Abstract
Microglia are the main reservoir for human immunodeficiency virus type 1 (HIV-1) in the central nervous system (CNS), and multinucleated giant cells, the result of fusion of HIV-1-infected microglia and brain macrophages, are the neuropathologic hallmark of HIV dementia. One potential explanation for the formation of syncytia is viral adaptation for these CD4+ CNS cells. HIV-1BORI-15, a virus adapted to growth in microglia by sequential passage in vitro, mediates high levels of fusion and replicates more efficiently in microglia and monocyte-derived-macrophages than its unpassaged parent (J. M. Strizki, A. V. Albright, H. Sheng, M. O'Connor, L. Perrin, and F. Gonzalez-Scarano, J. Virol. 70:7654–7662, 1996). Since the interaction between the viral envelope glycoprotein and CD4 and the chemokine receptor mediates fusion and plays a key role in tropism, we have analyzed the HIV-1BORI-15 env as a fusogen and in recombinant and pseudotyped viruses. Its syncytium-forming phenotype is not the result of a switch in coreceptor use but rather of the HIV-1BORI-15 envelope-mediated fusion of CD4+CCR5+ cells with greater efficiency than that of its parental strain, either by itself or in the context of a recombinant virus. Genetic analysis indicated that the syncytium-forming phenotype was due to four discrete amino acid differences in V1/V2, with a single-amino-acid change between the parent and the adapted virus (E153G) responsible for the majority of the effect. Additionally, HIV-1BORI-15 env-pseudotyped viruses were less sensitive to decreases in the levels of CD4 on transfected 293T cells, leading to the hypothesis that the differences in V1/V2 alter the interaction between this envelope and CD4 or CCR5, or both. In sum, the characterization of the envelope of HIV-1BORI-15, a highly fusogenic glycoprotein with genetic determinants in V1/V2, may lead to a better understanding of the relationship between HIV replication and syncytium formation in the CNS and of the importance of this region of gp120 in the interaction with CD4 and CCR5.
Multinucleated giant cells (MGC) are the most specific markers of human immunodeficiency virus (HIV) encephalitis, the pathological correlate of HIV dementia (HIVD), a progressive central nervous system (CNS) syndrome induced by HIV-1 infection (27, 43, 51, 68). MGC are thought to be the result of fusion of microglia and brain macrophages mediated by the HIV-1 glycoproteins, and in fact, the CNS is one of the few sites where syncytia, a common in vitro cytopathological effect of HIV infection, can be observed in vivo. Concomitantly, MGC express HIV antigens and RNA, as well as surface markers for microglia and macrophages (69). Microglia isolated from either adult or fetal brain can also be infected in vitro with certain HIV-1 strains (26, 30, 34, 42, 58), primarily those previously designated as macrophage tropic, and at least in adult microglia, this infection can be maintained for at least several weeks in culture. Depending on the specific HIV-1 isolate, microglia infected in vitro are also susceptible to giant cell formation (60, 67).
HIV-1 tropism and syncytium formation are closely reflected in the use of seven-transmembrane-domain G-protein coupled molecules, principally CXCR4 and CCR5, as coreceptors that in conjunction with CD4 mediate virus entry into primary cells. Microglia express both CXCR4 and CCR5, as well as other potential coreceptors, including CCR3 (1, 25). However, CCR5 appears to be the most important coreceptor for adult microglial cells (1, 58), although there may be some instances where CCR3 use predominates (30). Therefore, the formation of syncytia in microglial cells likely involves the interaction of HIV with those coreceptor molecules that are now considered to be the most important coreceptors in most cell types (76).
Additionally, sequence analysis of several HIV-1 genes has indicated that HIV strains within the CNS evolve somewhat independently of strains in other body compartments, such as the spleen (23, 37, 70), suggesting a degree of sequestration of HIV replication. Whether such sequestration has any specific effect on the development of HIVD or in syncytium formation is considerably more controversial; some investigators have suggested that specific sequence motifs are associated with CNS infection, whereas others have not found such relationships (18, 35, 49). However, it is reasonable to hypothesize that localized replication results in the adaptation of HIV-1 isolates to CNS cell types, and specifically microglia, which bear the brunt of the virus burden in the brain. To determine whether the process of adaptation to microglia could be mimicked in vitro, we sequentially passaged a primary, blood-derived isolate, HIV-1BORI (13), in microglial cultures and derived an isolate, HIV-1BORI-15, with greater capacity to replicate in these cells. Somewhat independently, this virus also resulted in marked enhancement of syncytium formation in microglia, less so in monocyte-derived macrophages (MDM), and not at all in peripheral blood mononuclear cells (PBMCs) (60).
Since the interactions resulting in syncytium formation in primary cells are potentially important in the development of HIVD, we have characterized in detail the differences between HIV-1BORI and HIV-1BORI-15. Using recombinant viruses, we determined that the HIV-1BORI-15 env sequences were sufficient to mediate syncytium formation, and those amino acids critical to fusion were identified. The results point to the importance of the V1/V2 regions in the interaction between these CCR5-using viruses and primary microglia. Additionally, we determined that fusion of microglia by recombinant viruses incorporating the relevant regions of HIV-1BORI-15 was not dependent on viral replication, and it was induced even in the presence of antiretroviral drugs, suggesting fusion from without (FFWO).
MATERIALS AND METHODS
Cells.
Microglia were prepared from fresh adult human brain tissue obtained from donors undergoing temporal lobectomy for medication-resistant epilepsy (67, 75). The microglia were cultured in Dulbecco's modified Eagle medium with 5% fetal calf serum, 5% Giant Cell Tumor Supernatant (Fisher), gentamicin (50 μg/ml), and sodium pyruvate (1 mM). U373-MAGI-CCR5E cells (66) and MAGI-CCR5 cells (9) (both provided by the AIDS Research and Reference Reagent Program) express CD4 and the chemokine receptor CCR5. These cells also contain an HIV-1–long terminal repeat–beta-galactosidase sequence resulting in beta-galactosidase activity following HIV infection. These cells were cultured in selective medium. 293T and U87 cells were cultured in Dulbecco's modified Eagle medium with 10% heat-inactivated fetal bovine serum (58), as were the quail QT6 cells.
HIV-1 isolates and env clones.
HIV-1BORI, which was obtained from an individual with primary HIV-1 infection, was a gift from G. Shaw (University of Alabama). HIV-1BORI-15 resulted from 15 sequential passages of the parental HIV-1BORI isolate in microglia (60). The env genes were cloned by PCR amplification and TA cloning (Invitrogen) from infected peripheral blood lymphocytes (for HIV-1BORI) or from infected microglia (HIV-1BORI-15) (58). Additional env clones were subsequently amplified from HIV-1BORI for further sequence comparison. env genes were sequenced with six primers that spanned the entire env sequence (Nucleic Acid Core Facility, Children's Hospital of Philadelphia).
Cell-to-cell fusion assay.
The cell-to-cell fusion assay (46) was performed as adapted by Rucker et al. (54). Briefly, 293T cells (effector cells) were infected with recombinant vaccinia virus expressing T7 polymerase (vTF1.1; a gift of B. Moss, National Institutes of Health) and transfected with envelope-expressing constructs, 89.6 env (15) (obtained from R. Collman, University of Pennsylvania), HIV-1BORI env (clone 11A), or HIV-1BORI-15 env (clone 4C) (58), by the calcium phosphate method. These cells were incubated overnight at 30°C in medium containing rifampin (100 μg/ml) to inhibit vaccinia virus replication. Quail QT6 cells (target cells) were transfected with plasmids expressing CD4 and/or the chemokine receptors CCR3 and CCR5 and then incubated overnight. The 293T effector cells were lifted with versene, resuspended in cold medium, and washed with cold phosphate-buffered saline. The cells were then resuspended in medium containing rifampin (0.1 mg/ml) and cytosine β-d-arabinofuranoside (10 μM) to inhibit vaccinia virus replication, overlaid on the transfected QT6 target cells, and incubated for 8 h at 37°C. The cells were then lysed in luciferase reporter lysis buffer (Promega), and luciferase activity was measured with a Wallac 1450 Microbeta Plus luminometer. Cell-to-cell fusion reactions were performed in triplicate, and the averages and standard deviations are shown.
Cloning env sequences into a common backbone virus.
The full-length provirus pIIIB (a derivative of HXB3) was originally used by Hwang et al. (33). Fragments of envelope clones from HIV-1BORI (clone 11A) and HIV-1BORI-15 (clone 4C) were first cloned into a shuttle vector containing the SalI-XhoI fragment of pIIIB by using KpnI and AvaI, and then a SalI-BamHI fragment from the shuttle vector was cloned into the full-length provirus pIIIB. The resulting chimeric viruses VH-Bori and VH-B15 contain the HIV-1BORI and HIV-1BORI-15 env sequences, respectively, with the exception of the first 39 and last 131 amino acids of env, which originate from pIIIB. The additional chimeric viruses VH-R4 and VH-R7 were constructed by using a BglII site which is located in the C2 region of env. The viruses were produced by calcium phosphate transfection of 293T cells, and the virus-containing supernatants were centrifuged (10 min at 1,750 × g on a Beckman centrifuge) for clarification and stored at −80°C. Envelope glycoprotein expression was assayed by Western blotting of transfected 293T cell lysates and virus-containing supernatants with a rabbit polyclonal anti-gp120 antibody (a gift from R. Doms, University of Pennsylvania) (32).
Fine mapping of amino acids involved in syncytium-forming phenotype.
The recombinant virus constructs were altered with PCR to include the full-length gp120 from HIV-1BORI or HIV-1BORI-15. First a fragment of pIIIB was amplified with the primers vpr-up and env-rev, and then a fragment of either HIV-1BORI or HIV-1BORI-15 env was amplified with the primers env1 and 3′-V3out. The fragments were purified and joined by PCR with the outer primers and cloned into VH-BORI or VH-B15 with SalI and BglII to give the recombinants VH-rBORI and VH-rB15. Site-directed mutants were generated by PCR with mutagenic oligonucleotides. Briefly, a fragment of env was amplified with env1 primer and a reverse mutagenic primer, and a second fragment of env was amplified with a forward mutagenic primer and the 3′-V3out primer. The fragments were joined with outer primers and cloned into VH-rBORI by using KpnI and BglII to yield individual mutations in V1/V2 loops in the background of VH-rBORI. VH-rV1/V2 was generated by cloning a KpnI-StuI fragment from HIV-1BORI-15 env into VH-rBORI to yield a virus that differs from VH-rBORI by only four amino acids in V1/V2. The primer sequences are as follows (with standard numbering positions relative to HXB2CG): vpr-up, ATGGAACAAGCCCCAGAAGACCAAGGGCCACA (5559 to 5590); env-rev, TCATTGCCACTGTCTTCTGCTCTTTCT (6228 to 6202); env1, AGAAAGAGCAGAAGACAGTGGCAATGA (6202 to 6228); 3′-V3out, AATTTCTGGGTCCCCTCCTG (7337 to 7318); 153-forw, GGGAGAAATGAGAGGAGGAATAAAAAAATGCTC (6657 to 6697); 153-rev, GAGCATTTTTTTATTCCTCCTCTCATTTCTCCC (6697 to 6657); 162-forw, GCTCTTTCAATGTCGCCACAAGAATAAG (6694 to 6721); 162-rev, CTTATTCTTGTGGCGACATTGAAAGAGC (6721 to 6694); 190-forw, GGTAATGGTAATACTAGATATAGGTTG (6777 to 6803); 190-rev, CAACCTATATCTAGTATTACCATTACC (6803 to 6777).
Infections.
Seven- to 10-day-old microglia were infected with 5 to 50 ng of p24gag of HIV-1 per well of a 96-well plate. The supernatant p24gag antigen concentrations were determined at regular intervals (58). For infection of U373-MAGI-CCR5E cells and MAGI-CCR5 cells, the cells were infected with VH-BORI or VH-B15 in the presence of 20 μg of DEAE dextran/ml for 2 h at 37°C (9, 29, 66).
Syncytium formation.
Microglia or MDM were cultured in either four- or eight-chamber Permanox tissue culture slides (Nunc) and infected as described above. Syncytium formation was assessed by staining the cells with a nuclear stain (1:50 dilution of Hoechst 33342 [bis-benzimide] fluorochrome; Molecular Probes Inc., Eugene, Oreg.) and with a ligand that labels microglia or MDM via their low-density lipoprotein receptors (1:100 dilution of DiI-Ac-LDL; Biomedical Technologies, Inc., Stoughton, Mass.). The U373-MAGI-CCR5E and MAGI-CCR5 cell lines were infected as indicated above and stained 48 h after infection either with Diff-Quik cell staining reagents (Baxter) or, for beta-galactosidase activity, with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). For the quantification of syncytia, digital photographs of random microscopic fields were obtained under a 10× objective and the nucleus/cell ratio was determined by direct counting. Approximately 400 to 800 nuclei were counted for each datum point.
Infection of cells transfected with different amounts of CD4.
env-pseudotyped luciferase reporter viruses were prepared by transfection of 293T cells as previously described (1, 16, 58). Basically, the cells were cotransfected with pNL-4-3-LucR+E− (a gift of N. Landau, Aaron Diamond AIDS Research Center) and envelope-expressing shuttle vectors used for cloning envelopes into pIIIB, and supernatants were collected 48 h later. 293T cells were transfected in six-well plates with various amounts of plasmid encoding CD4, CCR5-expressing plasmid, and control pcDNA3.1(−) (Invitrogen, San Diego, Calif.) in a total of 5 μg of DNA per well and infected 24 h later with pseudotyped viruses. The cells were lysed at 48 h postinfection, and luciferase activity was measured by mixing the lysates with Luciferase Assay Substrate (Promega) in a Wallac 1450 Microbeta luminometer detector (1).
Transfected 293T cells were stained for cell surface CD4 and CCR5 expression as described previously (1). The cells were stained with 5 μg of the anti-CD4 antibody no. 21 (a gift from J. Hoxie, University of Pennsylvania)/ml, the anti-CCR5 antibody 2D7 (71), or a control mouse immunoglobulin G, and with a secondary anti-mouse immunoglobulin G conjugated to fluorescein isothiocyanate. The cells were fixed with 2% paraformaldehyde and analyzed by flow cytometry (1).
RESULTS
Cell-to-cell fusion mediated by BORI envelopes is CD4 and CCR5 dependent.
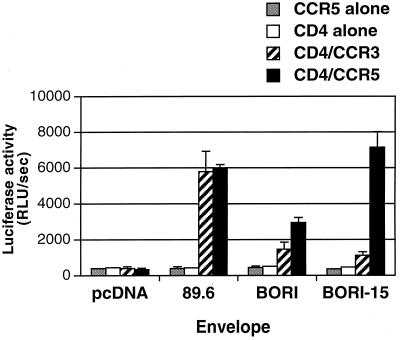
In comparison with the parental HIV-1BORI isolate, the virus derived by serial passage (HIV-1BORI-15) caused extensive syncytia in microglia (60). Previous experiments also showed that the envelope genes from HIV-1BORI and HIV-1BORI-15 utilize CCR5 as a coreceptor in conjunction with CD4 (58). Since several recent studies have demonstrated that coreceptors can be used as primary receptors (i.e., without CD4) by specific HIV-2 and HIV-1 isolates as well as by many simian immunodeficiency virus isolates (20–22, 31, 52), we performed additional cell-to-cell fusion assays with the HIV-1BORI and HIV-1BORI-15 env clones, paying particular attention to the use of coreceptors without CD4. The results (Fig. 1) indicated that the HIV-1BORI-15 envelope could not mediate fusion with target cells transfected with CCR5 only. However, we noted that in the presence of CCR5 and CD4, the HIV-1BORI-15 env demonstrated two- to threefold-higher levels of fusion than the HIV-1BORI env, although we could not rule out potential differences in glycoprotein expression at the cell surface.
FIG. 1.
Cell-to-cell fusion induced by HIV-1BORI and HIV-1BORI-15 glycoproteins. 293T effector cells expressing either env genes or a control plasmid (pcDNA) and the T7 polymerase were mixed with QT6 target cells that express CD4 and/or chemokine receptors (CCR3 or CCR5) together with the luciferase gene under the control of the T7 promoter. Cell-to-cell fusion was quantified by luciferase activity. Relative light units per second (RLU/sec) resulting from each experimental condition in one representative experiment are shown. Both HIV-1BORI and HIV-1BORI-15 envelopes mediated fusion with cells bearing CD4 with either CCR5 or CCR3, as did control HIV-189.6 envelope. However, these glycoproteins could not mediate fusion with cells expressing CCR5 without CD4. The error bars indicate standard deviations.
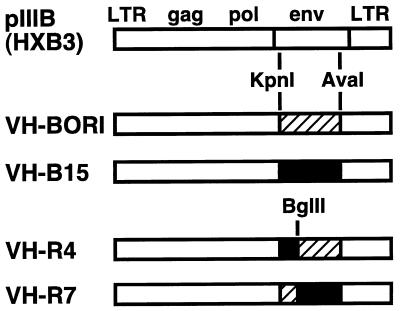
Generation of recombinant viruses with envelope sequences from HIV-1BORI and HIV-1BORI-15.
To determine whether the HIV-1BORI-15 env by itself could mediate the syncytium-forming phenotype, we cloned large fragments of env (KpnI-AvaI) from HIV-1BORI-15 or HIV-1BORI into a common full-length proviral backbone pIIIB (an HXB3-based clone) (33) to generate the recombinant viruses VH-BORI and VH-B15, respectively (Fig. 2) (see Materials and Methods). We also constructed additional recombinant viruses that contained chimeric env sequences (Fig. 2), either the upstream portion of env from HIV-1BORI-15 with the downstream portion of env from HIV-1BORI (VH-R4) or the upstream portion of the HIV-1BORI env with the downstream portion of the HIV-1BORI-15 env (VH-R7). There were no differences in the genes overlapping env in these constructs. To include as much of the gp160 sequence as possible in our recombinant viruses, we also replaced the pIIIB env sequences at the beginning of the env coding sequence of VH-BORI and VH-B15 with the corresponding env sequences from HIV-1BORI or HIV-1BORI-15 (constructs VH-rBORI and VH-rB15). Minor differences in vpu in these constructs are described below.
FIG. 2.
Recombinant viruses containing envelope sequences from HIV-1BORI and HIV-1BORI-15. HIV-1BORI or HIV-1BORI-15 envelope sequences (hatched and solid boxes, respectively) encompassing most of gp120 (amino acid 43 in gp120 to amino acid 213 in gp41 [HXB numbering system]) were cloned into a common provirus pIIIB (33) to generate VH-BORI and VH-B15, respectively. We also constructed VH-R4 and VH-R7, which contain chimeric HIV-1BORI or HIV-1BORI-15 envelope sequences, by using a BglII site (position 273) in C2 of gp120. LTR, long terminal repeat.
The fidelity of each recombinant provirus was confirmed by sequence analysis of env and its junctions, and viruses were produced by the transfection of 293T cells with plasmids followed by collection of virus-containing supernatants, as described in Materials and Methods. Western blotting revealed no differences in env expression and processing as judged by the generation of gp120 from gp160 in transfected 293T cell lysates and by the detection of gp120 in concentrated virus that had been normalized by p24gag content (data not shown).
Replication of VH-BORI and VH-B15 in peripheral blood lymphocytes.
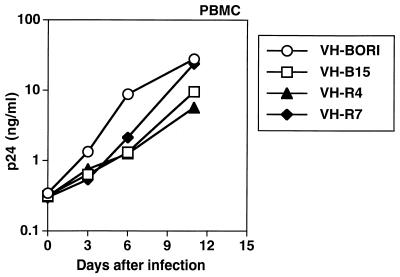
To confirm that the recombinant viruses were functional, we infected PBMCs with VH-BORI, VH-B15, VH-R4, and VH-R7 and monitored viral replication over time by determining p24gag antigen concentration (Fig. 3) at regular intervals. All of the recombinant viruses replicated efficiently in PBMCs, and there were no gross differences in the kinetics of viral replication.
FIG. 3.
Replication of viruses in PBMCs. Interleukin-2-stimulated PBMCs were infected with VH-BORI, VH-B15, VH-R4, and VH-R7, and virus replication was monitored over time by p24gag antigen concentration in the supernatant.
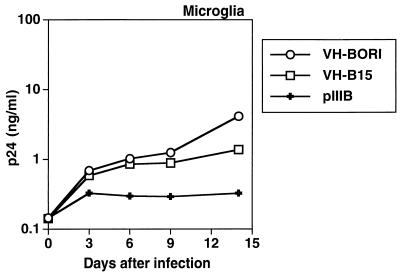
Syncytium formation in microglia due to VH-BORI and VH-B15.
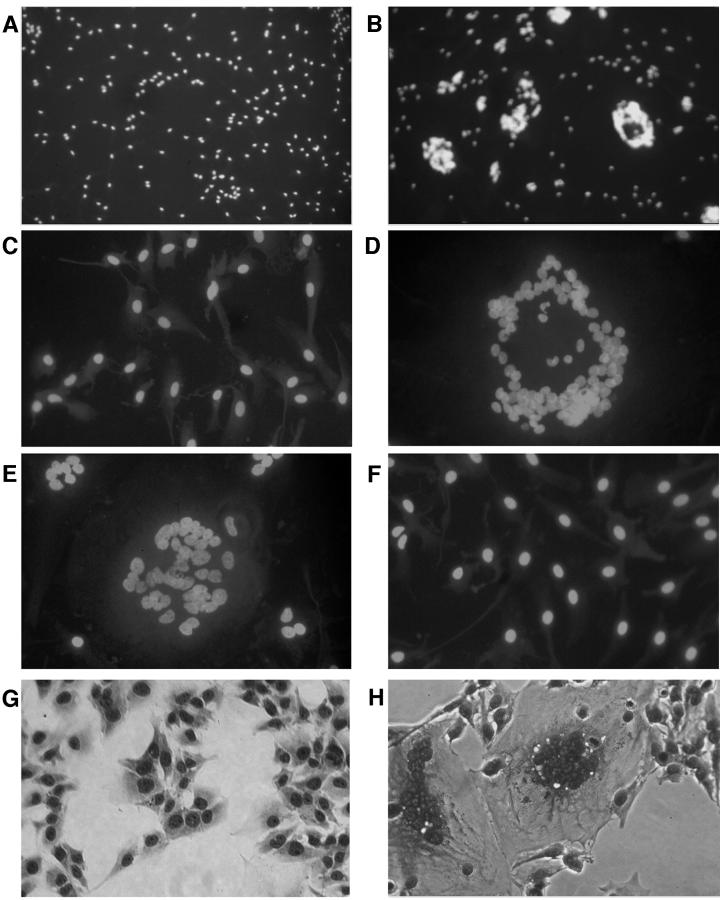
The recombinant viruses containing envelope sequences from HIV-1BORI or HIV-1BORI-15 were then assayed for syncytium formation in microglia. Equivalent p24gag antigen concentrations were used to infect the microglia, and the cultures were examined at several points for syncytium formation (Fig. 4). VH-BORI- and VH-B15-infected microglia demonstrated strikingly different syncytium formation phenotypes even under low-power magnification (Fig. 4A and B, respectively). VH-B15 induced classic syncytia, with focal aggregation of cells, often with circular arrangements of the corresponding nuclei. Under higher-power magnification (Fig. 4D), the large cells were easily identified as giant syncytia. In contrast to the results with VH-B15, VH-BORI did not induce syncytia (Fig. 4A and C).
FIG. 4.
Syncytium formation by recombinant viruses. Microglia in chamber slide plates were infected with 50 ng of p24gag of each recombinant virus and the cultures were observed at regular intervals for the formation of syncytia after being stained with a nuclear stain. (A to F) Representative photographs obtained 24 h after infection are shown. VH-B15 (B and D) and VH-R4 (E) demonstrated the ability to form extensive syncytia, while VH-BORI (A and C) and VH-R7 (F) did not. VH-B15 infection also induced extensive syncytia in U373-MAGI-CCR5E cells (H), which express CD4 and CCR5, whereas VH-BORI did not (G). A control virus (not shown) that contains the V3 loop of HIV-1Bal in the same proviral background replicates well in microglia but did not cause syncytium formation under the same conditions.
Surprisingly, we observed syncytia with VH-B15 within 24 h after infection. Subsequent experiments (not shown) indicated that the formation of syncytia by VH-B15 was not inhibited by pretreatment with zidovudine (50 μM), a concentration that was able to inhibit other viruses in microglia (A. V. Albright, S. Erickson-Viitanen, M. O'Connor, I. Frank, M. M. Rayner, and F. González-Scarano, unpublished results). This result, together with the time course of syncytium formation in microglia, was strong evidence that in this system fusion does not require viral replication.
To begin to map which regions of env are important in the syncytium-forming phenotype demonstrated by VH-B15, we tested recombinant viruses containing chimeric env sequences on microglia (Fig. 2). Infection with VH-R4 (with the upstream region of env from HIV-1BORI-15, which includes the V1/V2/C2 region) formed extensive syncytia, whereas VH-R7, containing the analogous upstream region from HIV-1BORI, did not fuse the cells.
To assess the role of coreceptors in the syncytium formation induced by VH-B15, we preincubated microglia with antibodies to chemokine receptors and infected them with VH-B15. The anti-CCR5 antibody 2D7 inhibited syncytium formation due to VH-B15, whereas antibodies against CCR3 (7B11) or CXCR4 (12G5) had no effect (data not shown).
We also tested whether the syncytium-forming phenotype could be demonstrated with other CD4+CCR5+ cells. As shown in Fig. 4, syncytium formation was evident in VH-B15-infected U373-MAGI-CCR5E cells but not in VH-BORI-infected cells. A similar syncytium-forming phenotype, although less dramatic, was observed in the HeLa-based MAGI-CCR5 cells and in MDM (data not shown). We were also able to quantify the infectivity of the recombinant viruses, since U373-MAGI-CCR5E cells express beta-galactosidase following infection. As shown in Table 1, both VH-B15 and VH-R4 demonstrated ∼5- to 20-fold-higher infectivity than VH-BORI or VH-R7. Thus, the envelope sequences of HIV-1BORI-15 were important in mediating high levels of both cell-to-cell fusion and virus-to-cell fusion.
TABLE 1.
Infectivities of recombinant HIV-1BORI and HIV-1BORI-15 viruses
See Fig. 2 for descriptions of recombinants.
Average of infections performed in triplicate with 10-fold dilutions of virus placed on U373-MAGI-CCR5E cells.
Replication of VH-BORI and VH-B15 in microglia.
We also determined whether VH-BORI and VH-B15 replicated in microglia. A representative experiment is shown in Fig. 5. Both VH-B15 and VH-BORI replicated slightly better than the X4 backbone virus pIIIB (HXB-3 env), which, as expected, did not replicate well in microglia. Interestingly, VH-B15 and VH-BORI replicated with similar kinetics and to similar peak levels, indicating that in the context of pIIIB the env sequences from HIV-1BORI-15 were not sufficient to mediate the high-replication phenotype observed with this isolate. Notably, the level of replication of VH-B15 was lower than that observed with the original microglia-passaged virus (HIV-1BORI-15) (60). Since it was formally possible that VH-B15-induced syncytium formation could inhibit virus replication, we infected microglia with 10- and 100-fold-lower inocula of VH-BORI and VH-B15, but we still did not observe differences in replication between VH-BORI and VH-B15 (data not shown). These data suggest that other regions of HIV-1BORI-15 are involved in its high replication in microglia.
FIG. 5.
Replication of recombinant viruses in microglia. Cultured microglia were infected with equivalent amounts of virus, as indicated in Materials and Methods. The culture medium was replaced at regular intervals and then assayed for p24gag antigen concentration. VH-BORI and VH-B15 replicated with similar kinetics and to similar levels.
Amino acid differences between HIV-1BORI and HIV-1BORI-15.
Comparison of the env sequences of the two isolates showed differences in eight amino acids (Table 2); seven were in gp120, and there was a single change in gp41. Furthermore, four of the differing amino acids were located in the V1/V2 loops of gp120, and two of these (T162A and S190R) encoded the loss of potential N-linked glycosylation sites. Most of the other changes were nonconservative. Using the chimeric data described above, it was clear that at most the first five amino acid differences could be important in mediating the VH-B15 syncytium-forming phenotype, since only those changes were incorporated into VH-R4, which mediated extensive syncytia (Fig. 2 and 4). We analyzed the V1/V2/C2 region of two other functional HIV-1BORI-15 env clones and one other functional HIV-1BORI env clone and confirmed that the amino acid differences were relatively conserved within different env clones from the same virus. The loss of the potential glycosylation site in V2 (S190R) was observed in all three HIV-1BORI-15 env clones; however, one HIV-1BORI env clone contained an R at position 190, even though infection with it did not result in extensive syncytium formation (data not shown). Changes in V3 (60) previously noted by PCR sequencing of the wild-type virus were variable when individual clones were examined (data not shown). There were no differences between the V3 domains of the env clones used in these experiments.
TABLE 2.
Amino acid differences between HIV-1BORI and HIV-1BORI-15 envelopesa
| Glycoprotein | Amino acid/region | Amino acid change |
|---|---|---|
| gp120 | 153/V1 | Glutamic acid (−) → glycine |
| gp120 | 162/V2 | Threonine → alanineb |
| gp120 | 166/V2 | Arginine (+) → glycine |
| gp120 | 190/V2 | Serine → arginine (+)b |
| gp120 | 229/C2 | Lysine (+) → asparagine |
| gp120 | 281/C2 | Threonine → isoleucine |
| gp120 | 336/C3 | Alanine → threonine |
| gp41 | 532 | Valine → methionine |
Charged amino acids are indicated. The amino acid numbers are based on the HXB2 sequence.
Change results in the loss of a potential glycosylation site.
Mapping of the amino acid differences involved in the syncytium-forming phenotype.
To exclude the possibility that the pIIIB env sequences at the beginning of the env coding sequence of VH-B15 were contributing to the observed fusogenicity, we replaced these pIIIB env sequences with the corresponding sequence from either HIV-1BORI-15 or HIV-1BORI. These additional recombinant viruses (VH-rBORI and VH-rB15) therefore contained full-length gp120 sequences from HIV-1BORI or HIV-1BORI-15 and demonstrated phenotypes similar to those of VH-B15 and VH-BORI in the syncytium-forming assay (Fig. 6). These constructions introduced a stop codon in the vpu reading frame, resulting in a slightly shorter deduced protein for VH-BORI. This did not have an effect on replication in PBMCs (data not shown). Furthermore, preincubation of microglia with the anti-CCR5 antibody (2D7) or an anti-CD4 antibody (no. 21; a gift of J. Hoxie) inhibited the syncytium formation due to VH-rB15 (Fig. 6). As with VH-B15, however, VH-rB15-induced syncytium formation was not inhibited by the reverse transcriptase inhibitors zidovudine and efavirenz (0.01 μM), indicating that viral replication was not required (data not shown).
FIG. 6.
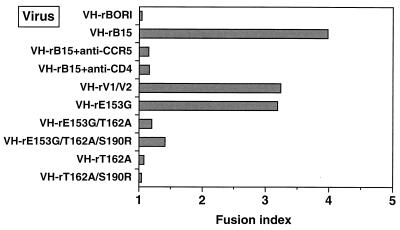
Quantification of syncytium formation induced by recombinant viruses with discrete differences. Microglia were infected with VH-rBORI, VH-rB15, VH-rBORI recombinant viruses with specific mutations in the V1/V2 loops (VH-rE153G, VH-rE153G/T162A, VH-rE153G/T162A/S190R, VH-rT162A, and VH-rT162A/S190R), or a virus containing the V1/V2 loops from HIV-1BORI-15 in the VH-rBORI background (VH-rV1/V2). Twenty-four hours later, the cells were stained and syncytia were quantified as described in Materials and Methods. The fusion index is the total number of nuclei divided by the total number of cells counted, and approximately 400 to 800 nuclei were counted for each datum point. Amino acid differences in the V1/V2 loops were primarily responsible for the syncytium-forming phenotype, but a mutation with a single-amino-acid change (E153G) also induced syncytium formation.
We then mutated individual residues on the VH-BORI background to the sequence of VH-B15 and also made a recombinant provirus (VH-rV1/V2) which placed the HIV-1BORI-15 V1/V2 in the background of the HIV-1BORI env (four amino acids were different). The viruses were generated, checked for infectivity in U373-MAGI-CCR5E, and assayed for the ability to form syncytia in microglia and in CCR5+ cell lines. As shown in Fig. 6, the virus containing only the four amino acid differences in V1/V2 of HIV-1BORI-15 on the background of HIV-1BORI env (VH-rV1/V2) was able to mediate extensive syncytium formation. Furthermore, the mutation of residue 153 (recombinant E153G) resulted in a virus that was almost as fusogenic as VH-rB15, whereas mutation T162A alone or in combination with S190R (the loss of two potential N-linked glycosylation sites) did not reproduce the wild-type phenotype. Interestingly, when the E153G mutation was combined with mutations that changed the glycosylation sites, there was less giant cell formation than when the E153G mutation was present by itself. Therefore, the effects of the amino acid alterations are context dependent, although those in V1/V2 are primarily responsible for the syncytium-forming phenotype.
Infection of cells with different amounts of CD4.
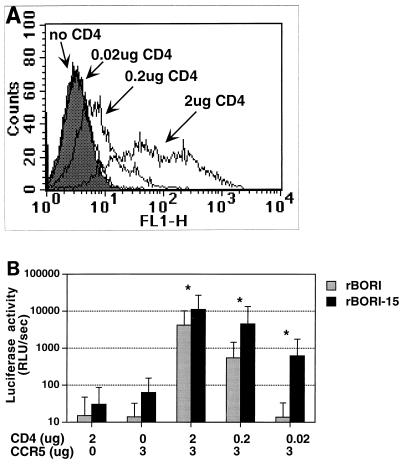
One possibility that might explain the mechanism of syncytium formation by HIV-1BORI-15 is adaptation to the relatively low levels of CD4 present in microglia. To test this, we performed an experiment where env-pseudotyped viruses were used to infect 293T cells transfected with a CCR5-expressing plasmid and with 10-fold-decreasing amounts of plasmid expressing CD4. When we stained transfected cells for CD4 and analyzed cell surface expression by flow cytometry (Fig. 7A), we noted a close relationship between the amount of CD4-expressing plasmid transfected and the levels of CD4 expression. As shown in Fig. 7B, pseudotyped viruses with envelopes from HIV-1BORI-15 and HIV-1BORI infected 293T cells transfected with large amounts of CD4 and CCR5; however, only pseudotypes with the env from HIV-1BORI-15 maintained the ability to infect cells transfected with smaller amounts of CD4. Similar results were observed in similarly transfected U87 cells (data not shown). These results suggest a number of potential mechanisms for the adaptation of HIV-1BORI-15 and will provide a framework for future studies.
FIG. 7.
Infection of cells transfected with different amounts of CD4-expressing plasmid. (A) 293T cells were transfected with the indicated amounts of CD4 and 3 μg of CCR5 plasmids, and then the cells were stained with an anti-CD4 antibody followed by a fluorescein-isothiocyanate-conjugated secondary antibody. The cells transfected with 2 μg of CD4-expressing plasmid demonstrated higher levels of fluorescence than those transfected with smaller amounts of plasmid. The cells transfected with 0.02 μg of plasmid had no detectable fluorescence in comparison with the control cells (no CD4). CCR5 expression was assessed with antibody 2D7, and it remained constant in all of the cells (not shown). FL1-H, fluorescence intensity. (B) 293T cells were transfected with the indicated amounts of CD4- and CCR5-encoding plasmids and with a control plasmid to equalize the total amount of DNA and were infected the next day with equivalent amounts (1.25 to 6 ng/ml of p24gag antigen) of luciferase reporter viruses pseudotyped with env from HIV-1BORI (rBORI) or from HIV-1BORI-15 (rBORI-15). Two days later the cells were lysed, and luciferase activity was measured in relative light units per second (RLU/sec). The results are expressed as the means of six independent experiments (each performed in triplicate) and standard deviations. ∗, Statistically significant differences between rBORI and rBORI-15-mediated infections were observed at each level of CD4 expression (Wilcoxon's rank sum test; P < 0.05).
DISCUSSION
MGC, the result of fusion between infected and uninfected microglia and macrophages, are the signature neuropathological finding in HIVD and are the main reservoir for HIV within the CNS (27, 63). Given the evidence of genetic sequestration within the CNS provided by postmortem studies (23, 37, 70), it is likely that a subpopulation of viruses replicates in microglia and adapts to them over an indeterminate period. While studies have documented the phenotype and to some extent the evolution of viruses in the cerebrospinal fluid (6, 38), because of the obvious sampling problem, few studies have looked at the functional evolution of virus in the CNS parenchyma, necessitating the design of in vitro studies.
We characterized a virus, HIV-1BORI-15, that forms extensive syncytia in microglia cultures and compared it to its parental primary isolate, HIV-1BORI. The syncytium-forming phenotype could not be explained by a simple coreceptor switch or by CD4-independent entry. Rather, the HIV-1BORI-15 envelope had a quantitatively greater ability to mediate fusion of several CD4+CCR5+ cell types. In the context of pIIIB backbone, envelope sequences from HIV-1BORI-15 (VH-B15) mediated a high level of syncytium formation in microglia in comparison with an equivalent construct with the HIV-1BORI env (VH-BORI), whereas both viruses replicated equivalently in PBMCs. Furthermore, VH-B15 infected U373-MAGI-CCR5E cells with greater efficiency than VH-BORI, and pseudotypes incorporating the HIV-1BORI-15 env constructs were less sensitive to decreases in the amount of CD4 on the cell surface when CCR5 levels were held constant.
Surprisingly, VH-B15-mediated fusion was independent of viral replication, since it occurred within 24 h of exposure to the cells and was not decreased by reverse transcriptase inhibitors, indicating FFWO. However, the timing of fusion with the uncloned HIV-1BORI-15, which occurred at 2 to 3 weeks after infection, did not suggest FFWO. FFWO, or fusion mediated by viral particles in the absence of infection, is a well-known phenomenon in other enveloped viruses with high fusion potential (2, 28, 56, 59) and has previously been described in HIV under certain circumstances (14). We hypothesize that the high fusion activity mediated by VH-B15 is due to the intrinsically greater fusogenicity of the HIV-1BORI-15 env. It is unlikely that the gp41 sequences from HXB-3 present in the VH-based recombinants played a major role in this fusion, since VH-BORI did not demonstrate any significant syncytium formation. We also found equivalent envelope expression among the recombinant viruses (data not shown). In any case, the syncytium-forming phenotype was clearly mapped to four amino acids in the V1/V2 region, with the bulk of the VH-B15 fusogenicity accounted for by a single-amino-acid difference, E153G. Surprisingly, the loss of two potential glycosylation sites in the same region, while perhaps associated with changes in neutralization (data not shown), had no major effect on syncytium formation when introduced independently. Moreover, the full phenotype depended on all four amino acid differences between VH-BORI and VH-B15, indicating that it is probably related to the overall conformation of the region.
Our results support the idea that envelope sequences play a major role in HIV-1 tropism for microglia, although it is probably not the exclusive determinant. Since HIV-1 isolates obtained from the CNS are predominantly R5 (i.e., use CCR5 as coreceptor), once a mechanism for its enhanced syncytium formation has been defined, the experiments with HIV-1BORI-15 will provide information regarding the interactions between HIV and these specialized cells. Specifically, there may be requirements for interaction with CD4 and CCR5 at the ratio present in microglial cells.
We believe that there are different interactions between the HIV-1BORI and the HIV-1BORI-15 envelopes and CD4 or CCR5 and that the V1/V2 loops are critical to this difference. Although the effects of V3 on tropism, particularly in determining coreceptor use, have received the widest attention (10, 12, 32, 57, 65), some investigators have found that sequences in V1/V2 can influence virus spread in MDM (64). There is also an extensive literature indicating that these loops are involved in neutralization (7, 11) and other cellular tropisms (5, 8, 36, 44, 45, 47, 62, 72). In the current model of HIV entry, the V1/V2 loops are thought to shield the coreceptor binding site (73). CD4 binding probably induces a conformational change involving the V1/V2 loops that exposes a conserved coreceptor binding site (53). This conformational change is detectable through increased binding of some antibodies, like 17b (61, 73, 74).
How could the HIV-1BORI-15 envelope influence this interaction? We can propose several potential scenarios. Firstly, the V1/V2 loops of HIV-1BORI-15 gp120 may favor the conformation triggered by CD4 binding, increasing the exposure of the chemokine receptor-binding site. On a membrane with few CD4 molecules, like those of microglia (17), this more stable conformation may be necessary to promote a gp120-CCR5 interaction. Along the same lines, the interaction between HIV-1BORI-15 gp120 and CD4 could be stronger, resulting in a similar outcome. Indeed, HIV-1 strains may have different affinities for the CD4-coreceptor complex and demonstrate variations in infectivity of cells with different amounts of receptor and coreceptor (39, 48). Alternatively the HIV-1BORI-15 gp120 could interact with CCR5 more efficiently, with the more basic V1/V2 region of the HIV-1BORI-15 gp120 (in comparison with HIV-1BORI) facilitating gp120 interaction with the acidic residues in the CCR5 amino terminus (19, 24). Interaction with different extracellular domains of CCR5 or different conformational states of CCR5 may also play a role (3, 4, 41, 55). Future studies with purified preparations of HIV-1BORI-15 gp120 should determine which of these possibilities is most relevant to its phenotype. If the results are generalized to other HIV strains, these findings could provide mechanistic information regarding the development of syncytia in this area of HIV pathogenesis. Indeed a recent report focused on the involvement of V1/V2 regions in HIVD (50).
We were surprised that, when placed in the context of the pIIIB backbone, the HIV-1BORI-15 env did not produce a virus with high replication in microglia, in comparison with VH-BORI. This may indicate that other regions of the HIV-1BORI-15 virus are involved in its neurotropism. Alternatively, the high fusogenicity exhibited by its envelope could have affected p24gag release or viral spread. Defining the mechanism of the enhanced replication will be an area for future experimentation.
Finally, the neutralization pattern of HIV-1BORI-15 may provide additional evidence of the importance of the conformation of env in generating an effective immune response. Preliminary studies with these viruses showed that HIV-1BORI-15 may be easier to neutralize than its parent. In light of recent findings that fusogenic intermediates of env may generate broadly cross-reactive antibody responses (40) and that a CD4-independent HIV-1 stably exposing its coreceptor binding site is more easily neutralized (31), this isolate may be a candidate for a better understanding of the potential role of V1/V2 in immunity.
ACKNOWLEDGMENTS
This work was supported by PHS grants NS-27405, NS-35743, and MH-58958 and by the Medical Scientist Training Program (J.T.C.S.).
We thank G. Shaw (University of Alabama) for the HIV-1BORI isolate and B. Moss (NIH) for vTF1.1. Bob Doms, Jim Hoxie, Trevor Hoffman, and Andrew Albright provided excellent advice and reagents. We also thank L. Shawver and W. Cao for technical assistance. A number of reagents were obtained through the NIH AIDS Research Reagent and Reference Program, including the U373-MAGI cell lines from M. Emerman and A. Geballe and the MAGI-CCR5 cell line from J. Overbaugh.
REFERENCES
- 1.Albright A V, Shieh J T C, Itoh T, Lee B, Pleasure D, O'Connor M J, Doms R W, Gonzalez-Scarano F. Microglia express CCR5, CXCR4, and CCR3, but of these, CCR5 is the principal coreceptor for human immunodeficiency virus type 1 dementia isolates. J Virol. 1999;73:205–213. doi: 10.1128/jvi.73.1.205-213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen K B. A domain of murine retrovirus surface protein gp70 mediates fusion, as shown in a novel SC-1 cell fusion system. J Virol. 1994;68:3175–3182. doi: 10.1128/jvi.68.5.3175-3182.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brew B J, Evans L, Byrne C, Pemberton L, Hurren L. The relationship between AIDS dementia complex and the presence of macrophage tropic and non syncytium inducing isolates of human immunodeficiency virus type 1 in the cerebrospinal fluid. J Neurovirol. 1996;2:152–157. doi: 10.3109/13550289609146877. [DOI] [PubMed] [Google Scholar]
- 7.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carillo A, Ratner L. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J Virol. 1996;70:1310–1316. doi: 10.1128/jvi.70.2.1310-1316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Brown A, Harouse J, Luciw P A, Mayer A J. Selection for neutralization resistance of the Simian/Human Immunodeficiency Virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J Virol. 1999;73:5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu J, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The b-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Clark S J, Saag M S, Decker W D, Campbell-Hill S, Roberson J L, Veldkamp P J, Kappes J C, Hahn B H, Shaw G M. High titers of cytopathic virus in plasma of patients with symptomatic primary HIV-1 infection. N Engl J Med. 1991;324:954–960. doi: 10.1056/NEJM199104043241404. [DOI] [PubMed] [Google Scholar]
- 14.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Dick A D, Pell M, Brew B J, Foulcher E, Sedgwick J D. Direct ex vivo flow cytometric analysis of human microglial cell CD4 expression: examination of central nervous system biopsy specimens from HIV-seropositive patients and patients with other neurological disease. AIDS. 1997;11:1699–1708. doi: 10.1097/00002030-199714000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Di Stefano M, Wilt S, Gray F, Dubois-Dalcq M, Chiodi F. HIV type 1 V3 sequences and the development of dementia during AIDS. AIDS Res Hum Retrovir. 1996;12:471–476. doi: 10.1089/aid.1996.12.471. [DOI] [PubMed] [Google Scholar]
- 19.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, Sakmar T P, Moore J P, Maddon P J. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggerty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 23.Epstein L G, Kuiken C, Blumberg B M, Hartman S, Sharer L R, Clement M, Goudsmit J. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology. 1991;180:583–590. doi: 10.1016/0042-6822(91)90072-j. [DOI] [PubMed] [Google Scholar]
- 24.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 25.Gabuzda D, He J, Ohagen A, Vallat A V. Chemokine receptors in HIV-1 infection of the central nervous system. Semin Immunol. 1998;10:203–213. doi: 10.1006/smim.1998.0133. [DOI] [PubMed] [Google Scholar]
- 26.Ghorpade A, Xia M Q, Hyman B T, Persidsky Y, Nukuna A, Bock P, Che M, Limoges J, Gendelman H E, MacKay C R. Role of the beta-chemokine receptors CCR3 and CCR5 in human immunodeficiency virus type 1 infection of monocytes and microglia. J Virol. 1998;72:3351–3361. doi: 10.1128/jvi.72.4.3351-3361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glass J D, Wesselingh S L, Selnes O A, McArthur J C. Clinical-neuropathological correlation in HIV-associated dementia. Neurology. 1993;43:2230–2237. doi: 10.1212/wnl.43.11.2230. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Scarano F, Pobjecky N, Nathanson N. La Crosse bunyavirus can mediate pH-dependent fusion from without. Virology. 1984;132:222–225. doi: 10.1016/0042-6822(84)90107-7. [DOI] [PubMed] [Google Scholar]
- 29.Harrington R D, Geballe A P. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 34.Ioannidis J P A, Reichlin S, Skolnik P R. Long-term productive human immunodeficiency virus-1 infection in human infant microglia. Am J Pathol. 1995;147:1200–1206. [PMC free article] [PubMed] [Google Scholar]
- 35.Keys B, Karis J, Fadeel B, Valentin A, Norkrans G, Hagberg L, Chiodi F. V3 sequences of paired HIV-1 isolates from blood and CSF cluster according to host and show variation related to the clinical stage of disease. Virology. 1993;196:475–483. doi: 10.1006/viro.1993.1503. [DOI] [PubMed] [Google Scholar]
- 36.Koito, A., G. Harrowe, J. A. Levy, and C. Cheng-Mayer. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 68:2253–2259. [DOI] [PMC free article] [PubMed]
- 37.Korber B T M, Kunstman K J, Patterson B K, Furtado M, McEvilly M M, Levy R, Wolinsky S M. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J T, Vinters H V, Chen I S Y. Dual infection of the central nervous system by AIDS virus with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 39.Kozak S L, Platt E J, Madani N, Ferro F E, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 41.Lee B, Sharron M, Blanpain C, Doranz B J, Vakili J, Setoh P, Berg E, Liu G, Guy H R, Drell S R, Parmentier M, Chang C N, Price K, Tsang M, Doms R W. Epitope mapping of CCR5 reveals multiple conformational states and distinct overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 42.Lee S C, Hatch C, Liu W, Kress Y, Lyman B D, Dickson D W. Productive infection of human fetal microglia by HIV-1. Am J Pathol. 1993;143:1032–1039. [PMC free article] [PubMed] [Google Scholar]
- 43.Lipton S A, Gendelman H E. Dementia associated with the acquired immunodeficiency syndrome. N Engl J Med. 1995;332:934–940. doi: 10.1056/NEJM199504063321407. [DOI] [PubMed] [Google Scholar]
- 44.Mondor I, Moulard M, Ugolini S, Klasse P J, Hoxie J, Amara A, Delaunay T, Wyatt R, Sodroski J, Sattentau Q J. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology. 1998;248:394–405. doi: 10.1006/viro.1998.9282. [DOI] [PubMed] [Google Scholar]
- 45.Morikita T, Maeda Y, Fujii S-I, Matsushita S, Obaru K, Takatsuki K. The V1/V2 region of Human Immunodeficiency Virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res Hum Retrovir. 1997;13:1291–1298. doi: 10.1089/aid.1997.13.1291. [DOI] [PubMed] [Google Scholar]
- 46.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5418. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer C, Balfe P, Fox D, May J C, Frederiksson R, Fenyo E-M, McKeating J A. Functional characterization of the V1V2 region of Human Immunodeficiency Virus type 1. Virology. 1996;220:436–449. doi: 10.1006/viro.1996.0331. [DOI] [PubMed] [Google Scholar]
- 48.Platt E J, Madani N, Kozak S L, Kabat D. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J Virol. 1997;71:883–890. doi: 10.1128/jvi.71.2.883-890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Power C, McArthur J C, Johnson R T, Griffin D E, Glass J D, Perryman S, Chesebro B. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J Virol. 1994;68:4643–4649. doi: 10.1128/jvi.68.7.4643-4649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Power C, McArthur J C, Nath A, Wehrly K, Mayne M, Nishio J, Langelier T, Johnson R T, Chesebro B. Neuronal death induced by brain-derived human immunodeficiency virus type 1 envelope genes differs between demented and nondemented AIDS patients. J Virol. 1998;72:9045–9053. doi: 10.1128/jvi.72.11.9045-9053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price R W, Brew B, Sidtis J, Rosenblum J, Scheck A C, Cleary P. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science. 1988;239:586–591. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- 52.Reeves J D, Schulz T F. The CD4-independent tropism of HIV-2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1997;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 54.Rucker J, Doranz B J, Edinger A L, Long D, Berson J F, Doms R W. Cell-cell fusion assay to study role of chemokine receptors in Human Immunodeficiency Virus type 1 entry. Methods Enzymol. 1997;288:118–133. doi: 10.1016/s0076-6879(97)88011-1. [DOI] [PubMed] [Google Scholar]
- 55.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in b-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 56.Saharkhiz-Langroodi A, Holland T C. Identification of the fusion-from-without determinants of Herpes Simplex Virus type 1 glycoprotein B. Virology. 1997;227:153–159. doi: 10.1006/viro.1996.8327. [DOI] [PubMed] [Google Scholar]
- 57.Sharpless N E, O'Brien W A, Verdin E, Kufta C V, Chen I S Y, Dubois-Dalcq M. Human immunodeficiency virus type 1 tropism for brain microglial cells is determined by a region of the env glycoprotein that also controls macrophage tropism. J Virol. 1992;66:2588–2593. doi: 10.1128/jvi.66.4.2588-2593.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shieh J T C, Albright A V, Sharron M, Gartner S, Strizki J, Doms R W, Gonzalez-Scarano F. Chemokine receptor utilization by HIV-1 isolates that replicate in microglia. J Virol. 1998;72:4243–4249. doi: 10.1128/jvi.72.5.4243-4249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siess D C, Kozak S L, Kabat D. Exceptional fusogenicity of Chinese hamster ovary cells with murine retroviruses suggests roles for cellular factor(s) and receptor clusters in the membrane fusion process. J Virol. 1996;70:3432–3439. doi: 10.1128/jvi.70.6.3432-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strizki J M, Albright A V, Sheng H, O'Connor M, Perrin L, Gonzalez-Scarano F. Infection of primary human microglia and monocyte-derived macrophages with human immunodeficiency virus type 1 isolates: evidence of differential tropism. J Virol. 1996;70:7654–7662. doi: 10.1128/jvi.70.11.7654-7662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan N, Thali M, Furman C, Ho D D, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67:3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi K, Wesselingh S L, Griffin D E, McArthur J C, Johnson R T, Glass J D. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann Neurol. 1996;39:705–711. doi: 10.1002/ana.410390606. [DOI] [PubMed] [Google Scholar]
- 64.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 65.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR5. Nature. 1996;14:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 66.Vodicka M A, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 67.Watkins B A, Dorn H H, Kelly W B, Armstrong R C, Potts B J, Michaels F, Kufta C V, Dubois-Dalq M. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science. 1990;249:549–552. doi: 10.1126/science.2200125. [DOI] [PubMed] [Google Scholar]
- 68.Wiley C A, Achim C L. Human immunodeficiency virus encephalitis is the pathological correlate of dementia in acquired immunodeficiency syndrome. Ann Neurol. 1994;36:673–676. doi: 10.1002/ana.410360422. [DOI] [PubMed] [Google Scholar]
- 69.Wiley C A, Schrier R D, Nelson J A, Lampert P W, Oldstone M B A. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong J K, Ignacio C C, Torriani F, Havlir D, Fitch N J S, Richman D D. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu Z, Kayman S C, Honnen W, Revesz K, Chen H, Vijh-Warrier S, Tilley S A, McKeating J, Shotton C, Pinter A. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J Virol. 1995;69:2271–2278. doi: 10.1128/jvi.69.4.2271-2278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 74.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yong V W, Antel J P. Culture of glial cells from human brain biopsies. In: Fedoroff S, Richardson A, editors. Protocols for neural cell culture. Totowa, N.J: Humana Press; 1992. pp. 81–96. [Google Scholar]
- 76.Zhang Y J, Moore J P. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J Virol. 1999;73:3443–3448. doi: 10.1128/jvi.73.4.3443-3448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]