Abstract
Our previous studies revealed that the expression of the 19‐kDa protein prenylated Rab acceptor 1 domain family, member 2 (PRAF2) is elevated in cancer tissues of the breast, colon, lung, and ovary, when compared to noncancerous tissues of paired samples. PRAF2 mRNA expression also correlated with several genetic and clinical features and is a candidate prognostic marker in the pediatric cancer neuroblastoma. The PRAF2‐related proteins, PRAF1 and PRAF3, play multiple roles in cellular processes, including endo/exocytic vesicle trafficking and glutamate uptake. PRAF2 shares a high sequence homology with these family members, but its function remains unknown. In this study, we examined PRAF2 mRNA and protein expression in 20 different human cancer types using Affymetrix microarray and human tissue microarray (TMA) analyses, respectively. In addition, we investigated the subcellular distribution of PRAF2 by immunofluorescence microscopy and cell fractionation studies. PRAF2 mRNA and protein expression was elevated in several cancer tissues with highest levels in malignant glioma. At the molecular level, we detected native PRAF2 in small, vesicle‐like structures throughout the cytoplasm as well as in and around cell nuclei of U‐87 malignant glioma cells. We further found that monomeric and dimeric forms of PRAF2 are associated with different cell compartments, suggesting possible functional differences. Importantly, PRAF2 down‐regulation by RNA interference significantly reduced the cell viability, migration, and invasiveness of U‐87 cells. This study shows that PRAF2 expression is elevated in various tumors with exceptionally high expression in malignant gliomas, and PRAF2 therefore presents a candidate molecular target for therapeutic intervention. (Cancer Sci 2010)
Malignant glioma (MG) is the most common adult brain tumor and is as yet almost incurable. Due to its invasive nature and resistance to chemotherapy, the median survival is only 14.6 months, even with the most advanced care.( 1 ) Therefore, there is an urgent need for new diagnostic markers and potential drug targets. Proteins which are over‐expressed in malignant tissues but not in the healthy tissue of origin, can provide important insights into understanding the underlying molecular mechanisms of malignant transformation, and also represent good candidate targets for drug development and intervention.
The prenylated Rab acceptor 1 domain family (PRAF) proteins have recently gained increasing attention in the study of cancer. In humans, three PRAF members exist (PRAF1–3), all of which have four transmembrane domains, a hydrophilic N‐ and C‐terminus, and a prenylated Rab acceptor (PRA) motif. PRAF1 and PRAF3 were shown to interact with membrane‐organizing Rab GTPases.( 2 , 3 , 4 , 5 ) In addition, they bind with proteins that regulate several well‐known cancer signaling pathways. For example, PRAF1 binds to and blocks the nuclear transport of β‐catenin( 6 ) and interacts with Ras and Rho GTPases,( 7 ) associated with the promotion of metastasis. PRAF1 also interacts with viral proteins of Epstein–Barr virus (EBV), influencing its oncogenic behavior.( 8 , 9 , 10 ) Similar to PRAF1, the PRAF3 protein is involved in various oncogenic processes. In the HeLa cervical cancer cell line, PRAF3 is essential for the anti‐proliferative effect of all‐trans retinoic acid (ATRA)( 11 ) and inhibits both F‐actin rearrangement and consequent cell migration via MAP kinase cascades.( 12 ) It is also known that the presence of point mutations in the human PRAF3 gene is associated with increased risk of various cancers.( 13 ) Finally, PRAF3 controls the endoplasmic reticulum (ER) exit of glutamate transporter EAAC1,( 14 , 15 ) and inhibits neuronal cysteine transport,( 16 ) thereby capable of reducing high intracellular glutathione (GSH) levels, a characteristic of cancer cells, which significantly contributes to their survival and drug resistance.
PRAF2 (previously known as JM4) is the least characterized member of the PRAF protein family. PRAF2 was initially identified as a human chemokine receptor CCR5‐interacting protein( 17 ) and also interacts with the human glycerophosphoinositol phosphodiesterase (GDE1/MIR16).( 18 ) Elevated protein levels were detected in cancer tissues of the breast, colon, lung, and ovary, when compared to noncancerous tissues of paired samples.( 19 ) More recently, we analyzed the expression pattern and localization of PRAF2 in the human brain and found evidence of PRAF2 accumulation in synaptic vesicles.( 20 ) We further discovered that elevated levels of PRAF2 correlated with unfavorable genetic and clinical features of the pediatric cancer neuroblastoma,( 21 ) suggesting that PRAF2 is a candidate prognostic marker. At the molecular level, PRAF2 primarily localized to endosome/vesicles in the cytoplasm of human neuroblastoma cells.( 21 )
In the present investigation, we have studied the expression of PRAF2 in tissue samples from 20 different human tumors including malignant glioma, where its expression was exceptionally high. At the molecular level, we identified the subcellular localization and distribution of PRAF2, which was associated with punctate structures in malignant glioma cells. We found that the monomeric/dimeric state of PRAF2 influences its localization within the cell environment and permanent down‐regulation of PRAF2 impaired the viability, migration, and invasiveness of glioblastoma cells.
Materials and Methods
Reagents and antibodies. Rabbit polyclonal anti‐PRAF2 peptide antibody (300 μg/mL) directed against the C‐terminus of PRAF2 and the PRAF2 blocking peptide was from QED Bioscience (San Diego, CA, USA). Peptide‐blocked PRAF2 antibody (PRAF2‐P) was prepared by saturating its binding sites with 5× excess (w/w) blocking peptide in PBS for 2 h at room temperature. Primary antibodies against neurexin 2α (ab34245), alpha‐tubulin (ab7291‐10), and calreticulin (ab14234‐50) were obtained from Abcam (Cambridge, MA, USA); and anti‐GFAP antibody (Z0334) was purchased from Dako (Glostrup, Denmark). Horseradish peroxidase (HRP)‐labeled anti‐rabbit secondary antibody (ECL #NA9340V) was purchased from GE Healthcare (Wauwatosa, WI, USA); and conjugated fluorescent secondary antibodies Alexa Fluor 555 goat anti‐mouse IgG (A21424), Alexa Fluor 647 goat anti‐chicken IgG (A21449), and Alexa Fluor 488 goat anti‐rabbit IgG (A11034) were from Invitrogen (Carlsbad, CA, USA).
Cell lines and culture conditions. The malignant glioma/glioblastoma cell line U‐87 was cultured in DMEM containing 10% heat‐inactivated fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Mediatech, Manassas, VA, USA). The malignant glioma cell line U‐251 was grown in 10‐cm Nunc Dishes (Fisher Scientific, Vastra Frolunda, Sweden) in DMEM medium supplemented with 10% FBS and 1% amino acids (Invitrogen) in the absence of antibiotics. Cultivation of cell lines included in the cell microarray (CMA) were performed as previously described.( 22 ) The PRAF2‐deficient or scrambled control U‐87 cell lines were generated by stable transfection with Lipofectamine2000 (Invitrogen) using pre‐designed 29‐mer HuSH small‐hairpin (sh)RNA vectors (pRS) against PRAF2 (shPRAF2; TI340825) or a non‐effective scrambled shRNA cassette (Origene; Rockville, MD, USA). Transfected cell lines were maintained as above in the presence of 0.2–1.0 μg/mL puromycin.
Affymetrix microarray analysis. Expression microarray analyses in this study were performed on the genome‐wide mRNA expression platform (Affymetrix HG‐U133 Plus 2.0, Santa Clara, CA, USA). All gene transcript levels were determined from data image files using GeneChip operating software (MAS5.0 and GCOS1.0; from Affymetrix). Samples were scaled by setting the average intensity of the middle 96% of all probe‐set signals to a fixed value of 100 for every sample in the dataset. Transcript levels could thus be compared between microarrays. The TranscriptView genomic analysis and visualization tool was used to certify that the probe‐set selected had an antisense position in an exon of the gene (http://bioinfo.amc.uva.nl/human‐genetics/transcriptview/). CEL data from the Affymetrix data‐sets in public GEO data‐sets on the NCBI website( 23 ) were downloaded and analyzed as previously described.( 24 ) Annotations and clinical data for the tissue samples analyzed are available from http://www.ncbi.nlm.nih.gov/geo/query/ through their GEO ID: GSE4290 (for the Sun‐153 glioma set( 25 )), GSE4536 (for the Lee‐101 glioma set( 26 )), GSE7307 (for the Roth‐504 set, unpublished), or GSE2109 (for all other sets: https://expo.intgen.org/expo/public).
Tissue microarray analysis (TMA). The human brain tissue samples were part of a large multi‐TMA analysis which was performed with over 8800 antibodies. Human brain tissue samples were collected from surgical specimens, in accordance with approval from the local ethics committee (Stockholm, Sweden). All antibodies were validated by western blotting and the validation strategy was previously published.( 27 ) Immunohistochemistry and quantification of stained tissues was in broad outline performed as previously described.( 20 , 28 , 29 ) Immunohistochemistry and quantification of stained CMAs was performed as previously described.( 22 , 30 , 31 ) All antibodies, validation data, and immunostainings are available at http://www.proteinatlas.org.
Immunofluorescence. A total of 10 000 cells were seeded in glass‐bottom, 96‐well Whatman plates (GE Healthcare, Stockholm, Sweden) coated with fibronectin. After 3 h of incubation, growth media was removed, and cells were washed in PBS and fixed for 15 min in ice cold 4% paraformaldehyde [pH 7.2] in growth medium supplemented with 10% FBS. Cells were then permeabilized for 3 × 5 min with 0.1% (v/v) Triton X‐100 in PBS, washed once in PBS, and labeled overnight at 4°C with a cocktail of primary antibodies against PRAF2 (1:150), calreticulin (1:1000), and alpha‐tubulin (1:1000) dissolved in PBS supplemented with 4% FBS. The same buffer was used for secondary antibodies (all used in 1:800 dilution) for 1.5 h at room temperature the next day after a thorough wash in PBS. Finally, cells were counterstained with 0.3 μM DAPI (D21490; Invitrogen) for 4 min, washed in PBS, and mounted in PBS supplemented with 78% glycerol. Images were acquired using an LSM 510 Meta confocal laser scanning microscope with a 63×/1.4 numerical aperture oil immersion objective (Carl Zeiss, Jena, Germany). Excitation lasers of 405, 488, 543, and 633 nm wavelengths, and emission filters BP 420‐480, BP 505‐550, BP 560‐615, and LP 650 were used to visualize DAPI, PRAF2, microtubules, and ER, respectively.
Gravity‐based subcellular and nuclear fractionation. All fractionation steps were carried out on ice or 4°C unless noted otherwise. Semi‐confluent U‐87 cells (2 × 107) grown in 15‐cm Petri dishes (Greiner, Monroe, NC, USA) were scraped and homogenized in a Dounce homogenizer (#357544; Wheaton, Millville, NJ, USA) in ice‐cold, detergent‐free buffer (20 mM Tris–HCl [pH 7.5], 1 mM EDTA, 5 mM MgCl2). Nuclei were sedimented at 800g, and the pellet was suspended in 20 mM Tris–HCl [pH 7.5], 255 mM sucrose, 10 mM MgCl2, and gently layered on a sucrose cushion (20 mM Tris–HCl [pH 7.5], 350 mM sucrose, 0.5 mM MgCl2) to remove non‐lysed cells. After 10‐min centrifugation at 1430g, the nuclear pellet was suspended and stored in membrane storage buffer (20 mM Tris–HCl [pH 7.5], 300 mM sucrose). To fractionate nuclei, four volumes of high ionic strength lysis buffer (10 mM Tris–HCl [pH 7.5], 0.2 mM MgCl2, 2 M NaCl) and 1% (v/v) 2‐mercaptoethanol was added( 32 ) and lysis was allowed to complete at 4°C for 15 min. After pelleting nuclear envelopes (1600g for 10 min), the supernatant representing the intranuclear content was saved, and the pellet was washed with high ionic strength lysis buffer, pelleted again, and suspended in membrane storage buffer. To separate membranous compartments (organelles and microsomes) from cytoplasm, the nucleus‐free sample was centrifuged with a MLS‐50 rotor at 163 000g in 5 mL volume on a Beckman Optima Max ultracentrifuge for 1 h. The membrane‐free supernatant (cytosol) fraction was saved and the pellet was dissolved in membrane storage buffer. All samples were volume‐adjusted and the relative amount of endogenous PRAF2 in each fraction compared. Every lane represents protein amounts corresponding to 0.5% of the total cell lysate volume.
Western blot analysis. Samples obtained from cell fractionation steps were mixed with equal volume of Laemmli buffer (Bio‐Rad, Hercules, CA, USA), incubated for 30 min at room temperature, resolved by 12% SDS‐PAGE, and electro‐transferred onto PVDF membrane (Pall Life Sciences, Port Washington, NY, USA) in 10 mM CAPS/10% (v/v) methanol transfer buffer. Membranes were air‐dried, reconstituted in methanol, and blocked in 0.1% TBST (TBS containing 0.1% (v/v) Tween‐20) containing 3% (w/v) BSA. PRAF2‐specific primary antibody or its peptide‐blocked version was then added in 1:5000 dilution, hybridized at 4°C overnight, washed, and incubated with rabbit secondary HRP‐antibody (1:5000) in 0.1% TBST for 1 h. After a final wash, the membrane was developed using the ECL plus kit (GE Healthcare). Membranes were stripped at 50°C for 30 min with ECL stripping buffer (62.5 mM Tris–HCl [pH 6.7], 2% (w/v) SDS, 100 mM 2‐mercaptoethanol) and sequentially probed.
Cell viability assay. The CellTiter 96 AQueous One Solution MTS Cell Proliferation Assay is a colorimetric method for determining the number of viable cells (Promega, San Luis Obispo, CA, USA) and was performed as previously described.( 33 )
Transwell migration and invasion assays. The permeable support was either uncoated (migration assay) or coated with reconstituted basement membrane extract (invasion assay) and 5 × 104 serum‐starved cells were seeded in the top chamber of a Transwell plate, in medium without FBS. Medium with FBS was added to the bottom chamber. Cells were incubated for 24 h, and maintained at 37°C in a CO2 incubator. The media was removed, both chambers washed, calcein‐AM was added to the bottom chamber, and incubated for 1 h at 37°C. The Relative Fluorescence Unit (RFU) was measured using a fluorescent plate reader (485 nm excitation/520 nm emission). A standard curve was used to determine the total number of cells that responded to serum‐induced migration or invasion.
Results
In our previous studies, we have described high PRAF2 protein expression in several human tumors (breast, colon, lung, ovary, and neuroblastoma).( 19 , 21 ) This prompted us to investigate the presence of PRAF2 in an extended set of normal and tumor tissues. First, we compared the expression level of PRAF2 mRNA in 20 tumor tissues of different origin. As shown in Figure 1(a), the expression of PRAF2 was higher in gliomas than in any other tumor type. To further expand our investigation, we determined the PRAF2 mRNA expression levels in various gliomas as well as normal tissues. The glioma samples showed higher PRAF2 expression, also when compared to the brain, where its expression was most prevalent. This seemed especially true for high‐grade glioma tumors, glioblastoma stage 4, in two different glioma sets (Fig. 1b). Statistical analysis revealed that PRAF2 mRNA expression in normal brain is significantly lower than in three different types of glioma samples (glioblastoma grade 4, commonly used glioma cell lines, and glioblastoma‐derived stem cells; Table S1).
Figure 1.
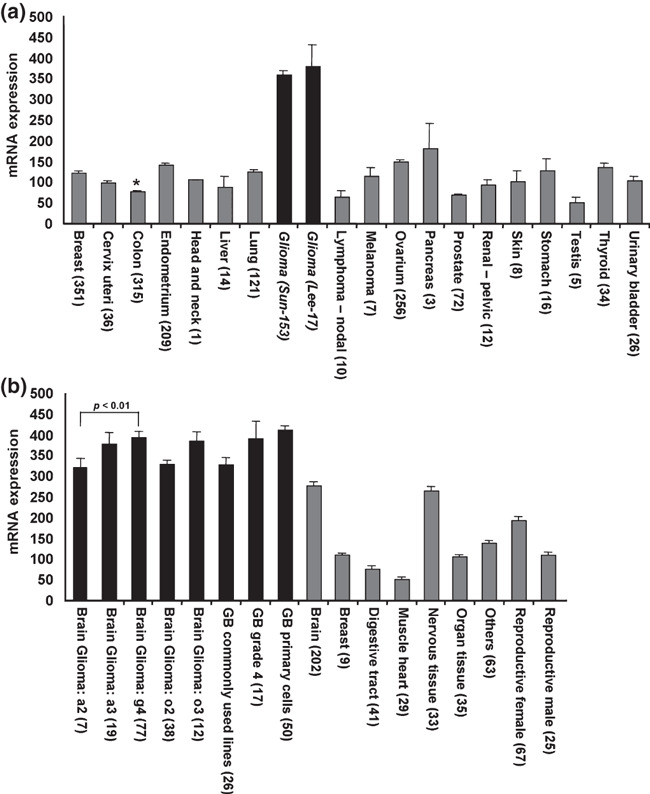
Prenylated Rab acceptor 1 domain family, member 2 (PRAF2) mRNA expression in malignant and healthy tissues. (a) PRAF2 mRNA level in a variety of human tumors. The expression of PRAF2 in two glioma tumor sets (black bars) was higher than in any other tumor tissue (gray bars). For comparison: average expressions of GAPDH and β‐actin in this dataset were 9240 and 11 890, respectively. *Expression data for other colon cancer sets are: 68.73 ± 6.63 for rectosigmoid tumors (n = 9) and 92.01 ± 10.25 (n = 38) for rectal tumors. Tumor sets are described in the Materials and Methods. (b) PRAF2 mRNA level is significantly higher in glioma (black bars) than in healthy tissues (gray bars) including the brain. Statistically significant differences in mRNA expression are shown above the bars. T‐tests were calculated in Excel. Datasets used: brain glioma Sun‐153, glioblastoma (GB) Lee‐101, healthy tissues Roth‐504. Error bars represent the SEM, numbers in parentheses refer to sample size.
To verify our findings at the protein level, we determined the PRAF2 expression in a TMA using the same tumor types. A total of 210 tissue samples were processed and graded based on PRAF2 signal strength following immune histochemical staining with a specific, PRAF2‐directed polyclonal peptide antibody. We found that malignant glioma tumors expressed the highest levels of PRAF2 among all tumor tissues investigated (Fig. 2a,b). PRAF2 expression was high in 11 out of 12 patient samples with predominant presence in the cytoplasm or membrane (Table 1 and Fig. S1). In contrast, renal tumor tissue samples expressed the lowest PRAF2 levels (Fig. 2a,b). As a control, the TMA was also stained with the astrocyte marker GFAP. GFAP was predominantly expressed in malignant glioma tumors and was only weakly present, or entirely absent, in all other tumor tissues including renal cancer (Fig. 2c,d).
Figure 2.
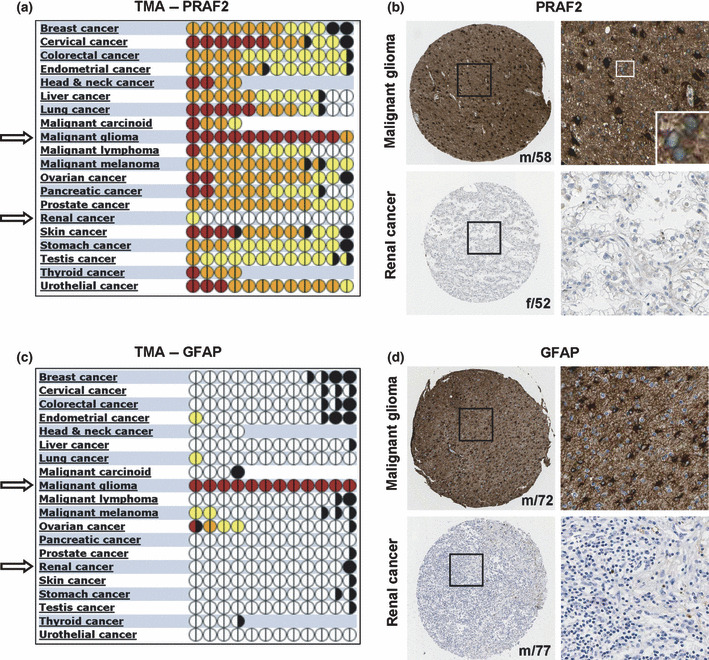
Expression of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) protein in tumor tissues. (a,c) Quantitative analysis of 220 tissue microarray (TMA) samples of various cancer types stained with PRAF2 antibody or astrocyte‐marker protein GFAP‐specific antibody. Each full circle represents one patient, and halves refer to staining performed in duplicates; color codes determine the strength/intensity of the immunohistochemical staining signal: white, no staining; yellow, light; orange, moderate; purple, strong; black, not evaluated. Tissue samples of malignant gliomas show extensive PRAF2 expression compared to other cancers. (b,d) Representative histological images of malignant glioma and renal cancer tissue sections stained with (b) anti‐PRAF2 or (d) anti‐GFAP antibody. In each case, a total of 12 tumor tissue samples were stained in duplicates (n = 12) and counterstained with hematoxylin. Patients’ gender and age are displayed in the lower right corner of each sample. f, female; m, male. Inset represents a higher magnification of PRAF2 perinuclear staining in malignant glioma.
Table 1.
Statistical evaluation of malignant glioma tumor tissues, stained for PRAF2
| Tumor cells/tissue data | Patient ratio | Percentage (%) |
|---|---|---|
| Intensity | ||
| Strong | 11/12 | 91.7 |
| Moderate | 1/12 | 8.3 |
| Weak | 0/12 | 0 |
| Negative | 0/12 | 0 |
| Quantity | ||
| >75% | 12/12 | 100 |
| 75–25% | 0/12 | 0 |
| <25% | 0/12 | 0 |
| Rare | 0/12 | 0 |
| Localization | ||
| Nuclear | 0/12 | 0 |
| Cytoplasm/Memb | 12/12 | 100 |
| Gender | ||
| Male | 9/12 | 75 |
| Female | 3/12 | 25 |
| Age | ||
| >60 years | 4/12 | 33.3 |
| >25, <60 years | 7/12 | 58.3 |
| <25 years | 1/12 | 8.4 |
| Cell type | ||
| Astrocytoma | 3/12 | 25 |
| Oligoastrocytoma | 1/12 | 8.3 |
| Oligodendroglioma | 3/12 | 25 |
| Glioblastoma | 5/12 | 41.7 |
PRAF2, prenylated Rab acceptor 1 domain family, member 2.
To compare the expression levels of PRAF2 in noncancerous brain tissue, we stained cross‐sections of the cerebral cortex. We found that PRAF2 primarily stained neuronal cells and was absent in non‐neuronal/glial cells of the cerebral cortex (Fig. 3a), consistent with our previous observations.( 20 ) In addition, tissue from the cerebral cortex was also stained for the neurexin 2α and GFAP marker proteins, confirming the staining of neuronal cells and glial cells, respectively, within the cerebral cortex (Fig. 3b,c).
Figure 3.
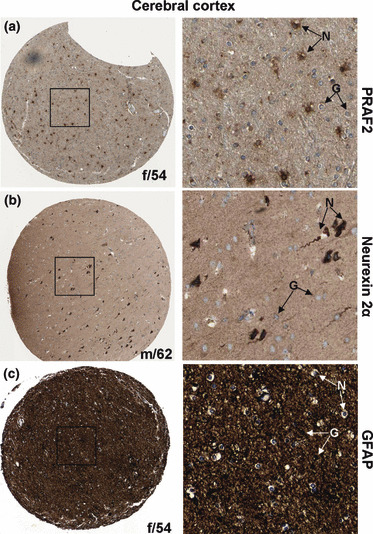
Immunohistochemical staining of normal cerebral cortex tissue samples with antibodies directed against proteins prenylated Rab acceptor 1 domain family, member 2 (PRAF2), neuronal marker neurexin 2α, and glial marker glial fibrillary acidic protein (GFAP). (a) Cerebral cortex tissue stained with PRAF2 shows weak PRAF2 presence in glial cells (G) and high expression in neurons (N). (b) Neuronal marker protein neurexin 2α localized to pyramidal neurons in normal brain sections and showed a similar staining pattern as observed for PRAF2. (c) Distribution of the glial marker GFAP in the cerebral cortex. GFAP is strongly expressed in glial cells and is absent in neurons. Each image is representative of three independent experiments.
Next, PRAF2 expression was determined in a total of 59 cancerous and noncancerous cell lines of various origins. A cluster of brain‐derived cell lines showed highest PRAF2 expression, especially two well‐characterized malignant glioma cell lines, U‐87 and U‐251 (Fig. S2), thus confirming our observation in malignant glioma tumor samples. In addition, high PRAF2 levels were detected in other cancer cell lines including HeLa, MCF‐7, RT‐4, A‐431, and F80 as well as two noncancerous cell types, HaCaT and PBMCs. In contrast, the leukemia (CML) cell line M11 expressed undetectable amounts of PRAF2.
To study the localization and distribution of PRAF2 within the cell environment, we chose the high PRAF2 protein‐expressing U‐251 malignant glioma cell line for further analysis by immunofluorescence using a confocal laser‐scanning microscope. As shown in Figure 4, the native PRAF2 protein was present in bright punctate structures throughout the cytoplasm and partially co‐localized with the ER. Prenylated Rab acceptor 1 domain family, member 2 (PRAF2) expression was further visible in and around the nucleus. The association of PRAF2 with distinct punctate structures was previously observed in neuroblastoma cells where PRAF2 was found enriched in endosomes.( 21 )
Figure 4.

Subcellular localization of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) in U‐251 malignant glioma cells. (a–d) PRAF2 (green) is expressed in punctuate structures throughout the cytoplasm and also in/around the nucleus (blue). The endoplasmic reticulum (ER) (yellow) and microtubules (red) are shown to further visualize the subcellular localization of PRAF2. (e) Superimposition by overlay of images (a b, and d). (f,g) Power images of (e), showing individual U‐251 cells. The images are representative of two independent experiments, and each experiment was performed in duplicate.
To further verify the subcellular distribution of PRAF2, we performed gravity‐based subcellular fractionation of U‐87 malignant glioma cells. U‐87 cell lysates (total) containing all cellular proteins revealed high levels of PRAF2 (Fig. 5a, lane 1), confirming the PRAF2 expression data shown in Figure S2. In addition to monomeric PRAF2 (19.3 kDa), we also detected a dimeric (SDS‐insoluble) form of PRAF2 (37 kDa), which was previously reported.( 20 , 21 ) Interestingly, both the membranous fraction as well as the nuclear fraction (including nuclear membranes) exclusively contained monomeric PRAF2 (Fig. 5a, lanes 2,4). In contrast, we detected the vast majority of dimeric PRAF2 in the cytosolic fraction (Fig. 5a, lane 3). We also detected a second, monomeric variant of PRAF2 (PRAF2‐v) which was slightly smaller in size (about 16–17 kDa, see lane 1) and which was present in the nuclear fraction only (lane 4). A peptide‐blocked PRAF2 control antibody (PRAF2‐P) was used to demonstrate the specificity of bands (data not shown). To learn more about the distribution of PRAF2 in the nucleus, isolated nuclei were lysed and nuclear membrane proteins (envelope) separated from soluble intra‐nuclear proteins. As expected, we found the majority of PRAF2 located in the membrane (envelope) fraction (Fig. 5b).
Figure 5.
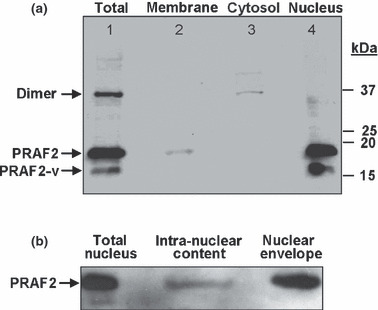
Subcellular distribution of native prenylated Rab acceptor 1 domain family, member 2 (PRAF2) in U‐87 malignant glioma cells. (a) Samples were prepared by gravity‐based fractional centrifugation of homogenized cells as described in the Materials and Methods. “Total” represents all cellular proteins prior to fractionation. While monomeric PRAF2 (19.3 kDa) and the alternatively spliced monomeric variant PRAF2‐v (16.7 kDa) are associated with membranous compartments, dimeric PRAF2 (37 kDa) is mainly detected in the membrane‐free cytosolic fraction. The specificity of all bands, including PRAF2‐v was confirmed by probing the stripped membrane with peptide‐blocked PRAF2 antibody (not shown). Molecular weights in kilodalton (kDa) are indicated. Lane 1, total cell lysate; lane 2, membranes; lane 3, cytosol; lane 4, nucleus. Data are representative of three independent experiments (n = 3). (b) Nuclear distribution of PRAF2 in U‐87 cells. Isolated nuclei were lysed, separated into intra‐nuclear and nuclear membrane fractions (envelope) and probed for PRAF2 by western blot analysis (n = 2).
To examine the potential function of PRAF2 in glioblastoma, a stable U‐87 shRNA PRAF2 knockdown cell line was generated using RNA interference (Fig. 6a). The cell viability of this phenotype was significantly reduced (Fig. 6b) and the migratory behavior and cell invasiveness was significantly impaired compared to controls (Fig. 6c,d).
Figure 6.

Functional analysis of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) knockdown in glioblastoma cells. (a) Stable transfection of U‐87 cells with shPRAF2 significantly reduced PRAF2 protein expression compared to controls (wild type and scrambled). Tubulin served as a loading control. (b) The cell viability was determined at 24, 48, and 72 h in triplicate wells in two separate experiments with similar results. (c,d) PRAF2 down‐regulation inhibits cell migration (c) and invasion (d). Five × 104 cells were seeded in the top chamber of Transwell plates. Serum‐induced migration/invasion was measured after 24 h. PRAF2 down‐regulation appeared to inhibit glioblastoma migration and invasion, compared to wild‐type and scrambled control glioblastoma cells. Three independent experiments were performed in triplicates and data are represented as mean ± SD (n = 9). WT, wild type.
Discussion
In this investigation, we examined the presence of PRAF2 in twenty types of tumor tissue at both mRNA and protein levels. In both cases, we found strong expression of PRAF2 in malignant glioma tumors and cell lines. We previously showed that PRAF2 is expressed in neuronal cells and enriched in synaptic vesicles.( 20 ) Similar to PRAF1 and PRAF3, we show here that PRAF2 is expressed at low levels in normal glia cells. Malignant glioma tissue samples, however, express excessive PRAF2, both in glioma tumor tissue and cell lines, even when compared to the PRAF2 levels of other cancer types. In contrast, PRAF1 and PRAF3 show only weak or slightly elevated expression in malignant glioma, respectively, comparable to that in other malignancies (data not shown). Therefore, the exceptionally high expression of PRAF2 in malignant glioma is a unique feature of this tumor type.
Strikingly, we found that PRAF2 appears in clear, bright punctate structures within the cell and localizes mainly to the cytoplasm of U‐251 malignant glioma cells. This finding is in conjunction with our previous observations with neuroblastoma cells.( 21 ) It is possible that the distinct structures represent endocytic vesicles, as these were shown to contain a high amount of endogenous PRAF2 in LAN‐1 neuroblastoma cells.( 21 ) The PRAF family members PRAF1 and PRAF3 were both shown to interact with a variety of proteins involved in endocytic secretory system including Rab GTPases and, therefore, PRAF2 may have similar responsibilities. Additionally, we have noticed PRAF2 inside or in close proximity to the nucleus. Subcellular fractionation data further support this observation, indicating the prevalent presence of PRAF2 in the nuclear membrane (envelope).
The few subcellular fractionation studies available for PRAF1 and PRAF3 were mostly conducted on cells over‐expressing PRAF proteins which does not necessarily mimic their native distribution under physiological conditions, and consequently, may give rise to contradictory data. For example, the monomeric form of over‐expressed PRAF1 in BHK cells localizes solely to membranes,( 34 ) whereas endogenous PRAF1 in PC12 cells is mostly cytosolic.( 35 ) Similarly, when overexpressed, the majority of PRAF3 localizes to the membranes,( 36 ) but it appears to be cytosolic in mouse brain homogenates.( 37 ) Here, we investigated the distribution of native PRAF2 in malignant glioma cells. The results revealed a distinct separation of monomeric and dimeric PRAF2 molecules among cellular compartments. The monomeric form of PRAF2 was strictly associated with membranous compartments, whereas the dimeric form was almost exclusively located in the membrane‐free cytosol. This sharp separation pattern can be well explained on the basis of the predicted PRAF2 structure. Due to the strong hydrophobic nature of their transmembrane domains, monomeric PRAF2 proteins are unlikely to be present in the aqueous environment and prefer the membranous environment. Indeed, computer prediction analyses and experimental data with PRAF1 and PRAF3 demonstrated their potential to form homodimers through their hydrophobic domains,( 38 , 39 ) and this was also observed with PRAF2.( 17 ) This is in support of our previous findings, where only monomers of PRAF2 could be detected in isolated synaptic vesicles.( 20 ) It is possible that dimer formation provokes a conformational change and camouflages the hydrophobic domains, thereby increasing the solubility of the complex by exposing only the hydrophilic N‐ and C‐termini to the cellular environment. Depending on their meric status, monomeric, homodimeric, and possibly heterodimeric PRAF proteins could function in different cellular compartments and fulfill multiple roles in diverse molecular processes.
Further of interest was the occurrence of a PRAF2 splice variant (PRAF2‐v) in malignant glioma cells. PRAF2‐v was not observed in our previous studies with neuroblastoma cells or in healthy tissues.( 19 , 21 ) In recent years, a growing number of splice variants have been determined that were expressed in a cancer‐specific manner( 40 ) with a role in tumorigenesis and metastasis.( 41 ) To elucidate the origin of the 16.7‐kDa PRAF2‐v monomer (Fig. 5a), we searched the expressed sequence tag (EST) library data set of PRAF2,( 42 ) which revealed the existence of numerous complete and incomplete splice variants, producing smaller PRAF2 molecules. Most variants lacked the characteristic C‐terminal sequence motif which contains the PRAF2 antibody recognition site, with the exception of one splice variant, PRAF2‐v. Although its 5′ sequence is incomplete in the database, the sequence‐based completion of the partial ORF resulted in a 158 amino acid residue protein with a predicted size of 16.7 kDa. This corresponds with the band size detected by western blotting (Fig. 5a), and suggests the existence of an alternatively spliced PRAF2 in malignant glioma cells. Alternative splicing can add or remove domains necessary for correct subcellular localization,( 43 ) influencing protein behavior and cellular signaling, sometimes even in a dominant‐negative manner. Our hydrophobicity profile analysis predicted that the PRAF2‐v splice variant retains three of four transmembrane domains (data not shown). Therefore, alternatively spliced PRAF2 could form a heterodimer with the full‐length protein and subsequently alter its function.
Malignant gliomas are among the most invasive of all tumors.( 44 ) Their diffuse nature contributes to poor surgical outcome and is thought to be one of the major causes of the infamously low survival rate.( 45 ) In recent years, numerous chemokine receptors were shown to contribute to cancer cell migration, including CCR3 and CCR5 as well as CXCR4 in glioblastoma.( 46 , 47 , 48 , 49 ) PRAF2 interacts with the intracellular C‐terminal tail of CCR5 and thereby might regulate the cell surface expression of CCR5.( 17 ) Here we showed that PRAF2 knockdown impairs the migration and invasiveness of U‐87 cells (Fig. 6). Together, this suggests a potential role for PRAF2 in tumor metastasis and invasion and renders it a novel target for therapeutic intervention.
In summary, we found strong PRAF2 expression in human malignant glioma tissues as well as cell lines, and comparably lower expression in normal tissues or other tumor types. PRAF2 is found in vesicle‐like, punctate structures throughout the cell suggesting its involvement in membrane trafficking. It is possible that PRAF2 contributes to the highly invasive nature of malignant gliomas either through its involvement in vesicular transport and/or by interaction with chemokine receptors. The present study suggests that PRAF2 is a candidate therapeutic target in malignant glioma.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Representative display of 12 malignant glioma tumor tissue samples immunohistochemically stainted with prenylated Rab acceptor 1 domain family, member 2 (PRAF2) antibody. Bar slices in the first rows represent the same 12 patients evaluated in Figure 2(a).Selected regions of all samples are magnified below. Cell‐type abbreviations; a, astrocytoma; oa, oligoastrocytoma; od, oligodendroglioma; g, glioblastoma. Numbers after cell type refer to tumor grade.
Fig. S2. Expression of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) protein in cultured cancer cell lines. The strength of PRAF2 expression in cultured cell lines is shown as detected by immunohistochemical staining with the PRAF2 antibody. Staining intensity is displayed relative to the strongest signal. Cell cultures with brain origin are among those with the highest PRAF2 expression, including malignant glioma cell lines U‐251 and U‐87 (circled).
Table S1. Statistical evaluation of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) mRNA expression differences between normal brain and gliomas.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Acknowledgments
The authors would like to thank Kate Jaremko and Lisette Yco for excellent technical support. This study was supported by institutional developmental funds from the Cancer Research Center of Hawaii to André S. Bachmann. The Human Protein Atlas (HPA) project is accessible at http://www.proteinatlas.org and is funded by the Knut & Alice Wallenberg foundation (Stockholm, Sweden). The Atlas is part of the HUPO Human Antibody Initiative (Montreal, QC, Canada).
References
- 1. Stupp R, Mason WP, Van den Bent MJ et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352: 987–96. [DOI] [PubMed] [Google Scholar]
- 2. Bucci C, Chiariello M, Lattero D, Maiorano M, Bruni CB. Interaction cloning and characterization of the cDNA encoding the human prenylated rab acceptor (PRA1). Biochem Biophys Res Commun 1999; 258: 657–62. [DOI] [PubMed] [Google Scholar]
- 3. Maier S, Reiterer V, Ruggiero AM et al. GTRAP3‐18 serves as a negative regulator of Rab1 in protein transport and neuronal differentiation. J Cell Mol Med 2009; 13: 114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab‐GDI complexes. Nature 2003; 425: 856–9. [DOI] [PubMed] [Google Scholar]
- 5. Compton SL, Kemppainen RJ, Behrend EN. Prenylated Rab acceptor domain family member 1 is involved in stimulated ACTH secretion and inhibition. Cell Signal 2009; 21: 1901–9. [DOI] [PubMed] [Google Scholar]
- 6. Kim JT, Cho MY, Choi SC et al. Prenylated Rab acceptor 1 (PRA1) inhibits TCF/beta‐catenin signaling by binding to beta‐catenin. Biochem Biophys Res Commun 2006; 349: 200–8. [DOI] [PubMed] [Google Scholar]
- 7. Figueroa C, Taylor J, Vojtek AB. Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J Biol Chem 2001; 276: 28219–25. [DOI] [PubMed] [Google Scholar]
- 8. Li LY, Shih HM, Liu MY, Chen JY. The cellular protein PRA1 modulates the anti‐apoptotic activity of Epstein‐Barr virus BHRF1, a homologue of Bcl‐2, through direct interaction. J Biol Chem 2001; 276: 27354–62. [DOI] [PubMed] [Google Scholar]
- 9. Compton SL, Behrend EN. PRAF1: a Golgi complex transmembrane protein that interacts with viruses. Biochem Cell Biol 2006; 84: 940–8. [DOI] [PubMed] [Google Scholar]
- 10. Liu HP, Wu CC, Chang YS. PRA1 promotes the intracellular trafficking and NF‐kappaB signaling of EBV latent membrane protein 1. EMBO J 2006; 25: 4120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mao WG, Liu ZL, Chen R, Li AP, Zhou JW. JWA is required for the antiproliferative and pro‐apoptotic effects of all‐trans retinoic acid in Hela cells. Clin Exp Pharmacol Physiol 2006; 33: 816–24. [DOI] [PubMed] [Google Scholar]
- 12. Chen H, Bai J, Ye J et al. JWA as a functional molecule to regulate cancer cells migration via MAPK cascades and F‐actin cytoskeleton. Cell Signal 2007; 19: 1315–27. [DOI] [PubMed] [Google Scholar]
- 13. Li CP, Zhu YJ, Chen R et al. Functional polymorphisms of JWA gene are associated with risk of bladder cancer. J Toxicol Environ Health A 2007; 70: 876–84. [DOI] [PubMed] [Google Scholar]
- 14. Lin CI, Orlov I, Ruggiero AM et al. Modulation of the neuronal glutamate transporter EAAC1 by the interacting protein GTRAP3‐18. Nature 2001; 410: 84–8. [DOI] [PubMed] [Google Scholar]
- 15. Ruggiero AM, Liu Y, Vidensky S et al. The endoplasmic reticulum exit of glutamate transporter is regulated by the inducible mammalian Yip6b/GTRAP3‐18 protein. J Biol Chem 2008; 283: 6175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Watabe M, Aoyama K, Nakaki T. A dominant role of GTRAP3‐18 in neuronal glutathione synthesis. J Neurosci 2008; 28: 9404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schweneker M, Bachmann AS, Moelling K. JM4 is a four‐transmembrane protein binding to the CCR5 receptor. FEBS Lett 2005; 579: 1751–8. [DOI] [PubMed] [Google Scholar]
- 18. Bachmann AS, Duennebier FF, Mocz G. Genomic organization, characterization, and molecular 3D model of GDE1, a novel mammalian glycerophosphoinositol phosphodiesterase. Gene 2006; 371: 144–53. [DOI] [PubMed] [Google Scholar]
- 19. Fo CS, Coleman CS, Wallick CJ, Vine AL, Bachmann AS. Genomic organization, expression profile, and characterization of the new protein PRA1 domain family, member 2 (PRAF2). Gene 2006; 371: 154–65. [DOI] [PubMed] [Google Scholar]
- 20. Koomoa DL, Go RC, Wester K, Bachmann AS. Expression profile of PRAF2 in the human brain and enrichment in synaptic vesicles. Neurosci Lett 2008; 436: 171–6. Epub Mar 15, 2008. [DOI] [PubMed] [Google Scholar]
- 21. Geerts D, Wallick CJ, Koomoa DL et al. Expression of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) in neuroblastoma: correlation with clinical features, cellular localization, and cerulenin‐mediated apoptosis regulation. Clin Cancer Res 2007; 13: 6312–9. [DOI] [PubMed] [Google Scholar]
- 22. Andersson AC, Stromberg S, Backvall H et al. Analysis of protein expression in cell microarrays: a tool for antibody‐based proteomics. J Histochem Cytochem 2006; 54: 1413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barrett T, Troup DB, Wilhite SE et al. NCBI GEO: archive for high‐throughput functional genomic data. Nucleic Acids Res 2009; 37: D885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Revet I, Huizenga G, Chan A et al. The MSX1 homeobox transcription factor is a downstream target of PHOX2B and activates the Delta‐Notch pathway in neuroblastoma. Exp Cell Res 2008; 314: 707–19. Epub Jan 16, 2008. [DOI] [PubMed] [Google Scholar]
- 25. Sun L, Hui AM, Su Q et al. Neuronal and glioma‐derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006; 9: 287–300. [DOI] [PubMed] [Google Scholar]
- 26. Lee J, Kotliarova S, Kotliarov Y et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum‐cultured cell lines. Cancer Cell 2006; 9: 391–403. [DOI] [PubMed] [Google Scholar]
- 27. Uhlen M, Bjorling E, Agaton C et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 2005; 4: 1920–32. Epub August 27, 2005. [DOI] [PubMed] [Google Scholar]
- 28. Kampf C, Andersson A, Wester K, Bjorling E, Uhlen M, Ponten F. Antibody‐based tissue profiling as a tool for clinical proteomics. Clin Proteomics 2005; 1: 285–99. [Google Scholar]
- 29. Nilsson P, Paavilainen L, Larsson K et al. Towards a human proteome atlas: high‐throughput generation of mono‐specific antibodies for tissue profiling. Proteomics 2005; 5: 4327–37. [DOI] [PubMed] [Google Scholar]
- 30. Stromberg S, Bjorklund MG, Asplund C et al. A high‐throughput strategy for protein profiling in cell microarrays using automated image analysis. Proteomics 2007; 7: 2142–50. [DOI] [PubMed] [Google Scholar]
- 31. Lundberg E, Gry M, Oksvold P et al. The correlation between cellular size and protein expression levels‐‐normalization for global protein profiling. J Proteomics 2008; 71: 448–60. [DOI] [PubMed] [Google Scholar]
- 32. Graham JM, Rickwood D. Subcellular Fractionation: A Practical Approach. USA: Oxford University Press, 1997. [Google Scholar]
- 33. Koomoa DL, Borsics T, Feith DJ et al. Inhibition of S‐adenosylmethionine decarboxylase by inhibitor SAM486A connects polyamine metabolism with p53‐Mdm2‐Akt/protein kinase B regulation and apoptosis in neuroblastoma. Mol Cancer Ther 2009; 8: 2067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang Z, Li G. Mouse prenylated Rab acceptor is a novel Golgi membrane protein. Biochem Biophys Res Commun 2000; 275: 509–16. [DOI] [PubMed] [Google Scholar]
- 35. Hutt DM, Da‐Silva LF, Chang LH, Prosser DC, Ngsee JK. PRA1 inhibits the extraction of membrane‐bound rab GTPase by GDI1. J Biol Chem 2000; 275: 18511–9. [DOI] [PubMed] [Google Scholar]
- 36. Abdul‐Ghani M, Gougeon PY, Prosser DC, Da‐Silva LF, Ngsee JK. PRA isoforms are targeted to distinct membrane compartments. J Biol Chem 2001; 276: 6225–33. [DOI] [PubMed] [Google Scholar]
- 37. Ikemoto MJ, Inoue K, Akiduki S et al. Identification of addicsin/GTRAP3‐18 as a chronic morphine‐augmented gene in amygdala. Neuroreport 2002; 13: 2079–84. [DOI] [PubMed] [Google Scholar]
- 38. Liang Z, Veeraprame H, Bayan N, Li G. The C‐terminus of prenylin is important in forming a dimer conformation necessary for endoplasmic‐reticulum‐to‐Golgi transport. Biochem J 2004; 380: 43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Akiduki S, Ikemoto MJ. Modulation of the neural glutamate transporter EAAC1 by the addicsin‐interacting protein ARL6IP1. J Biol Chem 2008; 283: 31323–32. [DOI] [PubMed] [Google Scholar]
- 40. Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem 2004; 37: 584–94. [DOI] [PubMed] [Google Scholar]
- 41. Yakushijin Y, Steckel J, Kharbanda S et al. A directly spliced exon 10‐containing CD44 variant promotes the metastasis and homotypic aggregation of aggressive non‐Hodgkin’s lymphoma. Blood 1998; 91: 4282–91. [PubMed] [Google Scholar]
- 42. Thierry‐Mieg D, Thierry‐Mieg J. AceView: a comprehensive cDNA‐supported gene and transcripts annotation. Genome Biol 2006; 7(Suppl 1): S12.1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang L, Duke L, Zhang PS et al. Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer. Cancer Res 2003; 63: 4724–30. [PubMed] [Google Scholar]
- 44. Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol 2003; 21: 1624–36. [DOI] [PubMed] [Google Scholar]
- 45. Maher EA, Furnari FB, Bachoo RM et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev 2001; 15: 1311–33. [DOI] [PubMed] [Google Scholar]
- 46. Muller A, Homey B, Soto H et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001; 410: 50–6. [DOI] [PubMed] [Google Scholar]
- 47. Kouno J, Nagai H, Nagahata T et al. Up‐regulation of CC chemokine, CCL3L1, and receptors, CCR3, CCR5 in human glioblastoma that promotes cell growth. J Neurooncol 2004; 70: 301–7. [DOI] [PubMed] [Google Scholar]
- 48. Meier R, Muhlethaler‐Mottet A, Flahaut M et al. The chemokine receptor CXCR4 strongly promotes neuroblastoma primary tumour and metastatic growth, but not invasion. PLoS ONE 2007; 2: e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L, Yeger H, Das B, Irwin MS, Baruchel S. Tissue microenvironment modulates CXCR4 expression and tumor metastasis in neuroblastoma. Neoplasia 2007; 9: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative display of 12 malignant glioma tumor tissue samples immunohistochemically stainted with prenylated Rab acceptor 1 domain family, member 2 (PRAF2) antibody. Bar slices in the first rows represent the same 12 patients evaluated in Figure 2(a).Selected regions of all samples are magnified below. Cell‐type abbreviations; a, astrocytoma; oa, oligoastrocytoma; od, oligodendroglioma; g, glioblastoma. Numbers after cell type refer to tumor grade.
Fig. S2. Expression of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) protein in cultured cancer cell lines. The strength of PRAF2 expression in cultured cell lines is shown as detected by immunohistochemical staining with the PRAF2 antibody. Staining intensity is displayed relative to the strongest signal. Cell cultures with brain origin are among those with the highest PRAF2 expression, including malignant glioma cell lines U‐251 and U‐87 (circled).
Table S1. Statistical evaluation of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) mRNA expression differences between normal brain and gliomas.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


