Abstract
The clinical significance of aberrant promoter methylation of the canonical Wnt pathway antagonist genes (sFRP1, sFRP2, sFRP4, sFRP5, Wif1, Dkk3, and Hdpr1) and also putative tumor‐suppressor gene Wnt5a, belonging to the non‐canonical Wnt signaling pathway, was investigated in a large series of 75 patients with Philadelphia chromosome‐positive acute lymphoblastic leukemia by methylation‐specific polymerase chain reaction. At least one methylated gene was observed in cells from 66% (49/75) of patients (methylated group). Disease‐free survival and overall survival at 9 years were 51 and 40%, respectively, for the unmethylated group and 3 and 2%, respectively, for the methylated group (both P < 0.0001). Multivariate analysis demonstrated that the Wnt methylation profile was an independent prognostic factor predicting disease‐free survival (P = 0.007) and overall survival (P = 0.039). Abnormal DNA methylation of promoter‐associated CpG islands in the Wnt signaling pathway is very common in Philadelphia chromosome‐positive acute lymphoblastic leukemia and potentially defines subgroups with distinct clinical characteristics. (Cancer Sci 2008; 99: 1865–1868)
The vast majority of patients with Philadelphia chromosome‐positive (Ph+) acute lymphoblastic leukemia (ALL) have a poor outcome when treated with chemotherapy alone, leading most investigators to favor allogeneic stem cell transplantation (SCT) for patients in first remission.( 1 ) Despite this overall dismal prognosis, it has been reported that patients with classic Ph+ ALL who receive extensively reinforced early chemotherapy show long‐term disease‐free survival (DFS), suggesting that a subset of Ph+ cases may be curable with intensive chemotherapy without SCT.( 2 ) However, specific risk markers that identify such patients are unknown.
Our group has demonstrated that the methylation of cytosine nucleotides in ALL cells is the most important way to inactivate cancer‐related genes in this disease. This epigenetic event can help to inactivate tumor‐suppressive apoptotic or growth‐arresting responses and has prognostic impacts in B‐ and T‐ALL.( 3 , 4 ) In fact, we have recently reported that epigenetic regulation is responsible at least in part for the activation of canonical Wnt signaling in ALL.( 5 ) We found that expression of the Wnt inhibitors sFRP1, sFRP2, sFRP4, sFRP5, Wif1, Dkk3, and Hdpr1 was downregulated due to abnormal promoter methylation in ALL cell lines and this event was associated with constitutive activation of the Wnt signaling pathway in ALL patients, as demonstrated by upregulation of the Wnt target genes Wnt16, Fz3, Tcf1, Lef1, and cyclin D1, and the nuclear localization of β‐catenin. In addition, we have also demonstrated that the putative tumor‐suppressor gene Wnt5a, belonging to the non‐canonical Wnt pathway, is silenced by methylation in ALL and associated with further upregulation of cyclin D1 expression.( 6 ) Interestingly, hypermethylation of the gene promoters of Wnt inhibitors and Wnt5a was observed in Ph+ ALL‐derived cell lines and was associated with downregulation of gene expression as demonstrated by restored gene expression after treatment with 5‐Aza‐2′‐deoxycytidine in these cell lines.( 5 , 6 ) Taken together, these data provide a rationale for the hypothesis that epigenetic disregulation of the Wnt signaling pathway is a potential mechanism that contributes to the pathogenesis and, probably, the clinical course of Ph+ ALL.
In the present study, we report that the methylation phenotype of Wnt pathway genes is able to redefine the prognostic impact of the Philadelphia chromosome in ALL patients.
Material and Methods
Patients. We studied 42 male and 33 female patients (total of 75) with classical de novo Ph+ ALL who were enrolled in successive multicenter studies of the ‘Programa para el estudio y tratamiento de las hemopatias malignas’ (PETHEMA) Spanish study group. All of these patients were referred to the Reina Sofia Hospital of Cordoba, Spain, from January 1989 to December 2004. The median age at diagnosis in the study population as a whole was 40 years (range 2–82 years). Of these patients, 12 were children (median age 8 years; range 2–14 years) and 63 presented with adult ALL (median age 46 years; range 19–82 years). The study was approved by the Investigational Review Board at Reina Sofia Hospital in accordance with the policies of the Department of Health and Human Services. Informed consent was obtained from the patient or the patient's guardians. Diagnosis was established according to standard morphological, cytochemical, and immunophenotypic criteria together with Philadelphia chromosome detection by cytogenetics or BCR‐ABL transcript detection by reverse transcription–polymerase chain reaction (PCR). Patients were entered in ALL protocols for high‐risk patients of the PETHEMA group. No patient showed additional chromosomal abnormalities. Forty‐four patients relapsed. Twenty‐four patients received SCT (1 autologous, 23 allogeneic) in the first (n = 14) or second (n = 10) complete remission (CR). There are 13 patients currently alive. The clinical characteristics of the patients are listed in Table 1.
Table 1.
Clinical characteristics and outcome of 75 Philadelphia‐positive acute lymphoblastic leukemia patients according to Wnt methylation status
| Feature | Non‐methylated (n = 26) | Methylated (n = 49) | P‐value |
|---|---|---|---|
| Age (n) | 0.002 | ||
| <15 years | 9 | 3 | |
| ≥15 years | 17 | 46 | |
| <55 years | 11 | 27 | |
| ≥55 years | 6 | 19 | |
| Sex (male/female) (n) | 15/11 | 27/22 | Not significant |
| White blood cells (n) | Not significant | ||
| <50 × 109/L | 16 | 25 | |
| ≥50 × 109/L | 10 | 20 | |
| Cell phenotype (n) | Not significant | ||
| Pro‐B | 1 | 3 | |
| Pre‐B | 7 | 8 | |
| Common | 16 | 37 | |
| Biphenotypic | 2 | 1 | |
| Stem cell transplantation (n) | 9 | 15 | Not significant |
| Best response (n) | |||
| Complete remission | 23 | 34 | 0.04 |
| Relapse (n) | 10 | 34 | <0.001 |
| Death (n) | 14 | 48 | <0.001 |
Methylation‐specific PCR. Bone marrow specimens were obtained from all patients at the time of diagnosis. High molecular weight DNA was prepared from mononuclear diagnostic marrow cells using conventional methods, frozen at –80°C, and analyzed retrospectively to assess the role of the methylation profile. In all cases, the diagnostic bone marrow sample contained at least 70% blast cells. Aberrant promoter methylation of the sFRP1, sFRP2, sFRP4, sFRP5, Wif1, Dkk3, Hdpr1, and Wnt5a genes was determined by methylation‐specific PCR after bisulfite treatment of DNA, as reported by Herman et al.( 7 ) Primer sequences of each gene for the unmethylated and methylated reactions have been reported elsewhere.( 5 , 6 )‘Hot start’ PCR was carried out for 30 cycles consisting of denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min, followed by a final 7‐min extension for all primer sets. The products were separated by electrophoresis on a 2% agarose gel. Bone marrow DNA from healthy donors was used as a negative control for methylation‐specific assays. Human male genomic DNA universally methylated for all genes (Intergen Company, Purchase, NY, USA) was used as a positive control for methylated alleles. Water blanks were included with each assay. The results were confirmed by repeat methylation‐specific PCR assays after an independent bisulfite treatment.
Statistical analysis. For statistical purposes, Ph+ ALL patients were classified into two different methylation groups according to the numbers of Wnt pathway genes methylated: the non‐methylated group (no methylated genes) and the methylated group (at least one methylated gene). This classification was derived after statistical analyses showing that the prognosis of patients with one to six methylated genes was similar with no significant differences (data not shown). P‐values for comparisons of continuous variables between groups of patients were two‐tailed and based on the Wilcoxon rank sum test. P‐values for dichotomous variables were based on the Fisher exact test. The remaining P‐values were based on the Pearson χ2‐test. Overall survival (OS) was measured from the day of diagnosis until death from any cause and was censored only for patients known to be alive at last contact. DFS was calculated from the date of first CR until the date of first relapse or the date of death in first CR. Patients alive and still in remission at the last follow‐up examination were censored in the analysis. Patients who underwent SCT were included but censored at the date of transplant. Distributions of OS and DFS curves were estimated by the method of Kaplan and Meier, with 95% confidence intervals calculated by means of Greenwood's formula. Comparisons of OS or DFS between groups were based on the log‐rank test. Univariate and multivariate analyses (logistic regression model) were used to determine the factors associated with DFS and OS. Stepwise modelling was carried out to screen potential variables for inclusion in the final model. The entry criterion for the multivariable Cox regression analysis was a P‐value less than 0.1 in univariate analysis. P‐values of no more than 0.05 were taken as the threshold for statistical significance in the final model. All relapse and survival data were updated in December 2007, and all follow‐up data were censored at that point.
Results and Discussion
Frequency of promoter methylation in ALL. Among the 75 Ph+ ALL, the methylation frequencies (in descending order) were as follows: 40% for Wnt5a; 23% for sFRP5; 21% for Dkk3; 20% for sFRP1; 13% for Hdpr1; 13% for Wif1; 12% for sFRP4; and 5% for sFRP2. No methylated genes (non‐methylated group) were found in 26 of 75 patients (34%) whereas most ALL (49 of 75 [66%]) had methylation of at least one gene (methylated group), ranging from one to six methylated genes.
Clinical outcome and promoter methylation profile. As shown in Table 1, aberrant methylation was observed more frequently in adults (73%) than in children (25%; P = 0.002). Among adults, elderly patients (≥55 years) showed the same level of methylation (76 vs 71%) as adolescents and young adults (<55 years). However, other features, including the number of patients who received SCT, were similarly distributed among both methylation groups. Table 1 also details the relapse history, CR rates, and mortality for patients included in the different methylation groups. The CR rates of patients in the non‐methylated and methylated groups were 88 and 69%, respectively (P = 0.04), suggesting that methylation profile correlates with response to remission induction therapy. Moreover, patients in the non‐methylated group had a lower relapse rate than patients in the methylated group (38 vs 69%; P < 0.001). Mortality rate was also lower for the non‐methylated group compared with the methylated group (54 vs 98%; P < 0.001).
We analyzed the DFS among patients who achieved CR according to the methylation profile. Estimated DFS at 9 years were 51 and 3% for the non‐methylated and methylated groups, respectively (P < 0.0001; Fig. 1a). The actual OS at 9 years calculated for all leukemic patients was 40% for non‐methylated patients and 2% for methylated patients (P < 0.0001; Fig. 1b).
Figure 1.
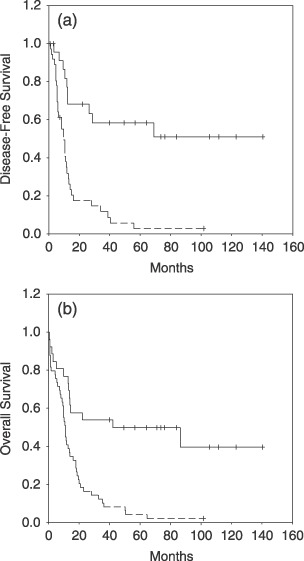
Kaplan–Meier survivor function for BCR‐ABL‐positive acute lymphoblastic leukemia patients. (a) Disease‐free survival and (b) overall survival (OS) curves for all of the patients enrolled in this study according to methylation profile. Solid lines, non‐methylated patients. Dashed lines, methylated patients. The Log‐rank test was significant for both DFS (P < 0.0001) and OS (P < 0.0001).
A multivariate analysis of potential prognostic factors demonstrated that the hypermethylation profile was an independent prognostic factor in predicting DFS (P = 0.007) as well as OS (P = 0.039; Table 2).
Table 2.
Multivariate Cox model for disease‐free survival and overall survival
| Feature | P‐value | |
|---|---|---|
| Univariate analysis | Multivariate analysis | |
| Disease‐free survival | ||
| Wnt methylation status | <0.001 | 0.007 |
| Age > 15 years | <0.001 | 0.02 |
| Sex | 0.1 | 0.09 |
| White blood cells > 50 × 109/L | 0.1 | 0.65 |
| Overall survival | ||
| Wnt methylation status | <0.001 | 0.039 |
| Age > 15 years | <0.001 | 0.006 |
| Sex | 0.1 | 0.18 |
| White blood cells > 50 × 109/L | 0.1 | 0.72 |
The paucity of Ph+ ALL long‐term survivors has made it difficult to recognize potential prognostic factors in order to distinguish good‐risk groups from poor responders. Two studies have demonstrated that in patients with early steroid response or showing a white blood cell count below 25 × 109/L the outcome is better.( 2 , 8 ) Moreover, variant forms of Ph+ chromosome or complex translocations confer a better prognosis than does classic rearrangement, whereas monosomy 7 is associated with dismal outcomes.( 9 , 10 , 11 , 12 ) However, the biological features of prognostic importance are not easily available in classical Ph+ ALL.
Our present findings indicate that classic t(9;22) ALL is a more heterogeneous disease than previously suspected, at least from an epigenetic point of view. Our data show that methylation in human Ph+ ALL cells participates in deregulation of the Wnt signaling pathway by inactivation of its antagonists Dkk3, sFRP1, sFRP2, sFRP5, Hdpr1, and Wif1 and also by activating non‐canonical signaling by silencing Wnt5a. Aberrant methylation of CpG islands is quantitatively different in individual tumors within the same tumor type, and this patient‐specific methylation profile provides important prognostic information in BCR‐ABL‐positive ALL patients treated with the same therapeutic protocol and with a long follow up. The presence in individual tumors of epigenetic events that affect the Wnt pathway is a factor of poor prognosis in this disease. Patients with methylation of genes had a poorer DFS and OS than patients with no methylated genes. Multivariate analysis confirmed that the methylation profile was associated with a shorter DFS and OS. Therefore, Wnt methylation profiling in BCR‐ABL‐positive ALL could provide important clinical information for: (i) guiding the selection of therapy; and (ii) providing a basis for developing novel therapies. Currently, the optimal treatment of Ph+ ALL patients requires the addition of BCR‐ABL tyrosine kinase inhibitors, such as imatinib. Combined with chemotherapy or as a single agent, it can produce high rates of CR. All of our patients were included in the protocol before the imatinib era, so the role of this therapy in the context of Wnt methylation could not be evaluated. However, our present results suggest that the use of demethylating agents like decitabine and 5‐aza‐2′‐deoxycytidine or specific inhibitors of Wnt pathway therapies such as quercetin and antagonists of the oncogenic Tcf–β‐catenin complex may be a useful therapeutic strategy in Ph+ ALL for whom SCT is either not feasible or refused.
Acknowledgments
The present study was supported by grants from Beca Ortiz de Landázuri 2006, Departamento de Salud‐Gobierno de Navarra, Fondo de Investigación Sanitaria (Spain) PI060285, PI070602, PI070608, PI060003, PI030141, PI030661, PI021299, and ISCIII‐RETIC RD06/0020, Junta de Andalucia 03/0143, 03/0144, 06/0356, and PI‐0004/2007 and funds from IMABIS (Malaga, Spain), Fundación de Investigación Médica Mutua Madrileña Automovilista; Asociacion Medicina e Investigacion and ‘UTE project CIMA’.
References
- 1. Apostolidou E, Swords R, Alvarado Y, Giles FJ. Treatment of acute lymphoblastic leukaemia: a new era. Drugs 2007; 67: 2153–71. [DOI] [PubMed] [Google Scholar]
- 2. Ribeiro RC, Broniscer A, Rivera GK et al . Philadelphia chromosome‐positive acute lymphoblastic leukemia in children: durable responses to chemotherapy associated with low initial white blood cell counts. Leukemia 1997; 11: 1493–6. [DOI] [PubMed] [Google Scholar]
- 3. Roman‐Gomez J, Jimenez‐Velasco A, Agirre X et al . Lack of CpG island methylator phenotype defines a clinical subtype of T‐cell acute lymphoblastic leukemia associated with good prognosis. J Clin Oncol 2005; 23: 7043–9. [DOI] [PubMed] [Google Scholar]
- 4. Roman‐Gomez J, Jimenez‐Velasco A, Castillejo JA et al . Promoter hypermethylation of cancer‐related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood 2004; 104: 2492–8. [DOI] [PubMed] [Google Scholar]
- 5. Roman‐Gomez J, Cordeu L, Agirre X et al . Epigenetic regulation of Wnt‐signaling pathway in acute lymphoblastic leukemia. Blood 2007; 109: 3462–9. [DOI] [PubMed] [Google Scholar]
- 6. Roman‐Gomez J, Jimenez‐Velasco A, Cordeu L et al . WNT5A, a putative tumour suppressor of lymphoid malignancies, is inactivated by aberrant methylation in acute lymphoblastic leukaemia. Eur J Cancer 2007; 43: 2736–46. [DOI] [PubMed] [Google Scholar]
- 7. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation‐specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996; 93: 9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schrappe M, Aricò M, Harbott J et al . Philadelphia chromosome‐positive (Ph+) childhood acute lymphoblastic leukemia: good initial steroid response allows early prediction of a favorable treatment outcome. Blood 1998; 92: 2730–41. [PubMed] [Google Scholar]
- 9. Rieder H, Ludwig WD, Gassmann W et al . Prognostic significance of additional chromosome abnormalities in adult patients with Philadelphia chromosome positive acute lymphoblastic leukaemia. Br J Haematol 1996; 95: 678–91. [DOI] [PubMed] [Google Scholar]
- 10. Thomas X, Thiebaut A, Olteanu N et al . Philadelphia chromosome positive adult acute lymphoblastic leukemia: characteristics, prognostic factors and treatment outcome. Hematol Cell Ther 1998; 40: 119–28. [PubMed] [Google Scholar]
- 11. Wetzler M, Dodge RK, Mrózek K et al . Additional cytogenetic abnormalities in adults with Philadelphia chromosome‐positive acute lymphoblastic leukaemia: a study of the Cancer and Leukaemia Group B. Br J Haematol 2004; 124: 275–88. [DOI] [PubMed] [Google Scholar]
- 12. Yanada M, Takeuchi J, Sugiura I et al . Karyotype at diagnosis is the major prognostic factor predicting relapse‐free survival for patients with Philadelphia chromosome‐positive acute lymphoblastic leukemia treated with imatinib‐combined chemotherapy. Haematologica 2008; 93: 287–90. [DOI] [PubMed] [Google Scholar]


