Abstract
Tob protein, when overexpressed, suppresses growth of NIH3T3 cells, presumably by regulating expression of various growth‐related genes. However, the molecular mechanisms underlying Tob‐mediated regulation of gene expression have been obscure. To address this issue we established stable Tob‐expressing cell lines and used a proteomics approach to identify Tob‐interacting proteins. We found that Tob associates with the CCR4‐NOT complex. The carboxyl‐terminal half of Tob interacted with Cnot1, a core protein of the CCR4‐NOT complex. We further showed that the deadenylase activity associated with the complex was suppressed in vitro by Tob. These results suggest that the antiproliferative activity of Tob is shown post‐transcriptionally by controlling the stability of the target mRNAs in addition to its involvement in transcriptional regulation, reported previously. (Cancer Sci 2008; 99: 755–761)
The Tob/BTG family comprises six proteins, Tob, Tob2, ANA, BTG1, BTG2/PC3/TIS21, and PC3B, that share a common amino‐terminal domain.( 1 ) All of these proteins, when overexpressed, suppress growth of NIH3T3 cells.( 2 , 3 , 4 , 5 , 6 , 7 , 8 ) There is evidence that the Tob/BTG family of proteins is involved in regulation not only of cell growth but also of differentiation and development.( 9 ) For instance, BTG1 is thought to be involved in myogenesis induced by triiodothyronine.( 10 ) Analysis of tob‐deficient mice revealed that Tob is involved in bone development.( 11 )
Although they lack DNA‐binding domains, the Tob/BTG proteins are generally viewed as transcriptional cofactors. For example, Tob interacts with Smad in BMP2 signaling,( 11 ) and in T‐cell anergy.( 12 ) BTG2 enhances Hoxb9‐mediated transcription.( 13 ) Both Tob and BTG2 reduce cyclin D1 expression,( 14 , 15 ) possibly by recruiting histone deacetylase to the cyclin D1 promoter,( 15 ) contributing to G0/G1 arrest. Furthermore, Cnot7, a partner of the Tob/BTG proteins, interacts with ERα and RXRβ and regulates their transcriptional activities,( 16 , 17 ) suggesting again that Tob/BTG proteins are involved in the regulation of gene expression through transcriptional mechanisms.
The CCR4‐NOT complex is conserved from yeast to humans and was initially described as a global regulator of transcription. The complex consists of Ccr4p/Cnot6, five NOT proteins (NOT1p‐NOT5p), Caf1p/Cnot7, Caf40p/Cnot9, and Caf130p/Cnot10.( 18 , 19 ) Caf1p and Ccr4p encode major yeast deadenylases, suggesting that this complex also plays a role in RNA degradation.( 19 ) Mammalian homologs of Ccr4p are Cnot6 and Cnot6L, and that of Caf1p is Cnot7 and Cnot 8. All are associated with deadenylase activity.( 20 , 21 ) Therefore, it appears that the human CCR4‐NOT complex has two functions in RNA metabolism, cytoplasmic function to regulate mRNA turnover and nuclear function to regulate mRNA synthesis. The mechanism by which these dual activities are coordinated remains largely elusive.
Intracellular signaling is mediated by protein–protein interaction as well as multiprotein complexes, and mass spectrometric analysis is commonly used to identify components of protein complexes. In the present study, we used a proteomics approach to identify components of the CCR4‐NOT complex as Tob‐interacting proteins. We provide evidence suggesting that Tob cooperates with the CCR4‐NOT complex to regulate the length of the poly(A) tail of mRNAs and thereby their translation.
Materials and Methods
Reagents. MG132, a proteasome inhibitor, was purchased from the Peptide Institute (Osaka, Japan). Phenylmethylsulfonyl fluoride (PMSF) was obtained from Sigma (St. Louis, MO). Aprotinin (Trazylol) was supplied by Bayer (Wayne, NJ). Anti‐Myc antibody (9E10) was from Santa Cruz Biotechnology (Santa Cruz, CA), and anti‐Flag antibody (M2) was from Sigma. Anti‐Tob antibodies were as described.( 22 ) Anti‐Cnot1 polyclonal antibodies were generated by immunizing rabbits with peptides corresponding to amino acids 21–40 of human Cnot1.
Expression plasmids. For construction of a full‐length cnot1 cDNA, a 5′‐proximal DNA fragment containing the translation start site was amplified by polymerase chain reaction from a HeLa cDNA library. We used the primers 5′‐CCGCTCGAG ATGAATCTTGACTCGCTCTCGCTGGCCTTG‐3′ and 5′‐GA GGGTCAATCTGAACAGGAGAAATGGCATCTAAG‐3′. The resulting 2149 bp polymerase chain reaction product was digested with XhoI and NheI then ligated into the NheI site of the KIAA1007 cDNA plasmid, which encodes the carboxyl terminus proximal region of cnot1 (Kazusa cDNA Bank). The sequence of the resulting full‐length cnot1 cDNA, which was re‐cloned into pME‐Flag( 23 ) vector, was confirmed with an ABI377 sequence analyzer from Applied Biosystems (Foster City, CA). The region between the NdeI site in the cnot1 cDNA and SpeI site on the vector or between the EcoRI site on the vector and the 3′ proximal PstI site in the cnot1 cDNA was deleted to generate a plasmid encoding the amino‐terminal 1700 amino acids or carboxyl‐terminal 700 amino acids of Cnot1, respectively. The resulting plasmids were termed pME‐Flag‐Cnot1(N) or pME‐Flag‐Cnot1(C), respectively. Expression plasmids for full‐length Tob (pME‐Flag‐Tob or pME‐Myc‐Tob), the amino‐terminal half of Tob (pME‐F‐TobN113), and the carboxyl‐terminal half of Tob (pME‐F‐TobC114) were as described.( 24 ) Expression plasmids for Cnot3 and Cnot6L (pcDNA‐Flag‐Cnot3 and pcDNA‐Flag‐Cnot6L) were as described.( 20 )
Cell culture. The HeLa S3 cell line was from the Riken Cell Bank (Tsukuba, Japan). HeLa S3 cells were maintained in suspension culture in Joklik's medium (JRH Biosciences, Lenexa, KS) containing 10% calf serum (HyClone, South Logan, UT). HeLa cells, HEK293T cells, and COS7 cells were cultured in Dulbecco's modified Eagles medium (Nissui, Tokyo, Japan) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA).
Establishment of stable HeLa S3 cell lines. Complementary DNA encoding Tob tagged with the Flag sequence at the carboxyl terminus was cloned in pME18S. The resulting expression plasmid, pME18S‐Flag‐Tob, and pApuro plasmid, containing the puromycin‐resistance cassette, were cotransfected into HeLa S3 cells with Lipofectamine Plus (Invitrogen) as described.( 20 ) Two days after transfection, puromycin was added to the culture medium to a final concentration of 1 µg/mL. Of the 81 isolated colonies, six were confirmed to stably express Tob‐Flag by Western blot analysis as described.( 25 )
Purification of the Tob complex and mass spectrometric analysis. Nuclear extracts were prepared from a total culture volume of 100 L according to the Dignam protocol.( 26 ) For batch purification of the Tob‐Flag‐containing protein complex, nuclear extracts were first incubated with 250 µL of M2 agarose beads (Sigma) for 4 h at 4°C with rotation. The precipitates were washed twice with 10 mL washing buffer 1 (500 mM KCl, 20 mM Tris‐HCl [pH 8.0], 5 mM MgCl2, 1 mM PMSF, 2 ng/mL aprotinin, and 50 µM MG‐132), 10 mL washing buffer 2 (300 mM KCl, 20 mM Tris‐HCl [pH 8.0], 5 mM MgCl2, 1 mM PMSF, 2 ng/mL aprotinin, and 50 µM MG‐132), and 10 mL washing buffer 3 (100 mM KCl, 20 mM Tris‐HCl [pH 8.0], 5 mM MgCl2, 10% glycerol, 0.1% Tween‐20, 10 mM β‐mercaptoethanol, 1 mM PMSF, 2 ng/mL aprotinin, and 50 µM MG‐132). Immunoprecipitated proteins were eluted from the M2 agarose beads in washing buffer 3 by competition with Flag peptide. The eluates were further incubated on a rotator for 2 h at 4°C with 20 µL Protein‐G Sepharose beads purchased from Amersham Pharmacia (Piscataway, NJ) conjugated to 3 µg of the anti‐Tob monoclonal antibody (4B1), and the beads were then precipitated. After the precipitates were washed five times with 1 mL washing buffer 3, precipitated proteins were eluted in 20 µL Laemmli buffer by incubation at 37°C for 30 min. Samples were separated on 5–20% sodium dodecyl sulfate–polyacrylamide gradient gels and stained with Coomassie Brilliant Blue. Bands were cut from the gel and digested in the gel with 1 pmol/µL trypsin (Promega, Madison, WI) for 12 h at 37°C. The eluted peptides were fractionated by reverse‐phase liquid chromatography. Fractions were mixed with α‐cyano‐4‐hydroxycinnamic acid and spotted onto a matrix‐assisted laser desorption ionization (MALDI) plate with a Probot (Applied Biosystems). The spotted peptides were sequenced with a MALDI–time of flight (TOF)/TOF mass spectrometer, ABI4700 (Applied Biosystems), and the fragment ion spectra generated were analyzed by MASCOT database search (Matrix Science, Boston, MA).
Co‐immunoprecipitation. Cells were transfected with the plasmids with FuGENE 6 transfection reagent (Roche, Basel, Switzerland). Twenty‐four hours after the transfection, cells were lyzed with TNE buffer (50 mM Tris‐HCl [pH 8.0], 100 mM NaCl, 1% Nonidet P‐40, 1 mM ethylenediaminetetraacetic acid, 1 mM PMSF, 2 ng/mL aprotinin, and 50 µM MG‐132). The insoluble fraction was removed by centrifugation. Supernatants were incubated with Protein‐G Sepharose beads and anti‐Flag antibody (M2) (Sigma) for 1 h at 4°C with rotation. Co‐immunoprecipitated proteins were subjected to Western blot analysis as described( 25 ) with anti‐Myc antibody (9E10), anti‐Flag antibody (M2), or anti‐Cnot1 antibodies.
Deadenylase assay. HEK293T cells were transfected with GFP‐Cnot6L expression plasmids with or without Tob expression plasmid. Forty‐eight hours after transfection, lysates were prepared from the cells and immunoprecipitated with anti‐GFP antibody. The precipitates were incubated at 37°C for 60 min with synthesized RNA substrate (5′‐UCUAAAUAAAA AAAAAAAAAAAAAAAA‐3′; final concentration, 0.1 µM) labeled with fluorescein‐isothiocyanate at the 5′‐end as described.( 20 ) The reaction products were fractionated on a 7 M urea–25% polyacrylamide denaturing gel. The gel was analyzed with an FLA‐5000 Fluorescence Imager (Fujifilm, Tokyo, Japan).
Results
Stable expression of Tob retards HeLa S3 growth. To obtain sufficient quantities of Tob‐interacting proteins, we transfected HeLa S3 cells with a Tob expression plasmid pME18S‐Flag‐Tob and pApuro, which contains a puromycin‐resistant gene. Most clones selected for puromycin resistance expressed Tob‐Flag, confirmed by Western blotting with an anti‐Flag antibody. Expression of Tob‐Flag in five selected clones is shown in Fig. 1a. Cells that stably expressed Tob‐Flag grew more slowly than the original HeLa S3 cells and mock‐transfected cells (Fig. 1b), as was expected from the antiproliferative activity of Tob.
Figure 1.
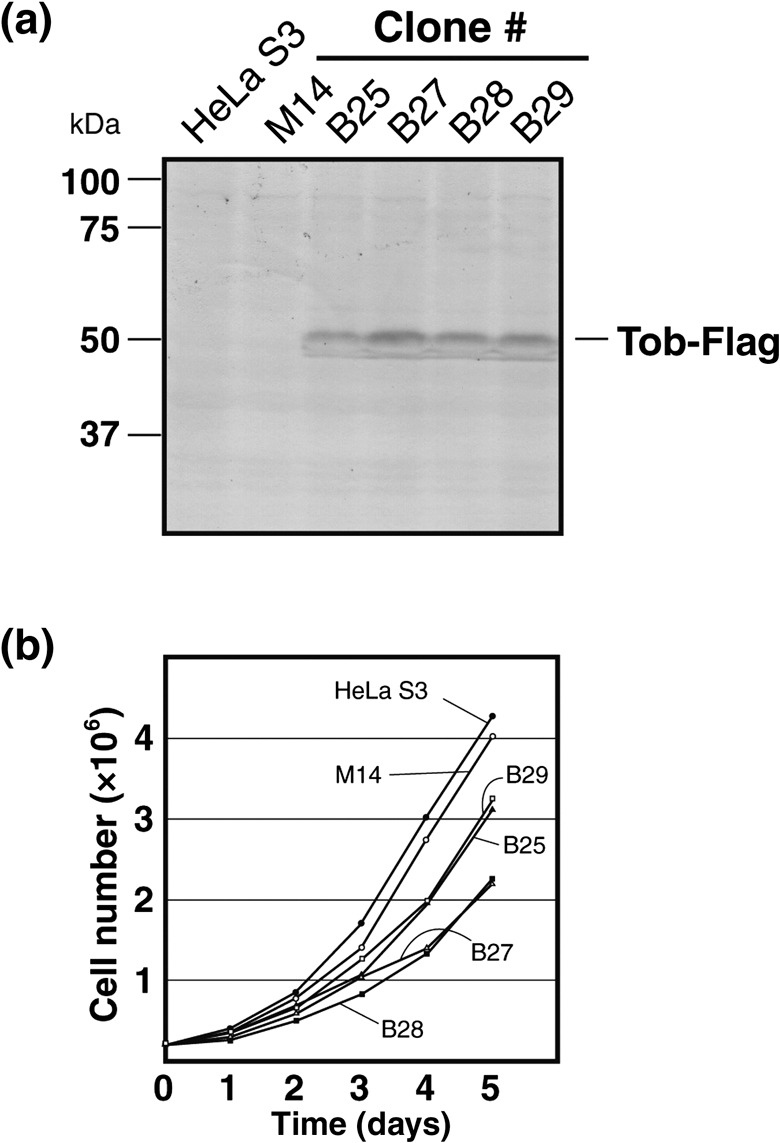
Establishment of HeLa S3 cell lines stably expressing Flag‐tagged Tob. (a) The expression of Tob‐Flag in HeLa S3 cell lines was confirmed by Western blotting with anti‐Flag antibody. (b) Each clone (2 × 105 cells) was seeded in five sets of 6‐cm cell culture dishes. After seeding, cells in each dish were trypsinized and counted for 5 consecutive days.
Identification of Tob‐associated proteins. To purify Tob‐associated proteins, we chose clone B29 from among the Tob‐Flag‐expressing HeLa S3 lines. After cells were cultured, their lysates were prepared and divided into nuclear and S100‐cytoplasmic fractions, then subjected to tandem‐affinity purification of Tob‐Flag with anti‐Flag and anti‐Tob monoclonal antibodies as illustrated in Fig. 2a. The purified proteins were separated by 5–20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and detected by Coomassie Brilliant Blue staining. Representative data for the nuclear fraction are shown in Fig. 2b. Proteins in bands indicated with lines were subjected to MALDI‐TOF/TOF mass spectrometry analysis, which identified components of the CCR4‐NOT complex, RNA helicases, RNA binding proteins, and tankyrase‐binding protein TAB182 as Tob‐interacting proteins (Table 1). Similarly, Cnot1, Cnot7, Cnot9, hnRNP H1, hnRNP H2, and TAB182 were identified in anti‐Flag immunoprecipitates of the S100‐cytoplasmic fraction (data not shown), suggesting that Tob forms similar, but not necessarily identical, protein complexes in the cytoplasm and nucleus.
Figure 2.
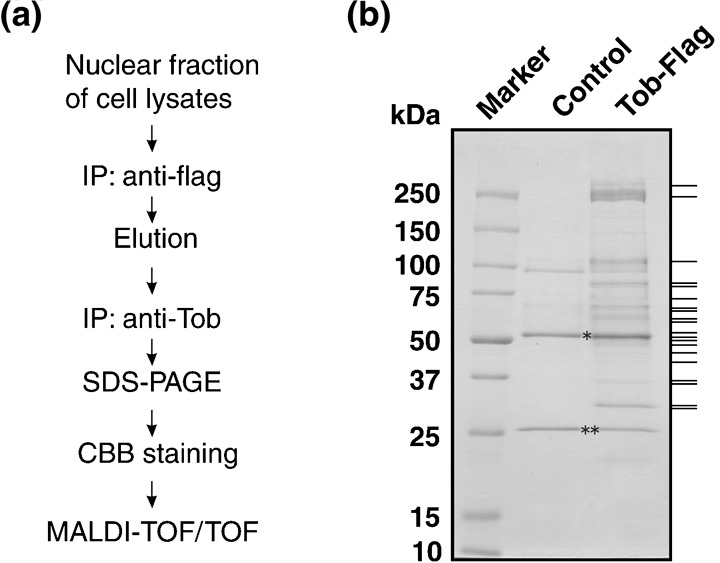
Purification of Tob‐containing protein complex. (a) Schematic diagram of the purification protocol. (b) Nuclear protein extracts were separated by 5–20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and stained with Coomassie Brilliant Blue (CBB), followed by in‐gel trypsin digestion. Proteins indicated by bars on the right were analyzed by matrix‐assisted laser desorption ionization–time of flight/time of flight (MALDI‐TOF/TOF) mass spectrometry. Immunoglobulin heavy chain and immunoglobulin light chain are indicated by * and **, respectively. IP, immunoprecipitation.
Table 1.
MASCOT scores and National Center for Biotechnology Information (NCBI) accession numbers of proteins in Tob‐containing complexes
| Identified proteins | Total score | Molecular weight (kDa) | Normalized score | NCBI accession number |
|---|---|---|---|---|
| Flag‐Tob | 431 | 38 131 | 565.0 | |
| CCR4‐NOT components | ||||
| Cnot1 (KIAA1007)§ | 1129 | 260 000 | 217.0 | NP_057368 |
| Cnot2 | 257 | 59 700 | 215.0 | NP_055330 |
| Cnot3 | 116 | 81 822 | 70.9 | NP_055331 |
| Cnot6L (CCR4 homolog) | 80 | 53 757 | 74.4 | NP_653172 |
| Cnot7 (Caf1) | 145 | 32 764 | 221.0 | Q9UIV1 |
| Cnot9 (Rcd1) | 117 | 33 160 | 176.0 | NP_005435 |
| Cnot10 | 116 | 82 258 | 70.5 | NP_056257 |
| RNA binding proteins | ||||
| Raver1 | 148 | 63 837 | 116.0 | NP_597709 |
| TLS/FUS | 166 | 53 323 | 156.0 | 1916411B |
| Sam68 | 156 | 48 197 | 162.0 | NP_006550 |
| TDP‐43 | 100 | 44 711 | 112.0 | NP_031401 |
| TIA‐1 | 42 | 42 934 | 48.9 | P31483 |
| hnRNPs | ||||
| hnRNP A2/B1 | 205 | 35 984 | 285.0 | NP_112533 |
| hnRNP F | 280 | 45 643 | 307.0 | NP_004957 |
| hnRNP H1 | 321 | 49 198 | 326.0 | NP_005511 |
| hnRNP H2 | 221 | 49 232 | 224.0 | NP_062543 |
| hnRNP H3 | 125 | 36 903 | 169.0 | NP_036339 |
| hnRNP K | 71 | 50 966 | 69.7 | NP_112553 |
| RNA helicase | ||||
| DDX3 | 118 | 73 198 | 80.6 | O00571 |
| DDX5 | 57 | 66 881 | 42.6 | AAB84094 |
| Other | ||||
| TAB182 (KIAA1741) | 1368 | 186 356 | 367.0 | BAB21832 |
Generated by the MASCOT database (Matrix Science, Boston, MA). Normalized score = (Total score) × 50 000/(Molecular weight).
Interaction of Tob with Cnot1, a scaffold protein of CCR4‐NOT. Because the yeast Not1 protein serves as a scaffold protein of the yeast CCR4‐NOT complex,( 27 ) we assumed that human Cnot1 might also act as a scaffold. Indeed our unpublished data suggested that depletion of Cnot1 destroys the formation of the CCR4‐NOT complex (data not shown). To substantiate the interaction between Tob and the CCR4‐NOT complex, we examined whether Cnot1 interacts with Tob. We transfected COS7 cells with pME‐Flag‐Cnot1 and pME‐Myc‐Tob and carried out co‐immunoprecipitation assays. The results showed that Myc‐Tob was present in Flag‐Cnot1 immunoprecipitates, suggesting either a direct or indirect interaction between the proteins (Fig. 3b). Furthermore, co‐imunoprecipitation assay with lysates prepared from COS7 cells transfected with pME‐Myc‐Tob together with pME‐Flag‐Cnot1(N) or pME‐Flag‐Cnot1(C) (Fig. 3a) revealed that both the amino‐terminal and carboxyl‐terminal sequences of Cnot1 participated in its association with Tob (Fig. 3b). We also carried out a reciprocal co‐immunoprecipitation assay using lysates of COS7 cells transfected with pME‐Flag‐Tob and found that endogenous Cnot1 was present in Flag‐Tob immunoprecipitates (Fig. 3c).
Figure 3.
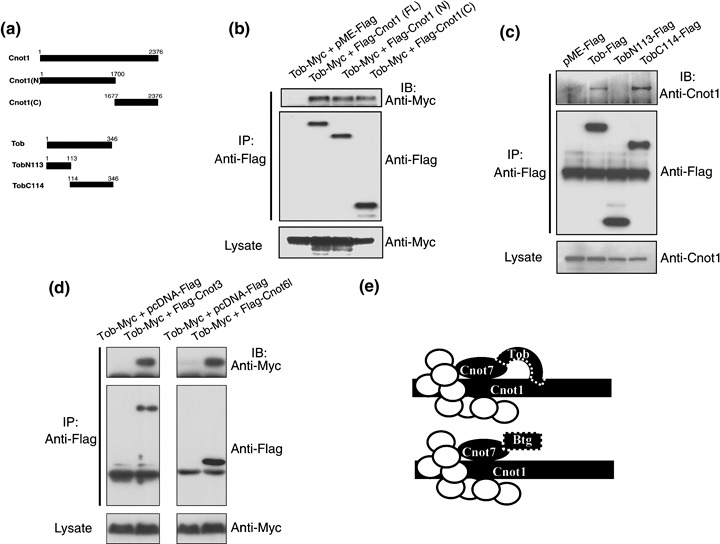
Domains relevant to the interaction between the Cnot1 scaffold protein and Tob. (a) Schematic diagram of Cnot1 and Tob expression constructs used in co‐immunoprecipitation assays. Cnot1(N), the amino terminal 1700 amino‐acid sequence of Cnot1; Cnot1(C), the carboxyl terminal 700 amino‐acid sequence of Cnot1; TobN113, amino terminal 113 amino‐acid sequence of Tob; TobC117, the carboxyl terminal 233 amino‐acid sequence of Tob. (b) The Flag‐tagged, full‐length Cnot1 (FL), Cnot1(N), or Cnot1(C) were exogenously expressed in COS7 cells with or without Myc‐tagged Tob. Proteins in the cell lysates were immunoprecipitated with anti‐Flag antibody, and the immunoprecipitates were separated by 7.5% (SDS‐PAGE) and subjected to Western blotting with the anti‐Myc (top panel) or anti‐Flag (middle panel) antibodies. Proteins in cell lysates were also separated by 7.5% SDS‐PAGE and subjected to Western blotting with the anti‐Myc antibody (bottom panel). (c) The Flag‐tagged, full‐length Tob, TobN113, or TobC114 were exogenously expressed in COS7 cells. Proteins in cell lysates were immunoprecipitated with anti‐Flag antibody. Western blot analyses of the cell lysates and the immunoprecipitates were carried out as in (b). (d) The Flag‐tagged, full‐length Cnot3, or Cnot6L in the pcDNA vector were exogenously expressed in COS7 cells with or without Myc‐tagged Tob. Proteins in the cell lysates were immunoprecipitated with anti‐Flag antibody. Western blot analyses of the cell lysates and the immunoprecipitates were carried out as in (b). (e) Schematic illustration of the interaction between Tob/BTG family proteins and the CCR4‐NOT complex. Subunits of the complex other than Cnot1 and Cnot7 are shown as white circles. Btg represents the family members except Tob. Tob is shown to be unique among the family proteins in that it likely interacts with the CCR4‐NOT complex through Cnot1 as well as through Cnot7.
Next, to examine the region of Tob responsible for the interaction with Cnot1, we transfected COS7 cells with pME‐F‐TobN113 encoding amino‐terminal 113 amino acids of Tob, or pME‐F‐TobC114 encoding carboxyl‐terminal 232 amino acids of Tob (residues 114–345) (Fig. 3a). Co‐immunoprecipitation assay revealed that Cnot1 bound the carboxyl‐terminal 232 amino‐acid sequence of Tob (Fig. 3c).
Because Cnot1 is thought to be a scaffold protein of the CCR4‐NOT complex, we wanted to confirm that other components of the complex interact with Tob. Using the lysates of the cells transfected with pME‐Tob‐myc and pcDNA‐Flag‐Cnot3 or pcDNA‐Flag‐Cnot6L, we showed that Tob was coprecipitated with Cnot3 and with Cnot6L (Fig. 3d).
Effects of Tob on deadenylase activity of CCR4‐NOT. We previously showed that anti‐Cnot6L immunoprecipitates contained components of the CCR4‐NOT complex, including Cnot1, Cnot2, Cnot3, Cnot8, and Cnot9, and were associated with deadenylase activities.( 20 ) To address the biologic significance of the interaction between Tob and the CCR4‐NOT complex, we examined whether the deadenylase activity of the CCR4‐NOT complex is affected by Tob using anti‐Cnot6L immunoprecipitates. HEK293T cells were transfected with green fluorescent protein (GFP)‐Cnot6L expression plasmid with or without pME‐Flag‐Tob. Twenty‐four hours after transfection, cell lysates were prepared, and Cnot6L and associated proteins were purified with anti‐GFP antibodies (Fig. 4a). We then incubated the purified proteins with a poly(A) RNA substrate and carried out the deadenylase assay. Analysis of reaction products on a denaturing sequencing gel revealed that cleavage of the poly(A) RNA substrate occurred as reported previously( 20 ). Importantly, anti‐GFP immunoprecipitates from the pME‐Flag‐Tob cotransfected cells contained the Flag‐Tob protein (Fig. 4a) and deadenylated the poly(A) substrates less efficiently (Fig. 4b). The data suggested that the in vitro deadenylase activity of the CCR4‐NOT complex was negatively regulated by Tob.
Figure 4.

Tob suppresses the deadenylase activity of Cnot6L in the CCR4‐NOT complex. (a) Green fluorescent protein (GFP)‐Cnot6L was expressed exogenously with or without Flag‐Tob in HEK293T cells. Proteins in the cell lysates were immunoprecipitated with anti‐GFP antibody, and the immunoprecipitates were subjected to immunoblot analysis with anti‐GFP (top panel) or anti‐Flag (bottom panel) antibody. (b) Anti‐GFP immunoprecipitates from HEK293T cells expressing GFP‐Cnot6L alone or GFP‐Cnot6L with Flag‐Tob were incubated in vitro with 5′‐fluorescein‐isothiocyanate‐labeled RNA substrate (RNA‐3′‐20 As) for 60 min. The labeled RNA was then analyzed on a denaturing sequencing gel.
Discussion
The CCR4‐NOT complex is well conserved in eukaryotes and consists of multiple core subunits.( 18 , 19 ) In mammals, the core subunits include Cnot1–4, Cnot6/CCR4a, Cnot6L/CCR4b, Cnot7/Caf1a, Cnot8/Caf1b, Cnot9/Caf40, and Cnot10/Caf130. Here we showed that Tob protein associates with the components of the CCR4‐NOT complex except Cnot4, Cnot6, and Cnot8. Because these three proteins are found in the CCR4‐NOT complex (see Table 2), we assume that Tob could associate with the whole CCR4‐NOT complex. The reason we failed to identify Cnot4, Cnot6, and Cnot8 in the Tob immunoprecipitates is unclear. Because Cnot7/Caf1 interacts not only with Tob, but also with BTG1, Tob2, and ANA,( 7 , 16 , 28 , 29 ) we assume that all Tob/BTG family proteins associate with the CCR4‐NOT complex. Tob and BTG1 can be cocrystallized with Cnot7 (data not shown). Therefore, their direct interactions with Cnot7 likely play an important role in their associations with the CCR4‐NOT complex. X‐ray crystallography‐based structural analysis as well as co‐immunoprecipitation analysis revealed that amino‐terminal amino acids of the B box and the flanking sequence are important for the Tob–Cnot7 interaction (data not shown and ( 16 )). In addition, we found that Tob interacts with two different regions of Cnot1, and that the carboxyl‐terminal domain of Tob, which is either missing or not conserved in the other family members, is responsible for the interaction with Cnot1 (Fig. 3). There are no sequences that are repeatedly present (even homologous to each other) at the two different regions of Cnot1. Accordingly, no sequence responsible for the interaction is identified. It is also not known whether this interaction is direct or indirect. It has been reported that yeast Caf1/Cnot7 directly binds to the conserved amino‐terminal domain of yeast Cnot1,( 30 ) suggesting that the interaction of Tob with Cnot1 might be mediated at least in part through Cnot7. In any event, Tob seems to be unique in its family of proteins in that it could associate with the CCR4‐NOT complex through its interaction with Cnot1, in addition to its interaction with Caf1/Cnot7 (Fig. 3e). It is possible that the Tob C‐terminal region can also associate with and that association of Tob and the CCR4‐NOT complex always occurs through Cnot7. Further studies are needed to unravel the exact mechanisms of the association of the components in the Tob–CCR4‐NOT complex.
Table 2.
Comparison of the CCR4‐NOT components identified in this study with those identified previously
| Present study | Previous report( 20 ) | Previous report( 18 ) |
|---|---|---|
| Tob | — | — |
| Cnot1 | Cnot1 | KIAA1007 (Cnot1) |
| Cnot2 | Cnot2 | Not2H (Cnot2) |
| Cnot3 | Cnot3 | — |
| — | Cnot6/CCR4a | KIAA1194 (Cnot6) |
| Cnot6L | Cnot6L/CCR4b | — |
| Cnot7 | Cnot7/Caf1a | CAF1 (Cnot7) |
| — | Cnot8/Caf1b | CALIF (Cnot8) |
| Cnot9 | Cnot9/Caf40 | RQCD1 (Cnot9) |
| Cnot10 | Cnot10 | AAH02928 (Cnot10) |
, not detect
The in vitro deadenylase activity of Cnot6L was partially suppressed by Tob (Fig. 4). Although the anti‐Cnot6L immunoprecipitates contained components of the CCR4‐NOT complex, including other deadenylases, Cnot6, Cnot7, and Cnot8, we assume that the deadenylase activity seen in Fig. 4 largely represents that of Cnot6L. Therefore, it is unclear whether the activities of other deadenylases, Cnot6, Cnot7, and Cnot8, are affected by Tob. Nevertheless, the data shown in Fig. 4 strongly suggest that Tob could be involved in control of the length of the poly(A) tail of some mRNAs, and thereby would influence their stability and translation efficiency. It may be worthy noting that Tob interacts with poly(A)‐binding proteins PABP and iPABP and is involved in translational control.( 24 ) Although Tob is predominantly localized in the nucleus, it can shuttle between the nucleus and cytoplasm, because it has both nuclear localization and nuclear exclusion sequences.( 31 ) It is possible that cytoplasmic Tob facilitates access of the CCR4‐NOT deadenylase complex to the poly(A) tails of mRNAs through its interaction with PABP and/or iPABP. The possible tripartite interaction of Tob, CCR4‐NOT, and PABP/iPABP would destabilize target mRNAs, resulting in suppression of translation. Ezzeddine et al. recently reported that Tob interacts with PABPC1 and Cnot6/Cnot7 simultaneously and regulates deadenylation positively.( 32 ) However, the scenario could be more complicated because, as we show in this report, Tob inhibits deadenylase activity in vitro, which could stabilize mRNA and enhance translation. Further studies are needed to clarify how the deadenylase activity of the CCR4‐NOT complex is regulated by Tob and/or PABP in vivo.
Finally, we found that Tob interacts not only with the CCR4‐NOT complex but also with RNA helicases (DDX3 and DDX5), RNA‐binding proteins (Raver1, TLS/FUS, Sam68, TDP‐43, TIA‐1, hnRNP A2/B1, hnRNP F, hnRNP H1, hnRNP H2, hnRNP H3, and hnRNP K) and TAB182 tankylase‐binding protein (Table 1). Some of these proteins are implicated in pre‐mRNA splicing, mRNA trafficking, and translation. For instance, α‐tropomyosin mRNA is a target of Raver1,( 33 ) cd44 mRNA of Sam68,( 34 ) and cftr of TDP‐43,( 35 ) for mRNA splicing. Cox‐2 expression is silenced at the translational level by the Au‐rich element‐binding protein TIA‐1.( 36 ) According to the HyNet database, an extensive human yeast two‐hybrid protein interaction network database (http://www.prolexys.com/public/Prolexys_Human_Interactome.pdf), Cnot1 binds directly to DDX5 and TLS/FUS. This is consistent with our observation that TLS/FUS RNA‐binding protein and CCR4‐NOT proteins are in the same Tob‐associated complex. It is unclear how the interactions of the proteins identified in the present study with Tob are regulated. Individual Tob molecules might interact with different proteins. However, because most of the identified proteins bind RNAs, our findings strongly suggest that the antiproliferative effects of Tob could be due to deregulation of not only transcription but also RNA metabolism that includes destabilization of mRNAs.
Acknowledgments
We thank L. Amy, J. Sawada, S. Kato, S. Takezawa, S. Ishii, M. Watanabe, and T. Nakamura for helpful discussions. This work was supported by grants‐in‐aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was supported in parts by grant‐in‐aid for priority area of cancer research from MEXT Japan, research fund from the Uehara Memorial Foundation, and Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms). MEXT, Japan. MM was supported by grant‐in‐aid from the Japan Society for the Promotion of Science (JSPS), Japan.
References
- 1. Tirone F. The gene PC3 (TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol 2001; 187: 155–65. [DOI] [PubMed] [Google Scholar]
- 2. Rouault JP, Rimokh R, Tessa C et al . BTG1, a member of a new family of antiproliferative genes. EMBO J 1992; 11: 1663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Matsuda S, Kawamura‐Tsuzuku J, Ohsugi M et al . Tob, a novel protein that interacts with p185erbB2, is associated with anti‐proliferative activity. Oncogene 1996; 12: 705–13. [PubMed] [Google Scholar]
- 4. Montagnoli A, Guardavaccaro D, Starace G, Tirone F. Overexpression of the nerve growth factor‐inducible PC3 immediate early gene is associated with growth inhibition. Cell Growth Differ 1996; 7: 1327–36. [PubMed] [Google Scholar]
- 5. Rouault JP, Falette N, Guehenneux F et al . Identification of BTG2, an antiproliferative p53‐dependent component of the DNA damage cellular response pathway. Nature Genet 1996; 14: 482–6. [DOI] [PubMed] [Google Scholar]
- 6. Yoshida Y, Matsuda S, Ikematsu N et al . ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene 1998; 16: 2687–93. [DOI] [PubMed] [Google Scholar]
- 7. Ikematsu N, Yoshida Y, Kawamura‐Tsuzuku J et al . Tob2, a novel anti‐proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin‐dependent kinases. Oncogene 1999; 18: 7432–41. [DOI] [PubMed] [Google Scholar]
- 8. Buanne P, Corrente G, Micheli L et al . Cloning of PC3B, a novel member of the PC3/BTG/TOB family of growth inhibitory genes, highly expressed in the olfactory epithelium. Genomics 2000; 68: 253–63. [DOI] [PubMed] [Google Scholar]
- 9. Jia S, Meng A. Tob genes in development and homeostasis. Dev Dynamics 2007; 236: 913–21. [DOI] [PubMed] [Google Scholar]
- 10. Rodier A, Marchal‐Victorion S, Rochard P et al . BTG1: a triiodothyronine target involved in the myogenic influence of the hormone. Exp Cell Res 1999; 249: 337–48. [DOI] [PubMed] [Google Scholar]
- 11. Yoshida Y, Tanaka S, Umemori H et al . Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 2000; 103: 1085–97. [DOI] [PubMed] [Google Scholar]
- 12. Tzachanis D, Freeman GJ, Hirano N et al . Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nature Immunol 2001; 2: 1174–82. [DOI] [PubMed] [Google Scholar]
- 13. Prevot D, Voeltzel T, Birot AM et al . The leukemia‐associated protein Btg1 and the p53‐regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem 2000; 275: 147–53. [DOI] [PubMed] [Google Scholar]
- 14. Guardavaccaro D, Corrente G, Covone F et al . Arrest of G(1)–S progression by the p53‐inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol 2000; 20: 1797–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshida Y, Nakamura T, Komoda M et al . Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Gene Dev 2003; 17: 1201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prevot D, Morel AP, Voeltzel T et al . Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J Biol Chem 2001; 276: 9640–8. [DOI] [PubMed] [Google Scholar]
- 17. Nakamura T, Yao R, Ogawa T et al . Oligo‐astheno‐teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nature Genet 2004; 36: 528–33. [DOI] [PubMed] [Google Scholar]
- 18. Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4‐NOT complex identifies two novel components of the complex. J Mol Biol 2001; 314: 683–94. [DOI] [PubMed] [Google Scholar]
- 19. Collart MA. Global control of gene expression in yeast by the Ccr4‐Not complex. Gene 2003; 313: 1–16. [DOI] [PubMed] [Google Scholar]
- 20. Morita M, Suzuki T, Nakamura T et al . Depletion of mammalian CCR4b deadenylase triggers elevation of the p27kip1 mRNA level and impairs cell growth. Mol Cell Biol 2007; 27: 4980–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Viswanathan P, Ohn T, Chiang YC et al . Mouse CAF1 can function as a processive deadenylase/3′‐5′‐exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem 2004; 279: 23988–95. [DOI] [PubMed] [Google Scholar]
- 22. Suzuki T, Kawamura‐Tsuzuku J, Ajima R et al . Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras‐mediated cell proliferation and transformation. Genes Dev 2002; 16: 1356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tokai‐Nishizumi N, Ohsugi M, Suzuki E, Yamamoto T. The chromokinesin Kid is required for maintenance of proper metaphase spindle size. Mol Biol Cell 2005; 16: 5455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T. Interaction of anti‐proliferative protein Tob with poly(A)‐binding protein and inducible poly(A)‐binding protein: implication of Tob in translational control. Gene Cell 2005; 10: 151–63. [DOI] [PubMed] [Google Scholar]
- 25. Nakazawa T, Watabe AM, Tezuka T et al . p250GAP, a novel brain‐enriched GTPase‐activating protein for Rho family GTPase‐activating protein for Rho family GTPases, is involved in N‐methyl‐D‐aspartate receptor signaling. Mol Biol Cell 2003; 14: 2921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nuclei Acid Res 1983; 11: 1475–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4‐NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol 1999; 19: 6642–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rouault JP, Prevot D, Berthet C et al . Interaction of BTG1 and p53‐regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem 1998; 273: 22563–9. [DOI] [PubMed] [Google Scholar]
- 29. Yoshida Y, Hosoda E, Nakamura T, Yamamoto T. Association of ANA, a member of the antiproliferative Tob family proteins, with a Caf1 component of the CCR4 transcriptional regulatory complex. Jpn J Cancer Res 2001; 92: 592–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu HY, Badarinarayana V, Audino DC et al . The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J 1998; 17: 1096–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawamura‐Tsuzuku J, Suzuki T, Yoshida Y, Yamamoto T. Nuclear localization of Tob is important for regulation of its antiproliferative activity. Oncogene 2004; 23: 6630–8. [DOI] [PubMed] [Google Scholar]
- 32. Ezzeddine N, Chang TC, Zhu Y et al . Human TOB, an anti‐proliferative transcription factor, is a PABP‐dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 2007; 27: 7791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gromak N, Rideau A, Southby J et al . The PTB interacting protein raver1 regulates alpha‐tropomyosin alternative splicing. EMBO J 2003; 22: 6356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matter N, Herrlich P, Konig H. Signal‐dependent regulation of splicing via phosphorylation of Sam68. Nature 2002; 420: 691–5. [DOI] [PubMed] [Google Scholar]
- 35. Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP‐43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 2001; 20: 1774–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dixon DA, Balch GC, Kedersha N et al . Regulation of cyclooxygenase‐2 expression by the translational silencer TIA‐1. J Exp Med 2003; 198: 475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]


