Abstract
Clinical proteomics using a large archive of formalin‐fixed paraffin‐embedded (FFPE) tissue blocks has long been a challenge. Recently, a method for extracting proteins from FFPE tissue in the form of tryptic peptides was developed. Here we report the application of a highly sensitive mass spectrometry (MS)‐based quantitative proteome method to a small amount of samples obtained by laser microdissection from FFPE tissues. Cancerous and adjacent normal epithelia were microdissected from FFPE tissue blocks of 10 squamous cell carcinomas of the tongue. Proteins were extracted in the form of tryptic peptides and analyzed by 2‐dimensional image‐converted analysis of liquid chromatography and mass spectrometry (2DICAL), a label‐free quantitative proteomics method developed in our laboratory. From a total of 25 018 peaks we selected 72 mass peaks whose expression differed significantly between cancer and normal tissues (P < 0.001, paired t‐test). The expression of transglutaminase 3 (TGM3) was significantly down‐regulated in cancer and correlated with loss of histological differentiation. Hypermethylation of TGM3 gene CpG islands was observed in 12 oral squamous cell carcinoma (OSCC) cell lines with reduced TGM3 expression. These results suggest that epigenetic silencing of TGM3 plays certain roles in the process of oral carcinogenesis. The method for quantitative proteomic analysis of FFPE tissue described here offers new opportunities to identify disease‐specific biomarkers and therapeutic targets using widely available archival samples with corresponding detailed pathological and clinical records. (Cancer Sci 2009; 100: 1605–1611)
Squamous cell carcinoma is the major histological type of oral cancer and develops in various anatomical locations within the oral cavity, including the tongue, bucca, oropharynx, gingiva, palate, lip, and floor of the mouth. Despite recent improvements in surgical techniques and chemo/radiotherapy,( 1 ) the overall 5‐year survival rate for patients with oral squamous cell carcinoma (OSCC) is still unsatisfactory.( 2 ) OSCC has a propensity for rapid local invasion and spread( 3 ) and is considered to be one of the most aggressive forms of squamous cell carcinoma of the head and neck region. Furthermore, the incidence of OSCC has been increasing among the young and middle‐aged.( 4 , 5 , 6 ) Therefore, there is an urgent need to develop new diagnostic and therapeutic modalities to improve the outcome of OSCC. Although there is considerable epidemiological evidence for a significant association of alcohol consumption, tobacco smoking, chronic mechanical stimulation, and betel quid chewing with the incidence of OSCC, the molecular mechanisms responsible for OSCC have not been fully elucidated. Overall gene expression in OSCC has been studied extensively over the past decade using microarray techniques. However, gene expression is not always correlated with the expression levels of the corresponding proteins.( 7 ) Although it is anticipated that protein expression reflects more directly the biological and pathological status of diseases, aberrations of protein expression during the course of oral carcinogenesis are largely unknown.
Although the use of fresh material is desirable for any analytical technology, human tissue samples are not always available in sufficient quantity. Formalin‐fixed paraffin‐embedded (FFPE) tissue blocks are routinely preserved and stored after pathological diagnosis, and such archived material may provide an ample alternative resource for research purposes. However, FFPE specimens have usually not been used for proteomic analyses, as formaldehyde‐induced intermolecular and intramolecular cross‐linking hinders the solubility of proteins and complicates the extraction of intact proteins from the samples.( 8 ) Recently, a method of extracting proteins from FFPE tissues in the form of tryptic peptides was developed, and the methodology is compatible with a variety of subsequent mass spectrometry (MS)‐based proteome applications.( 9 , 10 )
We previously developed a MS‐based quantitative proteome platform named 2DICAL (2‐dimensional image converted analysis of liquid chromatography and mass spectrometry)( 11 ) for quantitative comparison of large peptide datasets generated by nano‐flow liquid chromatography and mass spectrometry (LC‐MS). Owing to its simple procedure, 2DICAL is highly sensitive and reproducible: 60 000–160 000 peptides can be readily detected in a 1‐h LC run and accurately quantified without isotope labeling. In the present study we used 2DICAL for quantitative analysis of small samples of protein obtained from FFPE tissues by laser microdissection and searched for proteins that were differentially expressed between normal and cancerous epithelia of the oral cavity. Here we report the identification of transglutaminase 3 (TGM3) as an epigenetically silenced gene in OSCC cell lines.
Materials and Methods
Clinical samples and cell lines. FFPE tissues (n = 63) were collected from OSCC patients who underwent surgery at two medical institutions: the National Cancer Center Hospital (NCCH; Tokyo, Japan) between April 1997 and March 2006, and the Tokyo Medical and Dental University Hospital (TMDUH; Tokyo, Japan) between January 2001 and December 2006. All the patients were preoperatively diagnosed as having squamous cell carcinoma of the tongue. Surgically removed tongue tissues were routinely processed for pathological examination, fixed in formalin, embedded in paraffin, and stored at room temperature. Pathological examination confirmed the histology of invasive squamous cell carcinoma. None of the patients had received preoperative radiation, chemotherapy, or immunotherapy. The cases were followed up for at least 3 years after surgery. Fresh oral mucosa was donated by a volunteer who had no history of malignancy. The protocol of the study was reviewed and approved by the ethics committee boards of the NCC and TMDU.
Twelve OSCC cell lines (Ca9‐22, Ho‐1‐N‐1, Ho‐1‐u‐1, HSC‐2, HSC‐3, HSC‐4, HSC‐6, KON, KSOC‐2, KSOC‐3, SAS, SKN‐3) were obtained from the Japan Health Science Foundation (Osaka, Japan) and cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum.
A plasmid expressing TGM3 (namely pcDNA3.1/TGM3) was constructed by cloning the full‐length coding sequence of TGM3 cDNA into the pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA). pcDNA3.1/TGM3 or the control empty vector (pcDNA3.1) was transfected into cells using the Lipofectamine LTX reagent (Invitrogen).
Laser microdissection and peptide extraction. Paired tumor and adjacent normal epithelial cells were collected from the same FFPE tissues of the NCCH cases (n = 10). To recover cell populations of interest without contamination, we used laser microdissection. Ten‐micrometer‐thick FFPE sections were placed on DIRECTOR Laser Microdissection Slides (Expression Pathology, Gaithersburg, MD, USA), deparaffinized, and stained with hematoxylin–eosin (HE). Parts of the sections 3 mm2 in area (corresponding to approximately 10 000 cells) were then microdissected using a LMD6000 (Leica Microsystems, Wetzlar, Germany). Proteins were extracted in the form of tryptic peptides utilizing a Liquid Tissue MS Protein Partitioning Kit (Expression Pathology) in accordance with the manufacturer's protocol. In brief, the microdissected tissues were suspended in Liquid Tissue buffer, incubated at 95°C for 90 min, and then cooled on ice for 3 min. Trypsin (15–18 units) was added, and the samples were incubated at 37°C overnight. Dithiothreitol was added to a final concentration of 10 mM, and the samples were heated for 5 min at 95°C. The extracted peptide samples were stored at –80°C until analysis.
Liquid chromatography and mass spectrometry (LC‐MS). Twenty tissue samples (10 paired cancer and normal tissues) were blinded, randomized, and measured in triplicate with a linear gradient of 0–80% acetonitrile in 0.1% formic acid at a speed of 200 nL/min for 60 min using a nano‐flow high‐performance liquid chromatograph (HPLC) (NanoFrontier nLC; Hitachi High‐technologies, Tokyo, Japan) connected to an electrospray ionization quadrupole time‐of‐flight (ESI‐Q‐TOF) mass spectrometer (NanoFrontier LD, Hitachi High‐technologies) every second in the 400–1600 mass‐to‐charge ratio (m/z) range. MS peaks were detected, normalized, and quantified using in‐house 2DICAL software, as described previously.( 11 ) A serial identification (ID) number was applied to each detected MS peak, from ID1 to ID25018.
Protein identification by tandem mass spectrometry (MS/MS). MS/MS spectra were acquired from preparative LC. LC‐MS data were aligned with a tolerance of ±0.5 m/z and a retention time (RT) of ±0.4 min, and targeted MS/MS was performed. Peak lists were generated using the MassNavigator software package (Version 1.2; Mitsui Knowledge Industry, Tokyo, Japan) and searched against the NCBInr database (NCBInr_20070419.fast) using the Mascot software package (Version 2.2.1; Matrix Sciences, London, UK). Initial peptide tolerances in MS and MS/MS modes were ±0.05 Da and ±0.1 Da, respectively. Trypsin was designated as the enzyme, and up to one missed cleavage was allowed. The score threshold to achieve P < 0.05 is set by the Mascot algorithm, based on the size of the database used in the search.
Immunohistochemistry (IHC). FFPE sections of NCCH (n = 10) and TMDUH (n = 53) cases were used for IHC. Immunoperoxidase staining was performed using the avidin–biotin–proxidase complex method as described previously.( 12 , 13 ) Mouse monoclonal anti‐TGM3 (1:150; Abnova, Taipei, Taiwan), anti‐cytokeratin 4 (CK4) (1:200; Chemicon, Rosemont, IL, USA), anti‐cytokeratin 13 (CK13) (1:200; Abcam, Cambridge, UK), and anti‐annexin A1 (ANXA1) (1:200; BD Bioscience, Pharmingen, NJ, USA) antibodies and relevant secondary biotin‐conjugated antibodies (1:200; Vector Laboratories, Peterborough, UK) were used. Two investigators (K.H., A.N.) blinded to the clinical data reviewed the stained sections. Normal tongue epithelium in the same section served as an internal positive control. Cases in which 10% or more of the tumor cells were positively stained with anti‐TGM3 antibody were considered to be TGM3‐positive, while cases in which less than 10% of the tumor cells were TGM3‐positive were considered to be TGM3‐negative. If considerable tumor heterogeneity was present, staining was evaluated in the predominantly differentiated area of the tumor.
Immunoblot analysis. Cells were washed with phosphate‐buffered saline and lyzed in cell lysis buffer (50 mM Tris‐HCl pH 7.5, 1% Triton‐X100, 150 mM NaCl, 20 mM EDTA). The cell lysates were analyzed by immunoblotting, as described previously,( 13 ) using anti‐TGM3 (Abcam), CK14 (Thermo Scientific, Waltham, MA, USA), anti‐involucrin (Thermo Scientific), and anti‐β‐actin (Sigma, St Louis, MO, USA) antibodies.
Cytosine methylation analysis. The detection of CpG islands and design of PCR primers for amplification were performed by Methyl Primer Express Software v1.0 (Applied Biosystems, Foster City, CA, USA). Genomic DNA was extracted using DNeasy Blood and Tissue kits (Qiagen, Valencia, CA, USA). Bisulfite conversion was carried out using 2 µg of genomic DNA and the reagents provided in EpiTect Bisulfite Kit (Qiagen). The converted DNA was subjected to PCR using primer sets (5′‐GTTTAAATAAAGG TATTTGGTTTAGAG‐3′ and 5′‐CTTACCCATACTACTCATACC CAC‐3′). The PCR products were visualized by 3% agarose gel electrophoresis and subcloned into the TA vector using a TOPO TA Cloning Kit (Invitrogen). Eight colonies were sequenced using an ABI 3130 (Applied Biosystems).
Real‐time reverse‐transcription PCR. Cells were treated with 0, 2, or 5 µM 5′‐aza‐2′‐deoxycytidine (5Aza‐dC) for 5 days. Total RNA was prepared with an RNeasy Mini Kit (Qiagen). DNase‐I‐treated RNA was random‐primed and reverse‐transcribed using a High Capacity RNA‐to‐cDNA Kit (Applied Biosystems). The TaqMan universal PCR master mix and predesigned TaqMan Gene Expression probe and primer sets were purchased from Applied Biosystems. Amplification data measured as an increase in reporter fluorescence were collected using a PRISM 7000 Sequence Detection system (Applied Biosystems). The level of messenger RNA (mRNA) expression relative to the internal control (β‐actin) was calculated by the comparative threshold cycle (CT) method.( 14 )
Statistical analysis. Differences between subgroups were tested with paired t‐test. The clinicopathological variables pertaining to the corresponding patients were analyzed for statistical significance by Fisher's exact test. Statistical analyses were performed using an open‐source statistical language R (version 2.7.0) with the optional module design package.
Results
Identification of proteins differentially expressed in tongue cancer. Parts corresponding to an area of 3 mm2 (approximately 10 000 cells) were microdissected from cancerous and adjacent normal epithelia that lacked significant contamination with infiltrating inflammatory cells, stromal cells, muscular components, vascular components, and necrotic cells (Fig. 1a). Then, 20 paired protein samples were prepared from 10 tongue squamous cell carcinoma (TSCC) cases and analyzed for differential protein expression profiles using the 2DICAL proteome platform. A total of 25 018 MS peaks per sample were readily detected and quantified. Linear regression analysis demonstrated excellent linearity with a mean correlation coefficient (CC) of 0.9954 (0.9794–0.9989) for the entire 25 018 MS peaks between triplicates (Suppl Fig. S1), confirming the high reproducibility of 2DICAL. The MS peaks detected in a representative run are displayed with m/z along the x axis and RT along the y axis (Fig. 1b).
Figure 1.
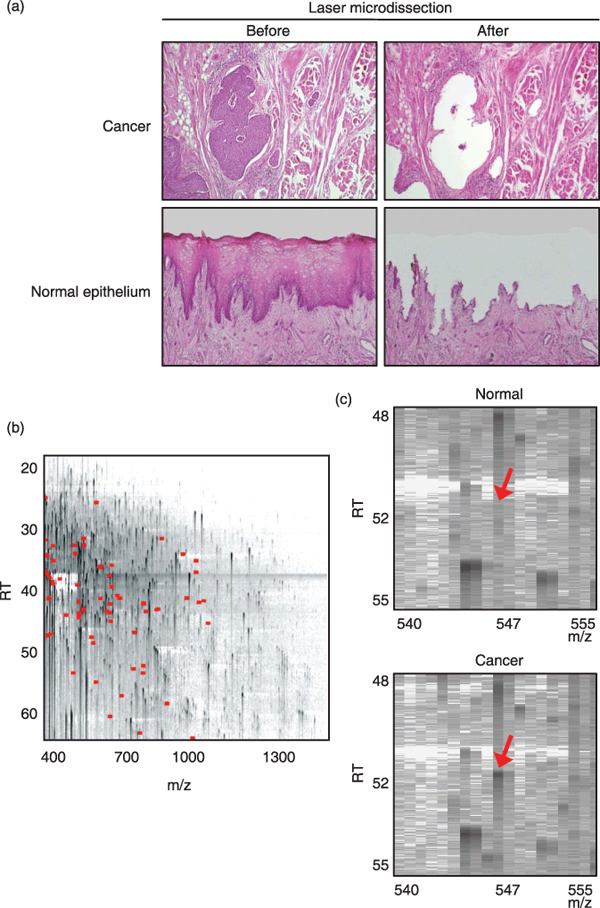
Identification of proteins differentially expressed in tongue squamous cell carcinoma (TSCC) by 2‐dimensional image‐converted analysis of liquid chromatography and mass spectrometry (2DICAL). (a) Selective removal of cancerous and normal tongue epithelia from formalin‐fixed paraffin‐embedded (FFPE) tissues. The microscopic appearances (HE staining) of cancer (top; magnification, ×100) and normal tongue (bottom; magnification, ×40) tissues before (left) and after (right) laser microdissection are shown. (b) Two‐dimensional display of all (>25 000) MS peaks of a representative sample with the m/z‐values (400–1600 m/z) along the horizontal (x) axis and retention time (RT) (20.0–63.0 min) along the vertical (y) axis. The 72 MS peaks whose average intensity of triplicates differed significantly between cancer and normal epithelia (P < 0.001 [paired t‐test]) are highlighted in red. (c) Close‐up view of a representative MS peak whose intensity differed significantly between normal (top) and cancerous (bottom) epithelia (indicated by red arrows).
Among the total of 25 018 MS peaks, we found 72 whose average intensity of triplicates differed significantly between cancer and normal epithelia as a relatively strict criterion (P < 0.001 [paired t‐test] and average peak intensity >100 [arbitrary unit] for either cancer or control samples) (Fig. 1b, indicated in red; Fig. 1c, indicated by arrows). The intensity of 10 peaks was increased in cancer and that of the remaining 62 peaks was decreased (data not shown). We further selected 12 peaks whose intensity was decreased in cancer by visually inspecting the aligned raw MS spectra (Fig. 2a, top, highlighted in green boxes) and the mean peak intensity (Fig. 2a, bottom) across the 20 samples. A database search using the NCBInr (NCBInr_20070419.fast) for the MS/MS spectra of the 12 peaks identified the amino acid sequences of seven proteins with significant confidence (P < 0.05) (Table 1 and Suppl Figs S2–5).
Figure 2.
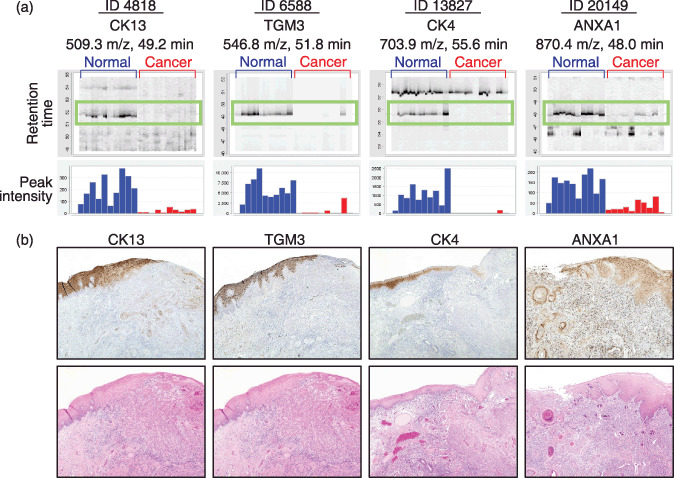
Identification and validation of differentially expressed proteins. (a) Gel‐like views of MS peaks with retention time (RT) along the vertical axes (top) and distribution of the mean peak intensity of triplicates (bottom) across 20 samples (60 liquid chromatography and mass spectrometry [LC‐MS] runs). Cancer tissues (red) exhibit reduced expression of the four indicated proteins compared with normal epithelia (blue). (b) Immunoperoxidase staining of CK13, TGM3, CK4, and ANXA1 proteins in tongue squamous cell carcinoma (TSCC) (top) (magnification, ×40). Images of corresponding HE‐stained serial sections are shown at the bottom.
Table 1.
List of peptides that differed between normal and cancerous tongue epithelia
| ID | M/Z | RT | Charge | Accession number | Protein description | Mascot score | Peptide sequence | Normal (mean ± SD) | Cancer (mean ± SD) | P‐values* |
|---|---|---|---|---|---|---|---|---|---|---|
| 4818 | 509.3 | 49.2 | 2 | gi|62897663 | Keratin 13 isoform a variant | 73.89 | VLDELTLSK | 6088 ± 2506 | 491 ± 149 | 2.56E‐04 |
| 13 827 | 703.9 | 55.6 | 2 | gi|109255249 | Keratin 4 | 59.37 | VDSLNDEINFLK | 1078 ± 633 | 23 ± 4 | 8.76E‐04 |
| 7777 | 572.9 | 44.2 | 2 | gi|62897663 | Keratin 13 isoform a variant | 54.31 | ILTATIENNR | 4338 ± 1645 | 233 ± 82 | 1.09E‐04 |
| 673 | 418.2 | 47.6 | 2 | gi|62897663 | Keratin 13 isoform a variant | 53.6 | LAVDDFR | 9241 ± 3859 | 848 ± 474 | 2.51E‐04 |
| 4572 | 504.3 | 46.2 | 2 | gi|62897663 | Keratin 13 isoform a variant | 47.73 | YENELALR | 5147 ± 2130 | 303 ± 95 | 1.77E‐04 |
| 6588 | 546.8 | 51.8 | 2 | gi|80478896 | Transglutaminase 3 (E polypeptide, protein‐ glutamine‐gamma‐ glutamyltransferase) | 42.36 | FSSQELILR | 209 ± 103 | 21 ± 15 | 6.36E‐04 |
| 20 149 | 870.4 | 48.0 | 2 | gi|442631 | Chain, annexin I | 41.86 | SEDFGVNEDLADSDAR | 141 ± 51 | 32 ± 19 | 5.41E‐04 |
| 1203 | 431.3 | 43.5 | 2 | gi|119568898 | hCG1643722, isoform CRA_b | 41.57 | VMLTELR | 9947 ± 4751 | 561 ± 163 | 4.78E‐04 |
| 5504 | 521.8 | 48.7 | 2 | gi|62897663 | Keratin 13 isoform a variant | 38.65 | VILEIDNAR | 9570 ± 2752 | 890 ± 302 | 3.04E‐05 |
| 12 366 | 673.4 | 55.1 | 2 | gi|11360071 | Hypothetical protein DKFZp434K0115.1 – human (fragment) | 38.49 | KTLEEQISEIR | 1757 ± 615 | 73 ± 9 | 3.40E‐05 |
| 434 | 412.3 | 42.9 | 2 | gi|6016414 | Keratin, type I cuticular Ha2 (hair keratin, type I Ha2) (keratin‐32) | 38.38 | LASYLTR | 5312 ± 2562 | 359 ± 91 | 5.88E‐04 |
Paired t‐test.
The decreased expression of four proteins, for which antibodies were available, was validated by IHC in 10 TSCC surgical specimens (NCCH) (Fig. 2b). Intense positive immunoreactivity for TGM3, CK13, CK4, and ANXA1 was detected in normal tongue epithelia. Cancerous lesions evidently demonstrated down‐regulation of these proteins. ANXA1 staining was observed in all the layers of normal epithelia, whereas only keratinized, but living, cancer cells showed moderate staining for ANXA1 (Fig. 2b). Among these proteins differentially expressed in TSCC, we decided to focus on gaining further insight into the characteristics of TGM3, whose roles in oral carcinogenesis have remained unclear.
Clinicopathological significance of TGM3. To assess the clinical significance of TGM3, its expression was evaluated by IHC in a larger cohort consisting of 53 TSCC cases (TMDUH) (Suppl Fig. S6). In normal tongue mucosa, TGM3 immunoreactivity was confined to the spinous and parakeratinized layers of epithelial cells (Suppl Fig. S6a). Strong nuclear staining for TGM3 was seen in the spinous layer, whereas cytoplasmic staining was detected in the parakeratinized layers. Only 12 out of 53 TSCC cases were positive for TGM3 expression (Table 2). Most cancer cells exhibited sparse immunoreactivity for TGM3 (Suppl Fig. S6c–e) and, when detected, the staining tended to be localized in the differentiated and keratinized area of TSCC (Suppl Fig. S6b). Statistical analysis of the correlation between TGM3 expression and clinicopathologic characteristics demonstrated that TGM3 expression was inversely correlated with loss of histological differentiation (P < 0.05, Fisher's exact test), but not with other clinicopathological variables including age, sex, and TNM classification (Table 2).
Table 2.
Correlation between clinical characteristics of TSCC and TGM3 protein expression
| Characteristics | Total | Number of TGM3‐positive cases (%) | Number of TGM3‐negative cases (%) | P‐values* |
|---|---|---|---|---|
| Total | 53 | 12 (22.6) | 41 (77.4) | |
| Age (years) | ||||
| ≤60 | 27 | 9 (33.3) | 18 (66.7) | 0.0994 |
| >60 | 26 | 3 (11.5) | 23 (88.5) | |
| Gender | ||||
| Male | 36 | 8 (22.2) | 28 (77.8) | 1.0000 |
| Female | 17 | 4 (23.5) | 13 (75.5) | |
| Stage | ||||
| I and II | 42 | 10 (23.8) | 32 (76.2) | 0.4705 |
| III and IV | 11 | 2 (18.2) | 9 (81.8) | |
| TNM classification | ||||
| T category | ||||
| T1 and T2 | 48 | 12 (25.0) | 36 (75.0) | 0.5765 |
| T3 and T4 | 5 | 0 (0) | 5 (100) | |
| N category | ||||
| N0 | 43 | 12 (27.9) | 31 (72.1) | 0.0932 |
| N1‐3 | 10 | 0 (0) | 10 (100) | |
| Histological grade | ||||
| Well | 22 | 9 (40.9) | 13 (59.1) | 0.0171 |
| Moderate or poor | 31 | 3 (9.7) | 28 (90.3) | |
P‐values were calculated by Fisher's exact test and considered statistically significant at <0.05 (two‐sided). TGM3, transglutaminase 3; TSCC, tongue squamous cell carcinoma.
Lack of TGM3 expression in OSCC cells and its restoration by 5Aza‐dC. We next examined TGM3 protein expression in 12 OSCC cell lines by immunoblot analysis. None of these cell lines expressed TGM3 protein. Some of the OSCC cell lines expressed differentiation‐associated markers of squamous epithelia, such as CK14 for the proliferative basal layer and involucrin for the upper differentiating layer (Fig. 3a). Real‐time PCR analysis to quantify the TGM3 mRNA expression of these cells gave results consistent with those obtained by immunoblot analysis (data not shown). In an attempt to investigate the molecular mechanism of the gene silencing of TGM3 in OSCC cells, we grew Ca9‐22 cells lacking TGM3 expression in the presence of a methyltransferase inhibitor, 5Aza‐dC, for 5 days. Real‐time PCR analysis revealed a dose‐dependent increase of TGM3 mRNA expression, and 5‐µM 5Aza‐dC increased the expression of TGM3 up to ~60 fold over untreated cells (Fig. 3b). Similar results were obtained in other OSCC cell lines treated with 5Aza‐dC (data not shown). Since histone deacetylation, which is catalyzed by the histone deacetylase family, is known to mediate transcriptional repression, we treated Ca9‐22 cells with a histone deacetylase inhibitor trichostatin A, with or without 5Aza‐dC. However, this had no effect on the restoration of TGM3 expression, indicating that histone deacetylation is not responsible for the transcriptional silencing of TGM3 in OSCC cell lines (data not shown).
Figure 3.
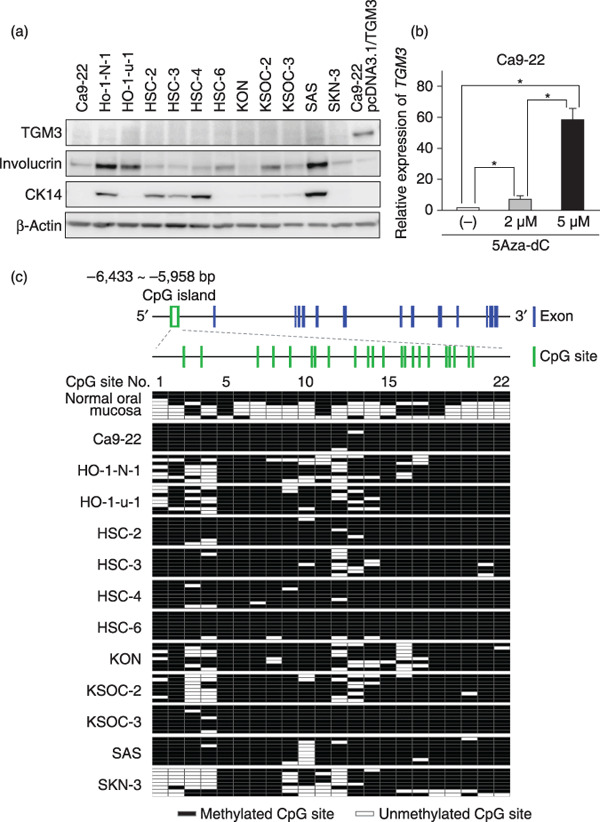
Epigenetic silencing of transglutaminase 3 (TGM3). (a) TGM3 protein expression in 12 oral squamous cell carcinoma (OSCC) cell lines and Ca9‐22 cells transfected with pcDNA3.1/TGM3 (positive control for TGM3). Total cell lysates (10 µg) were analyzed for expression of the indicated proteins by immunoblotting. Involucrin and CK14 were included as epithelial differentiation markers, and β‐actin as a loading control. (b) Restoration of TGM3 expression by 5‐aza‐2′‐deoxycytidine (5Aza‐dC). Ca9‐22 cells were untreated (–) or treated with 2 or 5 µM 5Aza‐dC for 5 days. Relative expression levels of the TMG3 gene were determined by real‐time RT‐PCR. Columns and bars represent mean ± SD. (c) Cytosine methylation of CpG sites in the upstream region of the TGM3 gene. Exon/intron structure and the CpG island in the upstream region of the TGM3 gene are presented schematically (Top). The 22 CpG sites in the CpG island are numbered 1–22 (middle). Genomic DNA was extracted from normal oral mucosa and OSCC cell lines and treated with sodium bisulfite. Eight independent clones per sample were sequenced (bottom). Clear green box, CpG island; vertical blue bars, exons; vertical green bars, CpG sites; clear squares, unmethylated CpG sites; solid squares, methylated CpG sites.
Methylation status of the CpG island located in the TGM3 gene upstream region. Reversal of gene silencing of TGM3 by treatment with 5Aza‐dC, but not with TSA, indicated that transcription of the TGM3 gene was regulated through hypermethylation of the TGM3 gene promoter and/or enhancer in OSCC cells. A database search of the approximately 20‐kb sequence of the human TGM3 gene including the 5′‐flanking region identified a CpG‐rich region –6433 to –5958 upstream of the transcription start site (Fig. 3c, top, indicated in a green box). This region was a 475‐bp‐long fragment containing 22 CpG sites (Fig. 3c, middle, indicated by green vertical bars) and had a GC content of 52.5%, thus meeting the proposed criteria for CpG islands.
To examine the DNA methylation status of these CpG sites in OSCC cells, the entire CpG island was sequenced after bisulfite conversion (Fig. 3c, bottom). All of the 22 CpG sites were recurrently methylated in seven cell lines (Ca9‐22, HSC‐2, HSC‐3, HSC‐4, HSC‐6, KSOC‐3, and SAS). The other five cell lines (Ho‐1‐N‐1, Ho‐1‐u‐1, KON, KSOC‐2, and SKN‐3) showed patchy methylation of CpG sites 1–4 and 9–16. The entire CpG island in normal oral mucosa was largely unmethylated in five out of eight clones examined. Taken together, these findings suggest that silencing of TGM3 in OSCC cells is likely attributable to the methylation of CpG sites 5–8 and 19–22.
Discussion
In the present study using our 2DICAL proteome platform, we identified CK4, CK13, TGM3, and ANXA1 as proteins whose expression was significantly decreased in microdissected FFPE tissue samples of TSCC. No protein was up‐regulated in TSCC on the basis of the current strict criterion (P < 0.001, paired t‐test), indicating the presence of genetic diversity even within such a small number (n = 10) of TSCC samples. IHC analysis of an independent cohort revealed that TGM3 expression was markedly decreased in 41 of 53 (77.4%) TSCC cases and that the reduced expression of TGM3 was clearly correlated with loss of histological differentiation. Consistent with these findings, recent studies utilizing a broad range of genomics and proteomics technologies have begun to reveal the importance of down‐regulation of TGM3 in SCC including laryngeal carcinoma,( 15 ) head and neck SCC,( 16 , 17 , 18 , 19 , 20 , 21 ) OSCC developing from leukoplakia,( 21 ) and esophageal SCC.( 22 , 23 ) Reduction or loss of TGM3 expression in SCC has been reportedly correlated with dedifferentiation,( 15 , 22 ) an increase in the invasive phenotype,( 18 ) a high incidence of lymph‐node metastasis,( 20 , 24 ) and poor prognosis.( 23 ) Together, these findings strongly implicate a crucial role of TGM3 in oral carcinogenesis.
Transglutaminases (TGMs) are a family of Ca2+‐dependent enzymes that catalyze protein cross‐linking through formation of intermolecular Nå‐(γ‐glutamyl) lysine isopeptide linkages.( 25 , 26 , 27 ) TGM3 is a zymogen, requiring activation by proteolytic cleavage, and is expressed predominantly in terminally differentiating stratified squamous epithelium.( 26 , 27 , 28 , 29 ) TGM3 is essential for cross‐linking cornified cell envelope (CCE) protein constituents and formation of the CCE.( 30 ) To date, nine members of the TGM gene family have been identified in the human genome, including TGM1–7, factor XIII, and erythrocyte band 4.2, a catalytically inactive homolog.( 26 ) Despite marked similarities in the genome structures, their distribution, localization, and mechanism for activation of TGM genes are highly variable.( 26 )
We found that transcription of the TGM3 gene is regulated by DNA methylation. A database search detected only one CpG island located approximately 6 kb upstream of the transcription start site of the TGM3 gene. Although the promoter of TGM3 was reported to be located –126 to –91 bp upstream of the transcripton start site,( 28 ) no CpG island was found in the proximal region. Among 22 CpG sites within the CpG island, methylation of CpG sites 5–8 (region 1) and 19–22 (region 2) was correlated with TGM3 silencing in the OSCC cell lines, reflecting the possibility that one or both of these two regions function as a distal enhancer for TGM3 transcription. It is intriguing to note that regions 1 and 2 encompass putative elements for transcription factors including GATA‐1 and GATA‐2, as well as GATA‐1, GATA‐2, AP‐1, and Ets, respectively (http://www.cbrc.jp/htbin/nph‐tfsearch). Transcription factors AP1 and Ets are known to positively regulate epidermal differentiation.( 31 ) It is plausible that tumor‐specific hypermethylation in region 2 prevents these transcription factors from binding to their recognition sequences, thereby inactivating transcription of TGM3.
Accumulating evidence indicates that TGMs are multifunctional proteins.( 26 ) In fact, some of their functions are even independent of their ability to mediate cross‐linking reactions, as exemplified by TGM2, which can function as a molecular switch for transducing cell signaling.( 26 ) TGM2 has been shown to be identical to an atypical high‐molecular‐weight G‐protein known as Ghα, which mediates the activation of phospholipase C through its ability to bind GTP and hydrolyze GTP to GDP.( 26 , 32 ) Analyses of the crystal structure of TGM3 have indicated that TGM3 possesses a GTP‐binding property similar to that of TGM2.( 33 ) TGM3 may also work as a molecular switch governing the cell signaling. Tgm3 knockout mice show an embryonic‐lethal phenotype( 34 ) indicating the non‐redundancy of TGM3. This observation also implies that TGM3 has one or more unique functions that cannot be compensated by other TGMs expressed in epithelia, such as TGM1 and TGM5.
Recent advances in proteomic technologies are being increasingly applied to studies of clinical samples in the search for diagnostic biomarkers and therapeutic targets. Here, we have demonstrated for the first time that the powerful combination of the 2DICAL quantitative proteomic high‐throughput platform and FFPE archival samples, for which matching clinicopathological information is available, is beginning to show promise.
Disclosure
None of the authors of this study have a conflict of interest.
Supporting information
Fig. S1. Reproducibility of 2‐dimensional image‐converted analysis of liquid chromatography and mass spectrometry (2DICAL). The horizontal (x) axis represents the distribution of the peak intensities of a liquid chromatography and mass spectrometry (LC‐MS) run (run 1), and the vertical (y) axis represents that of another run (run 2) for the same representative tongue squamous cell carcinoma (TSCC) sample. The average correlation coefficient (CC) of all corresponding 25 018 MS peaks between the duplicates was 0.9992. 94.5% (23 642/25 018) of the peaks are plotted within a two‐fold difference (blue lines), and 96.4% (24 117/25 018) within a three‐fold difference (red lines).
Fig. S2. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 4818, which matched the sequences of CK13.
Fig. S3. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 6588, which matched the sequences of transglutaminase 3 (TGM3).
Fig. S4. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 13827, which matched the sequences of CK4.
Fig. S5. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 20149, which matched the sequences of ANXA1.
Fig. S6. Expression of transglutaminase 3 (TGM3) in tongue squamous cell carcinoma (TSCC). The expression of TGM3 protein was evaluated by immunohistochemistry in 53 TSCC cases. Representative images of immunohistochemical staining for TGM3 (a–e) and corresponding HE staining (a′–e′) are aligned side by side (magnification, ×100). TGM3 immunoreactivity was localized predominantly in the cytoplasm and nuclei of non‐neoplastic tongue epithelial cells (a). TGM3 is detected in keratinized cancer pearls of some well‐differentiated TSCCs (b), but TGM3 immunoreactivity is hardly detectable in moderately (d) or poorly (e) differentiated TSCC.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgments
We thank Dr Norihiko Okada (Department of Diagnostic Oral Pathology, Tokyo Medical and Dental University) for advice regarding the design of this study. We also thank Ms Ayako Igarashi, Ms Yuka Nakamura, Ms Satoko Kouda, and Ms Kiyoko Nagumo for their technical assistance. This study was supported by the ‘Program for Promotion of Fundamental Studies in Health Sciences’ conducted by the National Institute of Biomedical Innovation of Japan, and by the ‘Third‐Term Comprehensive Control Research for Cancer’ and the ‘Research on Biological Markers for New Drug Development’ conducted by the Ministry of Health, Labor and Welfare of Japan.
References
- 1. Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med 2001; 345: 1890–900. [DOI] [PubMed] [Google Scholar]
- 2. Spitz MR. Epidemiology and risk factors for head and neck cancer. Semin Oncol 1994; 21: 281–8. [PubMed] [Google Scholar]
- 3. Franceschi D, Gupta R, Spiro RH, Shah JP. Improved survival in the treatment of squamous carcinoma of the oral tongue. Am J Surg 1993; 166: 360–5. [DOI] [PubMed] [Google Scholar]
- 4. Mackenzie J, Ah‐See K, Thakker N et al . Increasing incidence of oral cancer amongst young persons: what is the aetiology? Oral Oncol 2000; 36: 387–9. [DOI] [PubMed] [Google Scholar]
- 5. Annertz K, Anderson H, Biorklund A et al . Incidence and survival of squamous cell carcinoma of the tongue in Scandinavia, with special reference to young adults. Int J Cancer 2002; 101: 95–9. [DOI] [PubMed] [Google Scholar]
- 6. Schantz SP, Yu GP. Head and neck cancer incidence trends in young Americans, 1973–97, with a special analysis for tongue cancer. Arch Otolaryngol Head Neck Surg 2002; 128: 268–74. [DOI] [PubMed] [Google Scholar]
- 7. Yamaguchi U, Nakayama R, Honda K et al . Distinct gene expression‐defined classes of gastrointestinal stromal tumor. J Clin Oncol 2008; 26: 4100–8. [DOI] [PubMed] [Google Scholar]
- 8. Ahram M, Flaig MJ, Gillespie JW et al . Evaluation of ethanol‐fixed, paraffin‐embedded tissues for proteomic applications. Proteomics 2003; 3: 413–21. [DOI] [PubMed] [Google Scholar]
- 9. Patel V, Hood BL, Molinolo AA et al . Proteomic analysis of laser‐captured paraffin‐embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res 2008; 14: 1002–14. [DOI] [PubMed] [Google Scholar]
- 10. Hwang SI, Thumar J, Lundgren DH et al . Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin‐fixed paraffin‐embedded archival tissues. Oncogene 2007; 26: 65–76. [DOI] [PubMed] [Google Scholar]
- 11. Ono M, Shitashige M, Honda K et al . Label‐free quantitative proteomics using large peptide data sets generated by nanoflow liquid chromatography and mass spectrometry. Mol Cell Proteomics 2006; 5: 1338–47. [DOI] [PubMed] [Google Scholar]
- 12. Honda K, Yamada T, Hayashida Y et al . Actinin‐4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 2005; 128: 51–62. [DOI] [PubMed] [Google Scholar]
- 13. Shitashige M, Naishiro Y, Idogawa M et al . Involvement of splicing factor‐1 in β‐catenin/T‐cell factor‐4‐mediated gene transactivation and pre‐mRNA splicing. Gastroenterology 2007; 132: 1039–54. [DOI] [PubMed] [Google Scholar]
- 14. Huang L, Shitashige M, Satow R et al . Functional interaction of DNA topoisomerase IIα with the β‐catenin and T‐cell factor‐4 complex. Gastroenterology 2007; 133: 1569–78. [DOI] [PubMed] [Google Scholar]
- 15. He G, Zhao Z, Fu W, Sun X, Xu Z, Sun K. [Study on the loss of heterozygosity and expression of transglutaminase 3 gene in laryngeal carcinoma]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2002; 19: 120–3. (In Chinese.) [PubMed] [Google Scholar]
- 16. Gonzalez HE, Gujrati M, Frederick M et al . Identification of 9 genes differentially expressed in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 2003; 129: 754–9. [DOI] [PubMed] [Google Scholar]
- 17. Choi P, Jordan CD, Mendez E et al . Examination of oral cancer biomarkers by tissue microarray analysis. Arch Otolaryngol Head Neck Surg 2008; 134: 539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kondoh N, Ishikawa T, Ohkura S et al . Gene expression signatures that classify the mode of invasion of primary oral squamous cell carcinomas. Mol Carcinog 2008; 47: 744–56. [DOI] [PubMed] [Google Scholar]
- 19. Ye H, Yu T, Temam S et al . Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics 2008; 9: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mendez E, Fan W, Choi P et al . Tumor‐specific genetic expression profile of metastatic oral squamous cell carcinoma. Head Neck 2007; 29: 803–14. [DOI] [PubMed] [Google Scholar]
- 21. Ohkura S, Kondoh N, Hada A et al . Differential expression of the keratin‐4‐13‐14‐17 and transglutaminase 3 genes during the development of oral squamous cell carcinoma from leukoplakia. Oral Oncol 2005; 41: 607–13. [DOI] [PubMed] [Google Scholar]
- 22. Liu W, Yu ZC, Cao WF, Ding F, Liu ZH. Functional studies of a novel oncogene TGM3 in human esophageal squamous cell carcinoma. World J Gastroenterol 2006; 12: 3929–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uemura N, Nakanishi Y, Kato H et al . Transglutaminase 3 as a prognostic biomarker in esophageal cancer revealed by proteomics. Int J Cancer 2009; 124: 2106–15. [DOI] [PubMed] [Google Scholar]
- 24. Uchikado Y, Inoue H, Haraguchi N et al . Gene expression profiling of lymph node metastasis by oligomicroarray analysis using laser microdissection in esophageal squamous cell carcinoma. Int J Oncol 2006; 29: 1337–47. [PubMed] [Google Scholar]
- 25. Griffin M, Casadio R, Bergamini CM. Transglutaminases: nature's biological glues. Biochem J 2002; 368: 377–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol 2003; 4: 140–56. [DOI] [PubMed] [Google Scholar]
- 27. Esposito C, Caputo I. Mammalian transglutaminases. Identification of substrates as a key to physiological function and physiopathological relevance. FEBS J 2005; 272: 615–31. [DOI] [PubMed] [Google Scholar]
- 28. Lee JH, Jang SI, Yang JM, Markova NG, Steinert PM. The proximal promoter of the human transglutaminase 3 gene. Stratified squamous epithelial‐specific expression in cultured cells is mediated by binding of Sp1 and ets transcription factors to a proximal promoter element. J Biol Chem 1996; 271: 4561–8. [PubMed] [Google Scholar]
- 29. Hitomi K, Horio Y, Ikura K, Yamanishi K, Maki M. Analysis of epidermal‐type transglutaminase (TGase 3) expression in mouse tissues and cell lines. Int J Biochem Cell Biol 2001; 33: 491–8. [DOI] [PubMed] [Google Scholar]
- 30. Kim SY, Chung SI, Steinert PM. Highly active soluble processed forms of the transglutaminase 1 enzyme in epidermal keratinocytes. J Biol Chem 1995; 270: 18026–35. [DOI] [PubMed] [Google Scholar]
- 31. Mack JA, Anand S, Maytin EV. Proliferation and cornification during development of the mammalian epidermis. Birth Defects Res C Embryo Today 2005; 75: 314–29. [DOI] [PubMed] [Google Scholar]
- 32. Nakaoka H, Perez DM, Baek KJ et al . Gh: a GTP‐binding protein with transglutaminase activity and receptor signaling function. Science 1994; 264: 1593–6. [DOI] [PubMed] [Google Scholar]
- 33. Hitomi K, Ikura K, Maki M. GTP, an inhibitor of transglutaminases, is hydrolyzed by tissue‐type transglutaminase (TGase 2) but not by epidermal‐type transglutaminase (TGase 3). Biosci Biotechnol Biochem 2000; 64: 657–9. [DOI] [PubMed] [Google Scholar]
- 34. Kim SY, Jeitner TM, Steinert PM. Transglutaminases in disease. Neurochem Int 2002; 40: 85–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Reproducibility of 2‐dimensional image‐converted analysis of liquid chromatography and mass spectrometry (2DICAL). The horizontal (x) axis represents the distribution of the peak intensities of a liquid chromatography and mass spectrometry (LC‐MS) run (run 1), and the vertical (y) axis represents that of another run (run 2) for the same representative tongue squamous cell carcinoma (TSCC) sample. The average correlation coefficient (CC) of all corresponding 25 018 MS peaks between the duplicates was 0.9992. 94.5% (23 642/25 018) of the peaks are plotted within a two‐fold difference (blue lines), and 96.4% (24 117/25 018) within a three‐fold difference (red lines).
Fig. S2. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 4818, which matched the sequences of CK13.
Fig. S3. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 6588, which matched the sequences of transglutaminase 3 (TGM3).
Fig. S4. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 13827, which matched the sequences of CK4.
Fig. S5. Labeled tandem mass spectrometry (MS/MS) spectrum and peak list of ID 20149, which matched the sequences of ANXA1.
Fig. S6. Expression of transglutaminase 3 (TGM3) in tongue squamous cell carcinoma (TSCC). The expression of TGM3 protein was evaluated by immunohistochemistry in 53 TSCC cases. Representative images of immunohistochemical staining for TGM3 (a–e) and corresponding HE staining (a′–e′) are aligned side by side (magnification, ×100). TGM3 immunoreactivity was localized predominantly in the cytoplasm and nuclei of non‐neoplastic tongue epithelial cells (a). TGM3 is detected in keratinized cancer pearls of some well‐differentiated TSCCs (b), but TGM3 immunoreactivity is hardly detectable in moderately (d) or poorly (e) differentiated TSCC.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


