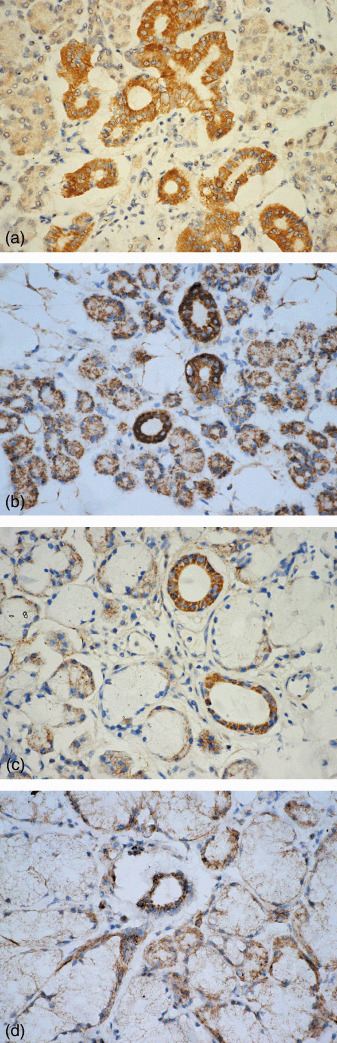
Nuclear and cytoplasmic localization of the runt‐related transcription factor‐3 (RUNX3) protein was noted in the ducal and acinous cells of (a) normal parotid, (b) submandibular, (c) sublingual, and (d) minor salivary glands. RUNX3 expression was also noted in (d) lymphoid cells (streptavidin‐peroxidase, SP; 400×).
Immunoreactivity against RUNX3 was detected mainly in the cytoplasm of ACC cells, but was not noted in the nucleus of those cells. In the tubular type of ACC, RUNX3 had higher immunopositivity in the inner layer of cells than in the outer layer (Fig. 4a). RUNX3 immunostained cells in cribriform structures lined cystic‐like spaces or were noted on the periphery of these cribriform arrangements (Fig. 4b). A diffuse pattern of immunostaining of RUNX3 was observed in the solid neoplasms (Fig. 4c). In case of perineural, intraneural, and perivascular invasion ACC, both tumor cells and nerve cells showed RUNX3 cytoplasmic immunopositivity (Fig. 4d). In gland invasion type, in contrast to nuclear and cytoplasmic location of RUNX3 protein in normal salivary gland, however, cytoplasmic mislocalization or even silence of RUNX3 expression was detected in invasive ACC cells (Fig. 4e).
Figure 4.
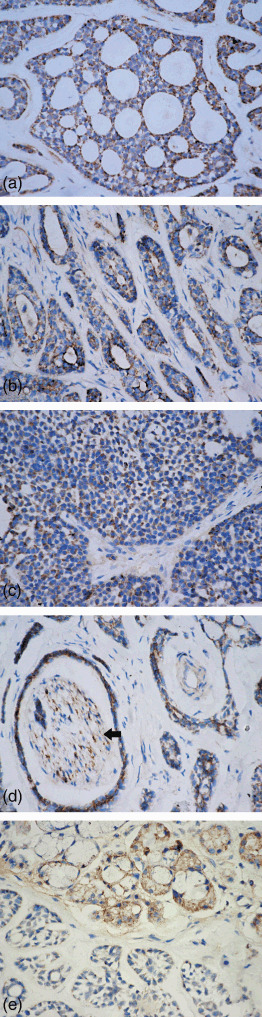
Runt‐related transcription factor‐3 (RUNX3) expressed in (a) cribriform, (b) tubular, and (c) solid patterns of ACC. (d) Tumor nest with perineural and intraneural invasion showed RUNX3 immunostaining in adenoid cystic carcinoma (ACC) cells and nerve cells (arrow). (e) In gland‐invasion type, RUNX3 protein is immunopositive in the normal salivary gland, but immunonegative in invasive ACC cells (streptavidin‐peroxidase, SP; 400×).
Correlation between RUNX3 expression and clinicopathological factors. Of the 73 patients with ACC included in this study, 43 were female (58.9%) and 30 were male (41.1%), giving a female‐male ratio of 1.4:1. The mean age of patients was 48.5 years (range, 23–79 years). Most tumors involved the minor salivary glands (60.3%), mainly in maxillary sinus (24.7%) and palate (20.5%), followed by the parotid (19.2%), submandibular gland (16.4%), and sublingual gland (4.1%). The detailed relationship between RUNX3 expression and clinicopathological profiles in the 73 salivary ACCs is shown in Table 1. The LI was 19.60 ± 4.26% in high group, 11.19 ± 2.42% in intermediate group, and 4.78 ± 1.59% in low group. The RUNX3 expression percentage was 13.69 ± 6.46% in the cribriform pattern, 11.78 ± 4.40% in the tubular pattern, and 8.86 ± 5.20% in the solid pattern. RUNX3 expression was significantly correlated with the histological patterns of the tumors (P = 0.025), whilst tumors with lower RUNX3 expression tended to have more frequent distant metastasis (P = 0.009), indicating the association between down‐regulation of RUNX3 expression and tumor progression. The LI was 13.65 ± 6.44% in stages T1–3 and 10.24 ± 5.19% in stage T4. There is only a weak tendency between the low expression of RUNX3 expression and stage T4 (P = 0.061). No statistical association between RUNX3 immunoexpression and other clinicopathologic parameters, such as age, gender, tumor site, perineural invasion, and lymph node involvement in ACCs (Table 1), was detected.
Survival analysis. Using univariate analysis (Cox's regression model), the following variables were found to be significantly associated with a worse prognosis, including low expression of RUNX3 protein (P = 0.012), lymph node involvement (P = 0.007), and distant metastasis (P < 0.001) (Table 2). The 5‐year survival rate was 90% in the 18 patients with high levels of RUNX3 expression (LI ≥ 14.91%), 75.4% in the 37 patients with moderate levels (6.94% < LI < 14.91%), and 48.27% in the 18 patients with low levels (LI ≤ 6.94%). Kaplan–Meier survival curves showed that the survival rates of patients with low expression of RUNX3 was significantly worse than that of patients with moderate or high RUNX3 expression (P = 0.022; log‐rank test) (Fig. 5a). Meanwhile, we observed that some other clinicopathogical parameters, including stage T4 (P = 0.003, Fig. 5b), lymph node involvement (P = 0.004, Fig. 5c), and distant metastasis (P < 0.001) were significantly related with decreased survival. However, no significant association between overall survival and solid histotype (P = 0.771) or perineural invasion (P = 0.554) was found. Multivariate survival analysis revealed that only distant metastasis was an independent significant prognostic factor (P = 0.043; Cox‐regression, Table 3). Stage T4 had a weak association with overall survival (P = 0.063; Cox‐regression, Table 3).
Table 2.
Univariate survival analysis of clinicopathologic data of 73 adenoid cystic carcinomas
| Variables | Hazard ratio (95% confidence interval) | P‐values |
|---|---|---|
| Age (<46/≥46) | 1.383 (0.490–3.903) | 0.540 |
| Gender (female/male) | 1.266 (0.467–3.428) | 0.643 |
| Site (Major/Minor) | 0.611 (0.221–1.687) | 0.341 |
| T‐phage (1–3/4) | 11.801 (1.545–90.117) | 0.017* |
| Histotypes (cribriform/tubular/solid) | 1.209 (0.692–2.111) | 0.506 |
| Distant metastasis (+/–) | 0.152 (0.055–0.421) | <0.001* |
| Perineural invasion (+/–) | 0.743 (0.276–1.999) | 0.556 |
| Lymph node involvement (+/–) | 0.215 (0.070–0.659) | 0.007* |
| RUNX3 (low/intermediate/high) † | 0.349 (0.154–0.794) | 0.012* |
Statistically significant.
Tertile cut‐off: low ≤ 6.94%; 6.94% < Intermediate < 14.91%; High ≥ 14.91%.
RUNX3, runt‐related transcription factor‐3.
Figure 5.
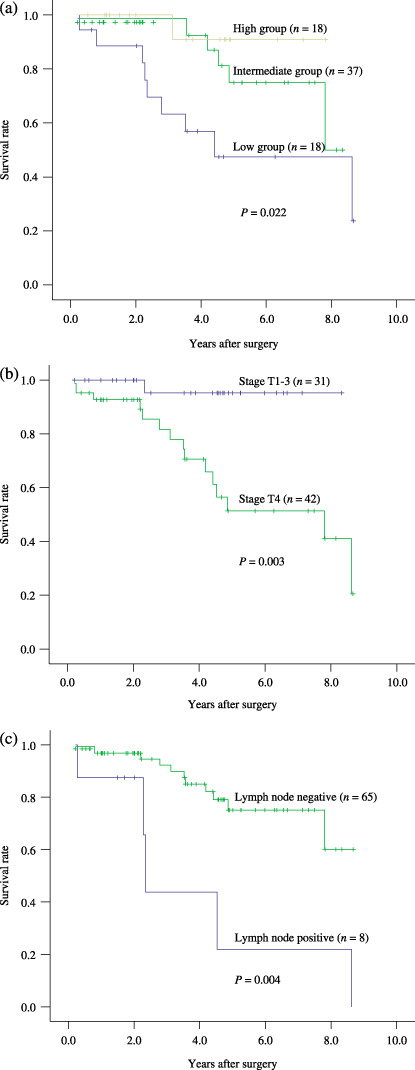
Kaplan–Meier survival curves for: (a) expression of runt‐related transcription factor‐3 (RUNX3), (b) clinical stage, and (c) lymph node involvement.
Table 3.
Multivariate survival analysis of clinicopathologic data of 73 adenoid cystic carcinomas
| Variables | Hazard ratio (95% confidence interval) | P‐values |
|---|---|---|
| Age (<46/≥46) | 0.819 (0.236–2.840) | 0.753 |
| Gender (female/male) | 2.904 (0.703–11.995) | 0.141 |
| Site (major/minor) | 0.996 (0.268–3.696) | 0.995 |
| T‐phage (1–3/4) | 7.679 (0.894–65.968) | 0.063 |
| Histotypes (cribriform/tubular/solid) | 0.804 (0.383–1.687) | 0.563 |
| Distant metastasis (+/–) | 0.203 (0.043–0.954) | 0.043* |
| Perineural invasion (+/–) | 0.615 (0.165–2.288) | 0.469 |
| Lymph node involvement (+/–) | 0.398 (0.077–2.072) | 0.274 |
| Runx3 (≤6.94%/>6.94%) | 0.989 (0.259–3.773) | 0.987 |
Statistically significant.
Discussion
This study clearly demonstrated RUNX3 expression in human normal salivary glands and salivary ACCs. The down‐regulation of RUNX3 mRNA in carcinoma tissues was detected by using relative quantitative RT‐PCR analysis and compared with relative RUNX3 levels in normal salivary glands. However, there was no significant different between the expression of RUNX3 gene in normal salivary glands and salivary ACCs. In conjunction with this result, Western blot analysis showed the expression of RUNX3 protein as a single band at 45 kDa both in normal salivary glands and ACCs. Nevertheless, there was no significant difference in RUNX3 protein expression between the ACCs and normal salivary glands, possibly owing to a result of the expression of the RUNX3 protein in non‐neoplastic cells, peripheral nerves, and lymphoid cells in salivary ACCs.
Immunohistochemistry revealed that RUNX3 expression was detected in ductal cells and serous or mucous acinous cells of normal salivary glands, in contrast to the lack of RUNX3 expression in the myoepithelial cells, suggesting that RUNX3 may play a specific functional role in ductal and acinous cells. We propose that the maintenance of the RUNX3 protein in acinous cells, but not in myoepithelial cells, might be expected to be a helpful tool in the identification of these cells. Furthermore, we found the nuclear as well as cytoplasmic expression of RUNX3 protein both in ductal and acinous cells in present study. It is thought that transient cytoplasmic localization of RUNX3 might be translocated to nuclei after stimulation by TGF‐β, or that RUNX3 has functions other than being a transcriptional factor.( 26 ) However, the details of biological RUNX3 functions in the human salivary glands are unclear. Therefore, further biochemical and biological studies are required to elucidate the detailed mechanisms.
In contrast to the nuclear localization of RUNX3 protein in normal salivary glands, weak expression of RUNX3 was observed apparently in the cytoplasm of tumor cells, which is concordant with some previous studies.( 20 , 22 , 27 , 28 , 29 ) Interestingly, in the normal salivary‐gland invasion type, the RUNX3 protein is detected in the nuclear and cytoplasm of normal salivary gland cells, but there can be cytoplasmic mislocalization or even silenced expression in the invasive ACC cells. It is possible that the cytoplasmic localization of RUNX3 could be an important mechanism whereby RUNX3 is inactivated in ACC. In many signaling pathways, signal transducers are transcription factors that are restricted by the nuclear envelope from gaining access to target genes. The cytoplasmic localization of RUNX3 protein was, possibly, inactive as a tumor suppressor protein, and resistant to TGF‐β‐inducible nuclear translocation of RUNX3 to induce cell apoptosis. To explain the potential mechanism for that, one possibility is that the COOH‐terminal region of RUNX3 protein might be modified to inhibit its translocation to the nucleus,( 28 ) for RUNX3 interacts with R‐Smads via its COOH‐terminal region to allow its nuclear translocation.( 16 ) Therefore the COOH‐terminally modified RUNX3 is retained in the cytoplasm because it cannot interact with Smads any more. The another possibility is that the cytoplasmic retention of RUNX3 protein may be due to a lack or inactivation of one or more components in the signaling pathway required for RUNX3 nuclear localization. It is reported that RUNX3 as a transcription factor regulates the expressions of numerous downstream genes, such as the cell‐cycle regulator and apoptosis‐inducing factor Bim, to induce cell apoptosis.( 17 , 22 , 30 , 31 ) Thus the cytoplasmic mislocalization of the RUNX3 protein cannot act as a transcription factor and does not elicit tumor‐suppressive effects.( 28 ) As reported in several previous studies, the cytoplasmic retention of RUNX3 protein in gastric cancer and breast cancer is thought to be a novel mechanism for inactivating RUNX3 function.( 27 , 28 ) Sakakura et al. report that the cytoplasmic or no RUNX3 expression in human esophageal squamous cell carcinomas was significantly associated with radiosensitivity and a worse prognosis.( 22 ) Hence, cytoplasmic retention of RUNX3 protein could be a potential valuable “molecular marker” for the detection of inactivity of RUNX3 as a tumor suppressor and impairment of the TGF‐β signaling pathway. Nevertheless, contrasting with the current understanding of RUNX3 as a tumor suppressor in several human carcinomas, it has been reported that RUNX3 might act as a putative oncogene in human basal cell carcinoma.( 25 ) Compared to the immunolocalization of RUNX3 protein in normal salivary glands, however, the reason for the cytoplasmic retention of RUNX3 protein in ACC cells is still unexplained at present. It is possible that cytoplasmic localization of RUNX3 in normal salivary glands and ACCs has different roles. Therefore, many other details remain to be elucidated.
In salivary ACC, RUNX3 expression was observed in all of the three histological patterns, namely cribriform, tubular, and solid type. The RUNX3 positive cells in the solid type (8.86 ± 5.20%) were significant lower than those in the cribiform (13.69 ± 6.46%) and tubular types (11.78 ± 4.40%) in this study. Additionally, as seen in Table 1, there was a significant correlation between the frequency of RUNX3 expression and histological patterns in the salivary ACCs (P = 0.025). However, in contrast to other reports,( 6 , 7 , 8 ) we did not find a correlation between solid histological subtype and poor prognosis. It is reported that the solid type ACC behaves much more aggressively than the tubular‐cribriform tumors without solid component.( 6 ) Meanwhile, RUNX3 is a tumor suppressor factor in gastric cancer and appears to be a crucial component of the TGF‐β‐induced tumor suppressor pathway.( 18 , 19 ) Li et al. reported that the RUNX3 protein has essential functions in both cell proliferation and differentiation in gastric epithelia and a tumor‐suppressor gene causally involved in gastric carcinogenesis.( 15 ) Thus, it might be considered that an unfavorable prognosis in human salivary ACC with a solid pattern is associated with a lower expression of RUNX3, indicating that it has a pivotal role in the tumor proliferation and apoptosis.
As RUNX3 is a downstream target of the TGF‐β‐induced apoptosis pathway, resistance to apoptosis involving the loss of RUNX3 expression is suspected in cases showing metastasis. In the present study, ACCs with low RUNX3 expression were biologically more aggressive and at an increased risk of distant metastasis leading to a fatal outcome. Sakakura et al. report that RUNX3 silencing in peritoneal metastasis of gastric cancers affects expression of important genes involved in aspects of metastasis including cell adhesion, proliferation, apoptosis, and promotion of the peritoneal metastasis of gastric cancer.( 23 ) It has been previously reported that down‐regulation of RUNX3 expression showed progressive silencing according to cancer stage, especially in stage IV, and that almost 100% of cases showed silencing of the RUNX3 protein.( 15 ) In this study, down‐regulation of RUNX3 expression only had a weak correlation with cancer stage. Albeit so, the lower expression of RUNX3 protein might indicate the progression and the risk of distant metastasis in salivary ACCs.
Additionally, on the basis of our results, it is worthwhile touching upon the prognostic significance of the RUNX3 expression. We found that the low RUNX3 immunoexpression was significantly related with overall survival (P = 0.022, log‐rank test; P = 0.012, Cox‐regression). However, in the Cox multivariate survival analysis, there was no significant association between RUNX3 expression and overall survival. Therefore, this trend warrants further investigation and statistical evaluation in a larger series of patients with a longer follow‐up period. It is significant, however, that a similar finding has been reported in a series of 98 lung adenocarcinomas.( 20 ) In this report, less than 10% of RUNX3 positive cells was a stronger predictor for worse patient survival. Meanwhile, the lower or lack of expression of the RUNX3 protein in esophageal squamous cell carcinomas is significantly associated with patient survival rates.( 22 ) Together, RUNX3 might be a candidate for prognostic marker and molecular therapeutic targets in salivary ACC.
In conclusion, in the present study, we have revealed the expression of RUNX3 both in human normal salivary glands and salivary ACCs. The cytoplasmic retention or even lack of expression of the RUNX3 protein might be a potential mechanism of salivary ACC development. Our results indicated that lower expression of RUNX3 is significantly correlated with histopathological growth pattern, distant metastasis, and patient survival. However, the function of the cytoplasmic localization of RUNX3 in normal salivary glands and ACC is still unclear. Therefore, further studies are needed to elucidate these questions and to clarify the value of adopting biological prognostic factors into clinical practice.
Acknowledgments
This work is supported by Zhejiang Provincial Natural Science Foundation of China (No. Y205660).
References
- 1. El‐Naggar AK, Huvos AG. Adenoid cystic carcinoma. In. Barnes JW, Evenson JW, Reichert P, Sidransky D, eds. World Health Organization Classification of Tumours: Pathology and Genetics of Head and Neck Tumours. Lyon: IARC Press, 2005: 221–2. [Google Scholar]
- 2. Van Der Wal JE, Becking AG, Snow GB, Van Der Waal I. Distant metastases of adenoid cystic carcinoma of the salivary glands and the value of diagnostic examinations during follow‐up. Head Neck 2002; 24: 779–83. [DOI] [PubMed] [Google Scholar]
- 3. Kokemueller H, Eckardt A, Brachvogel P, Hausamen JE. Adenoid cystic carcinoma of the head and neck – a 20 years experience. Int J Oral Maxillofac Surg 2004; 33: 25–31. [DOI] [PubMed] [Google Scholar]
- 4. Chummun S, McLean NR, Kellyt CG et al . Adenoid cystic carcinoma of the head and neck. Br J Plast Surg 2001; 54: 476–80. [DOI] [PubMed] [Google Scholar]
- 5. Garden AS, Weber RS, Morrison WH, Ang KK, Peters LJ. The influence of positive margins and nerve invasion in adenoid cystic carcinoma of the head and neck treated with surgery and radiation. Int J Radiat Oncol Biol Phys 1995; 32: 619–26. [DOI] [PubMed] [Google Scholar]
- 6. Szanto PA, Luna MA, Tortoledo ME, White RA. Histologic grading of adenoid cystic carcinoma of the salivary glands. Cancer 1984; 54: 1062–9. [DOI] [PubMed] [Google Scholar]
- 7. Perzin KH, Gullane P, Clairmont AC. Adenoid cystic carcinoma arising in salivary glands: a correlation of histologic features and clinical course. Cancer 1978; 42: 265–82. [DOI] [PubMed] [Google Scholar]
- 8. Da Cruz Perez DE, De Abreu Alves F, Nobuko Nishimoto I, De Almeida OP, Kowalski LP. Prognostic factors in head and neck adenoid cystic carcinoma. Oral Oncol 2006; 42: 139–46. [DOI] [PubMed] [Google Scholar]
- 9. Norberg‐Spaak L, Dardick I, Ledin T. Adenoid cystic carcinoma: use of cell proliferation, BCL‐2 expression, histologic grade, and clinical stage as predictors of clinical course. Head Neck 2000; 22: 489–97. [DOI] [PubMed] [Google Scholar]
- 10. Jia L, Esguerra RL, Tang X et al . Prognostic value of apoptosis and apoptosis‐associated proteins in salivary gland adenoid cystic carcinoma. Pathol Int 2004; 54: 217–23. [DOI] [PubMed] [Google Scholar]
- 11. Zhu QR, White FH, Tipoe GL. p53 oncoprotein accumulation in adenoid cystic carcinoma of parotid and palatine salivary glands. Pathology 1997; 29: 154–8. [DOI] [PubMed] [Google Scholar]
- 12. Levanon D, Negreanu V, Bernstein Y, Bar‐Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene‐family: cDNA structure, expression, and chromosomal localization. Genomics 1994; 23: 425–32. [DOI] [PubMed] [Google Scholar]
- 13. Ito Y. Molecular basis of tissue‐specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells 1999; 4: 685–96. [DOI] [PubMed] [Google Scholar]
- 14. Levanon D, Bernstein Y, Negreanu V et al . A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol 1996; 15: 175–85. [DOI] [PubMed] [Google Scholar]
- 15. Li QL, Ito K, Sakakura C et al . Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002; 109: 113–24. [DOI] [PubMed] [Google Scholar]
- 16. Hanai J, Chen LF, Kanno T et al . Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J Biol Chem 1999; 274: 31 577–82. [DOI] [PubMed] [Google Scholar]
- 17. Chi XZ, Yang JO, Lee KY et al . RUNX3 suppresses gastric epithelial cell growth by inducing p21 (WAF1/Cip1) expression in cooperation with transforming growth factor {beta}‐activated SMAD. Mol Cell Biol 2005; 25: 8097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF‐beta superfamily signaling. Curr Opin Genet Dev 2003; 13: 43–7. [DOI] [PubMed] [Google Scholar]
- 19. Zaidi SK, Sullivan AJ, Van Wijnen AJ, Stein JL, Stein GS, Lian JB. Integration of RUNX and Smad regulatory signals at transcriptionally active subnuclear site. Proc Natl Acad Sci USA 2002; 99: 8048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Araki K, Osaki M, Nagahama Y et al . Expression of RUNX3 protein in human lung adenocarcinoma: Implication for tumor progression and prognosis. Cance Sci 2005; 96: 227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanji Y, Osaki M, Nagahama Y, Kodani I, Ryoke K, Ito H. Runt‐related transcription factor 3 expression in human oral squamous cell carcinomas; implication for tumor progression and prognosis. Oral Oncol 2007; 43: 88–94. [DOI] [PubMed] [Google Scholar]
- 22. Sakakura C, Miyagawa K, Fukuda KI et al . Frequent silencing of RUNX3 in esophageal squamous cell carcinomas is associated with radioresistance and poor prognosis. Oncogene 2007; 26: 5927–38. [DOI] [PubMed] [Google Scholar]
- 23. Sakakura C, Hasegawa K, Miyagawa K et al . Possible involvement of RUNX3 silencing in the peritoneal metastases of gastric cancers. Clin Cancer Res 2005; 11: 6479–88. [DOI] [PubMed] [Google Scholar]
- 24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2 (‐Delta Delta C (T) Method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- 25. Salto‐Tellez M, Peh BK, Ito K et al . RUNX3 protein is overexpressed in human basal cell carcinoma. Oncogene 2006; 25: 7646–9. [DOI] [PubMed] [Google Scholar]
- 26. Osaki M, Moriyama M, Adachi K et al . Expression of RUNX3 protein in human gastric mucosa, intestinal metaplasia and carcinoma. Euro J Clin Invest 2004; 34: 605–12. [DOI] [PubMed] [Google Scholar]
- 27. Lau QC, Raja E, Salto‐Tellez M et al . RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res 2006; 66: 6512–20. [DOI] [PubMed] [Google Scholar]
- 28. Ito K, Liu Q, Salto‐Tellez M et al . RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res 2005; 65: 7743–50. [DOI] [PubMed] [Google Scholar]
- 29. Nagahama Y, Ishimaru M, Osaki M et al . Apoptotic pathway induced by transduction of RUNX3 in human gastric carcinoma cell line MKN‐1. Cancer Sci 2008; 99: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamamura Y, Lee WL, Inoue KI, Ida H, Ito Y. RUNX3 cooperates with FoxO3a to induce apoptosis in gastric cancer cells. J Biol Chem 2006; 281: 5267–76. [DOI] [PubMed] [Google Scholar]
- 31. Yano T, Ito K, Fukamachi H et al . The RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells undergoing transforming growth factor beta‐induced apoptosis. Mol Cell Biol 2006; 26: 4474–88. [DOI] [PMC free article] [PubMed] [Google Scholar]


