Abstract
Gastric carcinomas (GC) are classified into four phenotypes according to mucin expression. Previous studies revealed the association of distinct genetic profiles in GC with mucin phenotypic expression; however, the roles of epigenetic changes, such as DNA methylation, are poorly understood. We examined whether the phenotypic expression of GC was associated with DNA methylation of hMLH1, MGMT, p16 INK4a , RAR‐beta or CDH1. Expression of HGM, M‐GGMC‐1, MUC2, and CD10 was analyzed immunohistochemically in 33 advanced GC with differentiated histology. HGM was expressed in 14 (42.4%) cases, M‐GGMC‐1 in five (15.2%) cases, MUC2 in 15 (45.5%) cases and CD10 in 18 (54.5%) cases. DNA methylation was detected in five (15.2%) cases for hMLH1, 11 (33.3%) cases for MGMT, 13 (39.4%) cases for p16 INK4a , 17 (51.5%) cases for RAR‐beta and 14 (42.4%) cases for CDH1 by bisulfite‐polymerase chain reaction and methylation‐specific polymerase chain reaction. DNA methylation of hMLH1 occurred more frequently in MUC2‐negative GC than in MUC2‐positive GC (P = 0.0488, Fisher's exact test). In contrast, MGMT was more frequently methylated in MUC2‐positive GC than in MUC2‐negative GC (P = 0.0078, Fisher's exact test). There was no correlation between gastric or intestinal‐markers and methylation of the p16 INK4a , RAR‐beta and CDH1 genes. These results indicate that DNA methylation of specific genes, such as hMLH1 and MGMT, may be involved partly in the distinct phenotypic expression of GC. (Cancer Sci 2005; 96: 474 –479)
Gastric carcinoma (GC) is one of the most common malignancies worldwide. GC are often classified histologically into two major types: the differentiated and undifferentiated types described by Nakamura et al.( 1 ) or the Lauren intestinal and diffuse types( 2 ) based on glandular structure. Various genetic and epigenetic alterations are associated with GC; some are found in both the intestinal and diffuse types, whereas others are type specific.( 3 , 4 ) It was previously reported that GC can be subdivided according to mucin expression into four phenotypes:( 5 , 6 , 7 ) (i) gastric or foveolar phenotype (G type); (ii) intestinal phenotype (I type); (iii) intestinal and gastric mixed phenotype (GI type); and (iv) neither gastric nor intestinal phenotype (N type). Despite the usefulness of the Lauren classification, there are several variations of the intestinal‐type GC described by Lauren. To better understand the development of GC at the molecular level, it is important to analyze molecular alterations in intestinal‐type GC according to the mucin phenotype. Distinct genetic changes appear to be associated with I type and G type GC. p53 mutations and allelic deletions of the adenomatous polyposis coli (APC) gene are detected more frequently in I type GC than in G type GC,( 8 , 9 , 10 , 11 ) whereas microsatellite instability (MSI) is detected more frequently in G type GC than in I type GC.( 10 , 12 ) We reported previously that alterations of p73, including loss of heterozygosity and abnormal expression, play important roles in the genesis of G type GC.( 13 )
Several lines of evidence suggest that changes in DNA methylation patterns, such as hypermethylation of CpG islands, are common changes in human cancers.( 14 ) Hypermethylation of CpG islands in promoters is associated with silencing of some tumor‐related genes.( 15 , 16 , 17 ) We previously reported DNA methylation of the hMLH1,( 18 ) MGMT,( 19 ) p16 INK4a , RAR‐beta and CDH1 ( 20 ) genes. In contrast to the many studies of genetic alterations in G type and I type GC, epigenetic alterations in G type and I type GC are poorly understood. Associations between genetic and epigenetic alterations have been reported. DNA methylation of hMLH1 is associated with MSI,( 21 , 22 ) and DNA methylation of MGMT is associated with G to A mutations in the K‐ras ( 23 ) and p53 ( 24 ) genes. Because MSI occurs frequently in G type GC, it is possible that DNA methylation of hMLH1 may occur frequently in G type GC. In fact, it has been reported that DNA hypermethylation of hMLH1 occurs frequently in G type GC.( 25 ) Because p53 mutations are detected frequently in I type GC, it is possible that DNA methylation of MGMT occurs in I type GC. However, the association between DNA methylation and the mucin phenotypic expression of GC has been investigated only for hMLH1.
In the present study, we investigated the association between expression of gastric‐type and intestinal‐type markers and DNA methylation status of hMLH1, MGMT, p16 INK4a , RAR‐beta and CDH1 in differentiated‐type GC.
Materials and Methods
Tissue samples
Thirty‐three samples of differentiated‐type GC from 33 patients were examined. All GC samples were not early GC but advanced GC, that had invaded beyond the muscularis propria.( 26 ) Samples were obtained at time of surgery at Hiroshima University Hospital (Hiroshima, Japan) and affiliated hospitals. Tissue samples for molecular analyses were frozen immediately in liquid nitrogen and stored at −80°C until use. We confirmed microscopically that the tumor specimens consisted mainly of carcinoma tissue (> 50%). For immunohistochemical staining, tissues were fixed in 10% buffered‐formalin and embedded in paraffin. Tumor staging was carried out according to the tumor‐node‐metastasis stage grouping.( 27 ) Because written informed consent was not obtained, for strict privacy protection, all samples were dis‐identified before analyzing DNA methylation status. This procedure is in accordance with the Ethical Guidelines for Human Genome/Gene Research enacted by the Japanese Government.
Phenotypic analysis of gastric carcinomas
Tissue sections (4 µm thick) were prepared from paraffin blocks, and representative sections were immunostained for human gastric mucin (HGM), M‐GGMC‐1, MUC2 and CD10. Immunostaining was by the immunoperoxidase technique with a Histofine Simple Stain Kit (Nichirei Biosciences, Tokyo, Japan). Deparaffinized tissue sections were immersed in methanol containing 3% hydrogen peroxide for 15 min to block endogenous peroxidase activity. Microwave pretreatment in citrate buffer was carried out for 15–30 min to retrieve the antigenicity. The sections were then incubated with antibodies against gastric‐type markers HGM (NCL‐HGM‐45M1; Novocastra, Newcastle, UK; dilution 1:50) and M‐GGMC‐1 (HIK1083; Kanto Kagaku, Tokyo, Japan; dilution 1:50), and intestinal‐type markers MUC2 (Ccp58; Santa Cruz Biotechnology, Santa Cruz, CA, USA; dilution 1:200) and CD10 (NCL‐CD10‐270; Novocastra; dilution 1:50), for 1.5 h at 37°C followed by incubation with the secondary antibody for 30 min. The immunocomplexes were visualized with 3,3′‐diaminobenzidine. Sections were then counterstained with hematoxylin. GC were classified as G type, I type, GI type or N type. G type comprised those samples in which > 30% of the tumor cells were positive for gastric‐type markers and showed little staining with intestinal‐type markers. I type comprised those specimens in which > 30% of the tumor cells were positive for MUC2 or in which > 5% of the tumor cells were positive for CD10 and showed little staining with gastric‐type markers. GC that showed positive staining for both gastric‐type and intestinal‐type markers were classified as GI type, and those that showed no staining with those markers were classified as N type.
Genomic DNA extraction and methylation analysis
To examine DNA methylation patterns in the 5′ CpG islands of the hMLH1, MGMT, p16 INK4a , RAR‐beta and CDH1 genes, we extracted genomic DNA with a genomic DNA purification kit (Promega, Madison, WI, USA) and treated the genomic DNA with sodium bisulfite, as described previously.( 28 ) In brief, 2 µg of genomic DNA was denatured by treatment with NaOH and modified with 3 M sodium bisulfite for 16 h. DNA samples were purified with Wizard DNA purification resin (Promega), treated with NaOH, precipitated with ethanol and resuspended in 25 µL water. Aliquots (2 µL) were used as templates for methylation‐specific polymerase chain reaction (MSP) amplification of the MGMT, p16 INK4a , RAR‐beta and CDH1 genes. MSP primers for MGMT, p16 INK4a , RAR‐beta and CDH1 were described previously.( 28 , 29 , 30 ) For analysis of DNA methylation of hMLH1, we carried out bisulfite‐polymerase chain reaction (PCR) followed by restriction digestion as described previously.( 31 ) Primers and PCR conditions used for amplifying specific DNA fragments of various target genes are listed in Table 1. PCR products (15 µg) were loaded onto 8% non‐denaturing polyacrylamide gels, stained with ethidium bromide and visualized under UV light. According to the corresponding literature, CpG island hypermethylation in the regions examined revealed good correlation with epigenetic silencing of the respective target genes.( 31 , 32 , 33 , 34 , 35 )
Table 1.
Primer sequences for DNA methylation analysis
| Primer sequence | Primer sequence | Annealing temperature |
|---|---|---|
| hMLH1 | ||
| F: 5′‐TAGTAGTYGTTTTAGGGAGGGA ‐3′ | 55°C | |
| R: 5′‐TCTAAATACTCAACRAAAATACCTT‐3′ | ||
| MGMT (unmethylated) | ||
| F: 5′‐TTTGTGTTTTGATGTTTGTAGGTTTTTGT‐3′ | 59°C | |
| R: 5′‐AACTCCACACTCTTCCAAAAACAAAACA‐3′ | ||
| MGMT (methylated) | ||
| F: 5′‐TTTCGACGTTCGTAGGTTTTCGC‐3′ | 59°C | |
| R: 5′‐GCACTCTTCCGAAAACGAAACG‐3′ | ||
| p16 INK4a (unmethylated) | ||
| F: 5′‐TTATTAGAGGGTGGGGTGGATTGT‐3′ | 60°C | |
| R: 5′‐CCACCTAAATCAACCTCCAACCA‐3′ | ||
| p16 INK4a (methylated) | ||
| F: 5′‐TTATTAGAGGGTGGGGCGGATCGC‐3′ | 65°C | |
| R: 5′‐CCACCTAAATCGACCTCCGACCG‐3′ | ||
| RAR‐beta (unmethylated) | ||
| F: 5′‐TTAGTAGTTTGGGTAGGGTTTATT ‐3′ | 55°C | |
| R: 5′‐CCAAATCCTACCCCAACA‐3′ | ||
| RAR‐beta (methylated) | ||
| F: 5′‐GGTTAGTAGTTCGGGTAGGGTTTATC‐3′ | 64°C | |
| R: 5′‐CCGAATCCTACCCCGACG‐3′ | ||
| CDH1 (unmethylated) | ||
| F: 5′‐TAATTTTAGGTTAGAGGGTTATTGT‐3′ | 53°C | |
| R: 5′‐CACAACCAATCAACAACACA‐3′ | ||
| CDH1 (methylated) | ||
| F: 5′‐TTAGGTTAGAGGGTTATCGCGT‐3′ | 57°C | |
| R: 5′‐TAACTAAAAATTCACCTACCGAC‐3′ | ||
Statistical methods
Fisher's exact test was used for statistical analysis. P‐values less than 0.05 were regarded as statistically significant.
Results
Association between gastric‐type and intestinal‐type markers and DNA methylation
We carried out immunohistochemical analysis of 33 advanced differentiated‐type GC (Fig. 1). Of the 33 GC, expression of gastric and intestinal markers was detected in 14 (42.4%) cases for HGM, five (15.2%) cases for M‐GGMC‐1, 15 (45.5%) cases for MUC2 and 18 (54.5%) cases for CD10. Next, DNA methylation status was investigated. Representative data for bisulfite‐PCR followed by restriction digestion of the hMLH1 gene and MSP of the MGMT, p16 INK4a , RAR‐beta and CDH1 genes are shown in Fig. 2. Of the 33 GC, DNA hypermethylation was detected in five (15.2%) cases for hMLH1, 11 (33.3%) cases for MGMT, 13 (39.4%) cases for p16 INK4a , 17 (51.5%) cases for RAR‐beta and 14 (42.4%) cases for CDH1. Although recent evidence suggests that methylation of certain genes such as hMLH1 and CDH1 is associated with aging,( 36 , 37 ) there was no correlation between age and DNA methylation of a specific gene (Table 2). We compared DNA methylation status with each marker (3, 4, 5, 6). DNA methylation of hMLH1 was detected more frequently in MUC2‐negative GC (5/18, 27.8%) than in MUC2‐positive GC (0/15, 0.0%, P = 0.0488, Fisher's exact test). In contrast, DNA methylation of MGMT was detected more frequently in MUC2‐positive GC (9/15, 60.0%) than in MUC2‐negative GC (2/18, 11.1%, P = 0.0078, Fisher's exact test) (Table 5). There was no correlation between gastric and intestinal markers and methylation of the p16 INK4a , RAR‐beta and CDH1 genes.
Figure 1.
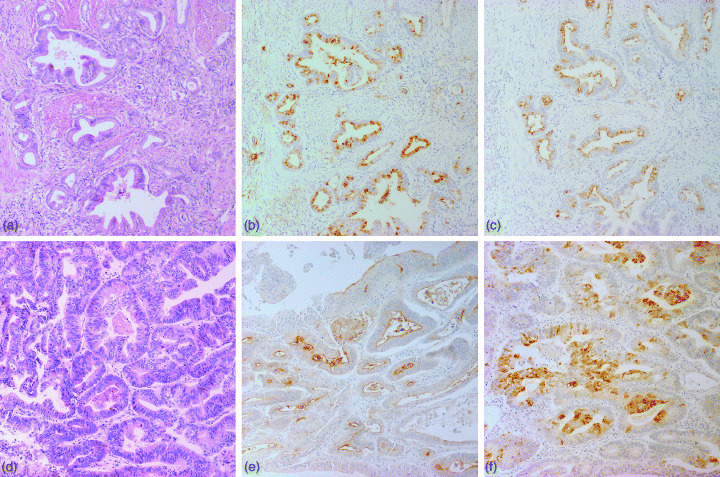
G type (case 3: a, b, c) and I type (case 10: d, e, f) gastric carcinomas. (a,d) Hematoxylin and eosin staining. (b) MUC5AC and (c) M‐GGMC‐1 were detected in the cytoplasm of cancer cells. (e) CD10 was expressed on the luminal surfaces of cancer cells. (f) MUC2 is positive in the cytoplasm of cancer cells. (Original magnification, ×100).
Figure 2.
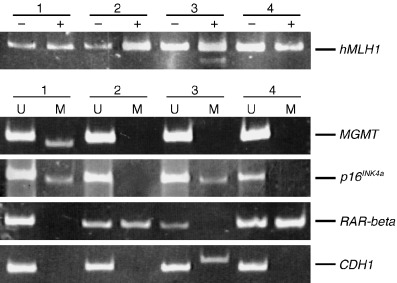
Bisulfite‐polymerase chain reaction followed by restriction digestion of the hMLH1 gene and methylation‐specific polymerase chain reaction of the MGMT, p16 INK4a , CDH1 and RAR‐beta genes. Methylated allele was detected in case 3 (hMLH1), case 1 (MGMT), cases 1 and 3 (p16 INK4a ), cases 2 and 4 (RAR‐beta) and case 3 (CDH1). M, methylated; U, unmethylated; +, after restriction enzyme digestion; –, before restriction enzyme digestion.
Table 2.
Association between age and DNA methylation
| Gene | Methylation status | Age | P‐value † | |
|---|---|---|---|---|
| > 61 | ≤ 60 | |||
| hMLH1 | Methylated | 5 (100.0%) | 0 | 0.5663 |
| Unmethylated | 22 (78.6%) | 6 | ||
| MGMT | Methylated | 7 (63.6%) | 4 | 0.1458 |
| Unmethylated | 20 (90.9%) | 2 | ||
| p16 INK4a | Methylated | 11 (84.6%) | 2 | 1.0000 |
| Unmethylated | 16 (80.0%) | 4 | ||
| RAR‐beta | Methylated | 13 (76.5%) | 4 | 0.6562 |
| Unmethylated | 14 (87.5%) | 2 | ||
| CDH1 | Methylated | 11 (78.6%) | 3 | 1.0000 |
| Unmethylated | 16 (84.2%) | 3 | ||
Fisher's exact test.
Table 3.
Association between human gastric mucin (HGM) expression and DNA methylation status
| Gene | Methylation status | HGM expression | P‐value † | |
|---|---|---|---|---|
| Positive | Negative | |||
| hMLH1 | Methylated | 2 (40.0%) | 3 | 1.0000 |
| Unmethylated | 12 (42.9%) | 16 | ||
| MGMT | Methylated | 5 (45.5%) | 6 | 1.0000 |
| Unmethylated | 9 (40.9%) | 13 | ||
| p16 INK4a | Methylated | 7 (53.8%) | 6 | 0.4720 |
| Unmethylated | 7 (35.0%) | 13 | ||
| RAR‐beta | Methylated | 8 (47.1%) | 9 | 0.7283 |
| Unmethylated | 6 (37.5%) | 10 | ||
| CDH1 | Methylated | 7 (50.0%) | 7 | 0.4969 |
| Unmethylated | 7 (36.8%) | 12 | ||
Fisher's exact test.
Table 4.
Association between M‐GGMC‐1 expression and DNA methylation status
| Gene | Methylation status | M‐GGMC‐1 expression | P‐value † | |
|---|---|---|---|---|
| Positive | Negative | |||
| hMLH1 | Methylated | 1 (20.0%) | 4 | 1.0000 |
| Unmethylated | 4 (14.3%) | 24 | ||
| MGMT | Methylated | 1 (9.1%) | 10 | 0.6431 |
| Unmethylated | 4 (18.2%) | 18 | ||
| p16 INK4a | Methylated | 1 (7.7%) | 12 | 0.6253 |
| Unmethylated | 4 (20.0%) | 16 | ||
| RAR‐beta | Methylated | 2 (11.8%) | 15 | 0.6562 |
| Unmethylated | 3 (18.8%) | 13 | ||
| CDH1 | Methylated | 3 (21.4%) | 11 | 0.6285 |
| Unmethylated | 2 (10.5%) | 17 | ||
Fisher's exact test.
Table 5.
Association between MUC2 expression and DNA methylation status
| Gene | Methylation status | MUC2 expression | P‐value † | |
|---|---|---|---|---|
| Positive | Negative | |||
| hMLH1 | Methylated | 0 (0.0%) | 5 | 0.0488 |
| Unmethylated | 15 (53.6%) | 13 | ||
| MGMT | Methylated | 9 (81.8%) | 2 | 0.0078 |
| Unmethylated | 6 (27.3%) | 16 | ||
| p16 INK4a | Methylated | 7 (53.8%) | 6 | 0.4928 |
| Unmethylated | 8 (40.0%) | 12 | ||
| RAR‐beta | Methylated | 8 (47.1%) | 9 | 1.0000 |
| Unmethylated | 7 (43.8%) | 9 | ||
| CDH1 | Methylated | 5 (35.7%) | 9 | 0.4824 |
| Unmethylated | 10 (52.6%) | 9 | ||
Fisher's exact test.
Table 6.
Association between CD10 expression and DNA methylation status
| Gene | Methylation status | CD10 expression | P‐value † | |
|---|---|---|---|---|
| Positive | Negative | |||
| hMLH1 | Methylated | 1 (20.0%) | 4 | 0.1523 |
| Unmethylated | 17 (60.7%) | 11 | ||
| MGMT | Methylated | 8 (72.7%) | 3 | 0.2659 |
| Unmethylated | 10 (45.5%) | 12 | ||
| p16 INK4a | Methylated | 7 (53.8%) | 6 | 0.7332 |
| Unmethylated | 11 (5.0%) | 9 | ||
| RAR‐beta | Methylated | 7 (41.2%) | 10 | 0.1663 |
| Unmethylated | 11 (68.8%) | 5 | ||
| CDH1 | Methylated | 6 (42.9%) | 8 | 0.3041 |
| Unmethylated | 12 (63.2%) | 7 | ||
Fisher's exact test.
Phenotypic expression of gastric carcinomas
On the basis of the combinations of expression of these four mucin markers, the 33 GC were classified phenotypically as five (15.2%) G type, 14 (42.4%) I type, 9 (27.2%) GI type and five (15.2%) N type. There was no apparent correlation between mucin phenotypic expression and clinicopathological findings (data not shown). No apparent association was observed between DNA methylation of a specific gene and phenotypic expression of GC (data not shown).
Discussion
Gastric carcinomas are classified into the G, I, GI, and N phenotypes according to gastric‐type and intestinal‐type markers. In this study, expression of HGM, M‐GGMC‐1, MUC2 and CD10 was investigated. We observed that hMLH1 was rarely methylated, whereas MGMT was frequently methylated in MUC2‐positive GC. Therefore, DNA methylation, especially of the hMLH1 and MGMT genes, may participate partly in the distinct phenotypic expression of GC. In fact, recent studies showed that the MUC2 gene is also a target of DNA methylation.( 38 ) Changes in genome‐wide DNA methylation may also affect DNA methylation of these genes. To our knowledge, there is no report regarding DNA methylation of HGM, M‐GGMC‐1 and CD10.
DNA hypermethylation of MGMT occurred frequently in MUC2‐positive GC. Previously reported data indicate that DNA hypermethylation of MGMT is associated with a G to A mutation in the K‐ras and p53 genes.( 23 , 24 ) Although we found no association between DNA methylation of MGMT and I type GC, MUC2 is a marker of intestinal epithelial cells. Thus, frequent p53 mutations in I type GC( 8 , 9 , 10 , 11 ) may be due to DNA methylation of MGMT.
The hMLH1 gene was rarely methylated in MUC2‐positive GC in this study. Endoh et al. reported that DNA hypermethylation of hMLH1 occurs frequently in G type GC,( 25 ) which does not express MUC2. Our findings support the notion that DNA methylation of hMLH1 occurs frequently in G type GC. On the other hand, MUC2‐positive GC were reported to show MSI more frequently than MUC2‐negative GC.( 39 ) DNA methylation of hMLH1 is associated with MSI, indicating that MUC2‐positive GC may have frequent DNA methylation of hMLH1. The reason for the discrepancy between our results and those of Lee et al. is unclear; however, the discrepancy may be due to differences in the samples analyzed.( 39 ) Lee et al. studied the MUC2 expression and MSI in both differentiated‐type and undifferentiated‐type GC, whereas we analyzed the phenotypic expression and DNA methylation in differentiated‐type GC only. Taken together, MUC2‐positive undifferentiated‐type GC may show frequent MSI and DNA methylation of hMLH1. In addition, because of a phenotypic shift from G type to I type expression in conjunction with tumor progression,( 7 ) G type early GC showing hMLH1 methylation may lose G type expression along with tumor progression.
There was no correlation between mucin marker expression and DNA methylation of p16 INK4a , CDH1 and RAR‐beta. Hypermethylation of the p16 INK4a gene is more common in differentiated‐type GC than in undifferentiated‐type GC, whereas CDH1 and RAR‐beta hypermethylation is observed more frequently in undifferentiated‐scattered‐type GC than in other types.( 20 ) Thus, DNA methylation of these three genes may be involved in histogenesis, but not in phenotypic expression of GC.
Although MUC2 expression was correlated with DNA methylation of hMLH1 and MGMT in this study, the number of cases we studied was too small to clarify correlation between phenotypic expression of GC and DNA methylation status. Additional studies are needed to obtain the definite association between DNA methylation and G and I phenotypes of GC.
In conclusion, our data show that DNA methylation of specific genes, such as hMLH1 and MGMT, may be associated with the distinct phenotypic expression of GC. Because DNA methylation of tumor‐related genes has been shown to occur in the early stages of stomach carcinogenesis( 40 ) and to increase in parallel with stomach carcinogenesis,( 41 ) the association between DNA methylation and GC phenotypes in early GC should be investigated.
Acknowledgments
We thank M. Takatani for excellent technical assistance and advice. This work was carried out with the kind cooperation of the Research Center for Molecular Medicine (RCMM), Faculty of Medicine, Hiroshima University. We thank the Analysis Center of Life Science, Hiroshima University, for the use of their facilities. This work was supported, in part, by Grants‐in‐Aid for Cancer Research from the Ministry of Education, Culture, Science, Sports, and Technology of Japan, and from the Ministry of Health, Labor, and Welfare of Japan.
References
- 1. Nakamura K, Sugano H, Takagi K. Carcinoma of the stomach in incipient phase: its histogenesis and histological appearances. Gann 1968; 59: 251–8. [PubMed] [Google Scholar]
- 2. Lauren P. The two histological main types of gastric carcinoma. Diffuse and so‐called intestinal type carcinoma: an attempt at histological classification. Acta Pathol Microbiol Scand 1965; 64: 31–49. [DOI] [PubMed] [Google Scholar]
- 3. Yasui W, Yokozaki H, Fujimoto J, Naka K, Kuniyasu H, Tahara E. Genetic and epigenetic alterations in multistep carcinogenesis of the stomach. J Gastroenterol 2000; 35: 111–15. [PubMed] [Google Scholar]
- 4. Oue N, Hamai Y, Mitani Y et al. Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res 2004; 64: 2397–405. [DOI] [PubMed] [Google Scholar]
- 5. Fiocca R, Villani L, Tenti P et al. Characterization of four main cell types in gastric cancer: foveolar, mucopeptic, intestinal columnar and goblet cells. An histopathologic, histochemical and ultrastructural study of ‘early’ and ‘advanced’ tumours. Pathol Res Pract 1987; 182: 308–25. [DOI] [PubMed] [Google Scholar]
- 6. Tatematsu M, Ichinose M, Miki K, Hasegawa R, Kato T, Ito N. Gastric and intestinal phenotypic expression of human stomach cancers as revealed by pepsinogen immunohistochemistry and mucin histochemistry. Acta Pathol Jpn 1990; 40: 494–504. [DOI] [PubMed] [Google Scholar]
- 7. Tatematsu M, Tsukamoto T, Inada K. Stem cells and gastric cancer: role of gastric and intestinal mixed intestinal metaplasia. Cancer Sci 2003; 94: 135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uchino S, Noguchi M, Ochiai A, Saito T, Kobayashi M, Hirohashi S. p53 mutation in gastric cancer: a genetic model for carcinogenesis is common to gastric and colorectal cancer. Int J Cancer 1993; 54: 759–64. [DOI] [PubMed] [Google Scholar]
- 9. Kushima R, Muller W, Stolte M, Borchard F. Differential p53 protein expression in stomach adenomas of gastric and intestinal phenotypes. Possible sequences of p53 alteration in stomach carcinogenesis. Virchows Arch 1996; 428: 223–7. [DOI] [PubMed] [Google Scholar]
- 10. Endoh Y, Sakata K, Tamura G et al. Cellular phenotypes of differentiated‐type adenocarcinomas and precancerous lesions of the stomach are dependent on the genetic pathways. J Pathol 2000; 191: 257–63. [DOI] [PubMed] [Google Scholar]
- 11. Wu LB, Kushima R, Borchard F, Molsberger G, Hattori T. Intramucosal carcinomas of the stomach: phenotypic expression and loss of heterozygosity at microsatellites linked to the APC gene. Pathol Res Pract 1998; 194: 405–11. [DOI] [PubMed] [Google Scholar]
- 12. Shibata N, Watari J, Fujiya M, Tanno S, Saitoh Y, Kohgo Y. Cell kinetics and genetic instabilities in differentiated type early gastric cancers with different mucin phenotype. Hum Pathol 2003; 34: 32–40. [DOI] [PubMed] [Google Scholar]
- 13. Yokozaki H, Shitara Y, Fujimoto J, Hiyama T, Yasui W, Tahara E. Alterations of p73 preferentially occur in gastric adenocarcinomas with foveolar epithelial phenotype. Int J Cancer 1999; 83: 192–6. [DOI] [PubMed] [Google Scholar]
- 14. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3: 415–28. [DOI] [PubMed] [Google Scholar]
- 15. Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev 1991; 55: 451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merlo A, Herman JG, Mao L et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995; 1: 686–92. [DOI] [PubMed] [Google Scholar]
- 17. Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet 1997; 13: 444–9. [DOI] [PubMed] [Google Scholar]
- 18. Oue N, Oshimo Y, Nakayama H et al. DNA methylation of multiple genes in gastric carcinoma: association with histological type and CpG island methylator phenotype. Cancer Sci 2003; 94: 901–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oue N, Shigeishi H, Kuniyasu H et al. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer 2001; 93: 805–9. [DOI] [PubMed] [Google Scholar]
- 20. Oue N, Motoshita J, Yokozaki H et al. Distinct promoter hypermethylation of p16INK4a, CDH1, and RAR‐beta in intestinal, diffuse‐adherent, and diffuse‐scattered type gastric carcinomas. J Pathol 2002; 198: 55–9. [DOI] [PubMed] [Google Scholar]
- 21. Fleisher AS, Esteller M, Wang S et al. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res 1999; 59: 1090–5. [PubMed] [Google Scholar]
- 22. Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC. hMLH1 promoter methylation and lack of hMLH1 expression in sporadic gastric carcinomas with high‐frequency microsatellite instability. Cancer Res 1999; 59: 159–64. [PubMed] [Google Scholar]
- 23. Esteller M, Toyota M, Sanchez‐Cespedes M et al. Inactivation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K‐ras in colorectal tumorigenesis. Cancer Res 2000; 60: 2368–71. [PubMed] [Google Scholar]
- 24. Esteller M, Risques RA, Toyota M et al. Promoter hypermethylation of the DNA repair gene O(6)‐methylguanine‐DNA methyltransferase is associated with the presence of G : C to A : T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 2001; 61: 4689–92. [PubMed] [Google Scholar]
- 25. Endoh Y, Tamura G, Ajioka Y, Watanabe H, Motoyama T. Frequent hypermethylation of the hMLH1 gene promoter in differentiated‐type tumors of the stomach with the gastric foveolar phenotype. Am J Pathol 2000; 157: 717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hohenberger P, Gretschel S. Gastric cancer. Lancet 2003; 362: 305–15. [DOI] [PubMed] [Google Scholar]
- 27. Sobin LH, Wittekind C, eds. TNM Classification of Malignant Tumors, 6th edn. New York: Wiley‐Liss, 2002. [Google Scholar]
- 28. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation‐specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996; 93: 9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999; 59: 793–7. [PubMed] [Google Scholar]
- 30. Widschwendter M, Berger J, Hermann M et al. Methylation and silencing of the retinoic acid receptor‐beta2 gene in breast cancer. J Natl Cancer Inst 2000; 92: 826–32. [DOI] [PubMed] [Google Scholar]
- 31. Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age‐related CpG island methylation in ulcerative colitis. Cancer Res 2001; 61: 3573–7. [PubMed] [Google Scholar]
- 32. Oue N, Sentani K, Yokozaki H, Kitadai Y, Ito R, Yasui W. Promoter methylation status of the DNA repair genes hMLH1 and MGMT in gastric carcinoma and metaplastic mucosa. Pathobiology 2001; 69: 143–9. [DOI] [PubMed] [Google Scholar]
- 33. Shim YH, Kang GH, Ro JY. Correlation of p16 hypermethylation with p16 protein loss in sporadic gastric carcinomas. Lab Invest 2000; 80: 689–95. [DOI] [PubMed] [Google Scholar]
- 34. Hayashi K, Yokozaki H, Goodison S et al. Inactivation of retinoic acid receptor beta by promoter CpG hypermethylation in gastric cancer. Differentiation 2001; 68: 13–21. [DOI] [PubMed] [Google Scholar]
- 35. Nakayama S, Sasaki A, Mese H, Alcalde RE, Tsuji T, Matsumura T. The E‐cadherin gene is silenced by CpG methylation in human oral squamous cell carcinomas. Int J Cancer 2001; 93: 667–73. [DOI] [PubMed] [Google Scholar]
- 36. Nakajima T, Akiyama Y, Shiraishi J et al. Age‐related hypermethylation of the hMLH1 promoter in gastric cancers. Int J Cancer 2000; 94: 208–11. [DOI] [PubMed] [Google Scholar]
- 37. Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E‐cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol 2002; 161: 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mesquita P, Peixoto AJ, Seruca R et al. Role of site‐specific promoter hypomethylation in aberrant MUC2 mucin expression in mucinous gastric carcinomas. Cancer Lett 2003; 189: 129–36. [DOI] [PubMed] [Google Scholar]
- 39. Lee HS, Choi SI, Lee HK et al. Distinct clinical features and outcomes of gastric cancers with microsatellite instability. Mod Pathol 2002; 15: 632–40. [DOI] [PubMed] [Google Scholar]
- 40. Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res 2001; 61: 2847–51. [PubMed] [Google Scholar]
- 41. Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along the multistep pathway of gastric carcinogenesis. Lab Invest 2003; 83: 635–41. [DOI] [PubMed] [Google Scholar]


