Abstract
Overexpression of the olfactomedin 4 (OLFM4 GW112,/hGC−1 ) gene was recently reported to inhibit various apoptotic pathways and promote proliferation of cancer cells, suggesting that OLFM4 might serve as a diagnostic marker for human cancers. Therefore, we examined cancer‐specific OLFM4 overexpression. OLFM4 mRNA was highly expressed in cancerous tissues obtained from the colon, breast and lung. Positivity for OLFM4 mRNA, defined as the mean + 2 SD in non‐cancerous colon and breast tissues, was observed in 68 and 50% of the studied colon and breast cancer tissues. OLFM4 mRNA expression was not detected in non‐cancerous lung tissues but was evident in 62% of the lung cancer tissues. On comparing paired samples, the expression of OLFM4 mRNA was observed to be elevated in 90, 69 and 85% of colon, breast and lung cancer tissues, respectively. OLFM4 mRNA expression was observed even in the early stages of each cancer type. The expression of OLFM4 mRNA did not correlate with that of the antiapoptotic molecule survivin, indicating that it can be used independently in cancer diagnosis. Combining OLFM4 and survivin resulted in higher positivity. Thus, OLFM4 mRNA might be a useful tool to support the diagnosis of cancer, irrespective of the clinical stages. (Cancer Sci 2007; 98: 315–320)
Antiapoptotic molecules represented by the bcl‐2 family( 1 , 2 ) and the inhibitor of apoptosis protein (IAP) family are expressed strongly by cancer cells.( 3 , 4 , 5 , 6 , 7 ) Therefore, measuring the gene or protein expression of these molecules could be useful for the diagnosis of cancer and for identifying potential targets for cancer treatment. Olfactomedin 4 (OLFM4GW112/hGC−1) was recently reported as a novel antiapoptotic molecule by Zhang et al.( 8 ) This olfactomedin‐related glycoprotein was originally cloned from human hematopoietic precursor cells and identified as human G‐CSF‐stimulated clone‐1 (hGC‐1).( 9 ) Other studies have also reported that this molecule is expressed by GW112 in inflammatory tissues.( 10 ) Olfactomedin was originally identified as a glycoprotein expressed in the mucus layer of the olfactory neuroepithelium of bullfrogs.( 11 ) Olfactomedin‐related proteins have a variable N terminus and a more conserved C terminus, called the olfactomedin domain. One olfactomedin‐related protein, Noelin, is expressed in the brain and is involved in regulating the development of neural crest cells, neurogenesis and neuronal differentiation.( 12 ) Mutations in the olfactomedin domain of the myocilin gene may lead to juvenile open‐angle glaucoma, and another olfactomedin‐related protein, optimedin, is reported to interact with myocilin.( 13 )
OLFM4 localized in the nucleus and mitochondria inhibits activation of the caspase cascade at the level of cytochrome c release.( 8 ) This protein also binds to GRIM‐19, a potent mediator of apoptosis,( 8 , 14 , 15 ) to inhibit its function. Thus, OLFM4 can prevent apoptosis induced by various cytostresses. Zhang et al. further demonstrated the facilitation of tumor growth in mice bearing transplanted prostate cancer cells when tumors overexpressed OLFM4 as a result of gene transfection.( 8 ) These findings have led to speculation that OLFM4 gene expression may be upregulated in human cancers, but the investigation has not yet yielded conclusive results.
Therefore, we quantitated OLFM4 mRNA expression in colon, breast and lung cancer tissues and compared the results with those of non‐cancerous tissues obtained from the same resection specimen. We had previously reported the abundant expression of survivin, a member of the IAP family, in various human tumor tissues; its expression was extremely low in most of the normal tissues.( 3 , 4 , 5 , 7 ) Similarly, other studies have demonstrated that survivin may be a useful diagnostic and prognostic marker that will assist in devising anticancer drug treatments.( 16 , 17 , 18 ) Given the differences in mechanisms leading to apoptotic signal inhibition,( 8 , 19 , 20 ) we considered the possibility that the OLFM4 expression profile could differ from that of survivin. Therefore, we examined the correlations between OLFM4 and survivin mRNA expression in various tissues to determine whether OLFM4 could be an independent marker for cancer diagnosis.
Materials and Methods
Tissue samples and cell culture. Cancerous and non‐cancerous tissues of the colon, breast and lung were obtained from patients undergoing surgery at Sapporo Medical University Hospital (Sapporo, Japan) and Sapporo Breast Surgical Clinic (Sapporo, Japan). As for non‐cancerous tissues of the colon, tissues excepting those from hyperplastic polyp and adenoma were used. Prior to the acquisition of these tissues, informed consent explaining the investigational nature of the study was obtained from the patients. The tissues were frozen immediately and stored in liquid nitrogen. Some of the tissues were stained with hematoxylin/eosin and examined by experienced pathologists. The clinicopathological factors were evaluated according to the criteria set by the Japanese Society of Colon, Breast, and Lung Cancer.( 6 , 21 , 22 )
The human lung cancer cell line SBC‐1 (LC817) was cultured in RPMI‐1640 (Sigma‐Aldrich, St Louis, MO, USA) supplemented with 10% heat‐inactivated fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and grown at 37°C in a humidified atmosphere of 5% CO2.
Small interfering RNA transduction. A small interfering RNA (siRNA) was designed to target the coding region of the OLFM4 gene (GenBank accession no. NM‐006418, which was originally expressed as hGC‐1,( 9 ) or GW112,( 8 , 10 ) nucleotides 299–321, relative to the start codon), and prepared by using QIAGEN (Tokyo, Japan). Single‐stranded RNA was annealed by incubating each strand in siRNA suspension buffer for 1 min at 90°C, followed by incubation for 1 h at 37°C. The siRNA duplexes used in this study were as follows: OLFM4, 5′‐CGCUUGGAAUUCACAGCUCUU‐3′ and 3′‐UUGCGAACCUUAAGUGUCGAG‐5′. As a negative control for siRNA, non‐silencing control RNA (QIAGEN) was used. The transduction was carried out using a HyPerFect transfection reagent (QIAGEN) according to the manufacturer's protocol. Briefly, 1 × 105 SBC‐1 cells were cultured in six‐well culture plates (Costar, Tokyo, Japan) in 2.3 mL RPMI supplemented with 10% FBS, and incubated for 24 h. On the day of transduction, the siRNA and the HyPerFect transfection reagent in a total volume of 100 µL RPMI were mixed by vortexing. After 10 min of incubation at room temperature, the siRNA mixture was added to the cells to give a final siRNA concentration of 20 nM. After 72 h of transduction, the cells were harvested and the mRNA expression level was estimated.
Reverse transcription–polymerase chain reaction. The expression of OLFM4 mRNA was determined by quantitative reverse transcription (RT)–polymerase chain reaction (PCR) using an ABI PRISM 7700 sequence detector system (Applied Biosystems, Foster City, CA, USA). Isolation of total RNA was carried out using the ISOGEN reagent (Nippon Gene, Toyama, Japan). The gene‐specific primers and fluorescent hybridization probes used in quantitative PCR were as follows: OLFM4 forward primer, 5′‐CTGCCAGACACCACCTTTCC‐3′; reverse primer, 5′‐CTCGAAGTCCAGTTCAGTGTAAG‐3′; and probe, 5′‐(FAM)CGCTTGGAATTCACAGCTCATGTTCTTTC(TAMRA)‐3′. Survivin forward primer, 5′‐AAG AAC TGG CCC TTC TTG GA‐3′; reverse primer, 5′‐CAA CCG GAC GAA TGC TTT T‐3′; and probe, 5′‐(FAM) CCA GAT GAC GAC CCC ATA GAG GAA CA (TAMRA)‐3′. Quantitative RT‐PCR was carried out using a TaqMan Universal PCR Master Mix (Applied Biosystems) and TaqMan One‐Step RT‐PCR Master Mix reagents (Applied Biosystems). LS180 cells were used as control cells, and representative amplification plots and the standard curve for OLFM4 are shown in Fig. 1. To compare the amounts of mRNA encoding OLFM4 in different samples, the quantity of specific mRNA was normalized as a ratio to the amount of 18S rRNA, which was determined using TaqMan Ribosomal RNA Control reagents (Applied Biosystems) according to the manufacturer's protocol.
Figure 1.
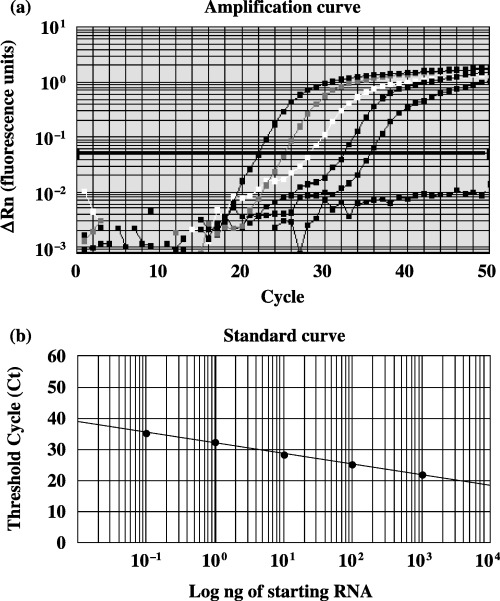
(a) Amplification plots of olfactomedin 4 (OLFM4) mRNA. Five different dilutions (0.1–1000 ng) of the total RNA from LS180 cells were tested. Amplification plots were used to determine the threshold cycle (Ct). In these x–y plots the polymerase chain reaction cycle number is represented on the x‐axis and the ΔRn values (fluorescence units) on the y‐axis. To determine the optimum Ct value, the optimum ΔRn (y = 0.05) was selected in the exponential phase of the amplification plots for each dilution of total RNA. (b) Standard curves of OLFM4. RNA in the LS180 cells was quantitated using FAM‐labeled probes. The standard curves on these plots represent the ng of total starting RNA on a logarithmic x‐axis and the Ct values on the y‐axis. Equations were derived from the lines of the standard curves. The equation for Ct is y = −3.41x + 32.05 (r = 1.00), where x = log (ng total starting RNA). The Ct values of a sample were substituted as y into the equation for OLFM4 to yield the level of OLFM4 expression.
Statistical analysis. Paired Student's t‐test was used for the analysis of mRNA expression and cell numbers in siRNA experiments. The Mann–Whitney U‐test was used for the comparison of OLFM4 mRNA expression in various groups. Pearson's correlation coefficiency was calculated to examine the relationship between the expression of OLFM4 and survivin mRNA.
Results
OLFM4 mRNA expression in various cancers. To examine OLFM4 gene expression in various cancers, we quantified OLFM4 mRNA expression in human colon, breast and lung cancers, comparing its expression to that in non‐cancerous tissues. In general, OLFM4 mRNA was expressed strongly in many cancerous tissues. The relative expression in colon and breast cancer tissues was significantly higher than that in corresponding non‐cancerous tissues (Table 1; 2, 3). All of the non‐cancerous tissues resected in continuity with the lung cancer tissues contained no detectable OLFM4 mRNA (Fig. 4).
Table 1.
Olfactomedin 4 mRNA expression in various cancer patients
| Organs | Tissue | Case no. | Median | Interquartile range | P value |
|---|---|---|---|---|---|
| Colon | Non‐cancerous | 27 | 0.0043 | 0.0061 | |
| Cancerous | 28 | 0.0404 | 0.0834 | 0.000017 | |
| Breast | Non‐cancerous | 32 | 0 | 0.0014 | |
| Cancerous | 32 | 0.0359 | 0.3711 | 0.000027 | |
| Lung | Non‐cancerous | 13 | 0 | 0 | |
| Cancerous | 39 | 0.0005 | 0.0396 | 0.00095 |
Figure 2.
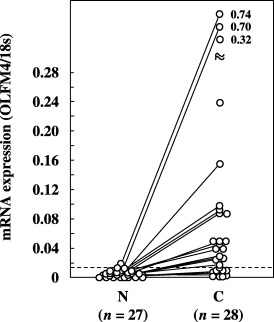
Expression of olfactomedin 4 (OLFM4) mRNA in non‐cancerous and cancerous tissues from colonic cancer patients. OLFM4 mRNA expression was quantitated as a value relative to 18S rRNA expression in each sample. The dotted line indicates the mean + 2 SD (0.015) for the amount of mRNA in non‐cancerous tissues. Data are the mean values for three measurements in identical experiments. A solid line indicates a paired sample. C, cancerous tissues; N, non‐cancerous tissues.
Figure 3.
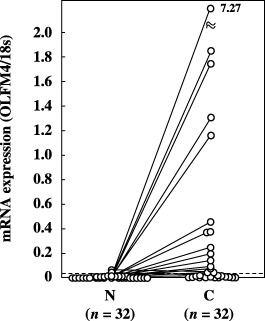
Expression of olfactomedin 4 (OLFM4) mRNA in non‐cancerous and cancerous tissues from breast cancer patients. OLFM4 mRNA expression was quantitated as a value relative to 18S rRNA expression in each sample. The dotted line indicates the mean + 2 SD (0.038) for the amount of mRNA in non‐cancerous tissues. Data are the mean values for three measurements in identical experiments. A solid line indicates paired sample. C, cancerous tissues; N, non‐cancerous tissues.
Figure 4.
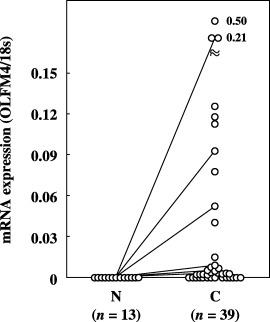
Expression of olfactomedin 4 (OLFM4) mRNA in non‐cancerous and cancerous tissues from lung cancer patients. OLFM4 mRNA expression was quantitated as a value relative to 18S rRNA expression in each sample. The dotted line indicates the mean + 2 SD for the amount of mRNA in non‐cancerous tissues. Data are the mean values for three measurements in identical experiments. A solid line indicates paired samples. None of the non‐cancerous tissues showed detectable mRNA expression. C, cancerous tissues; N, non‐cancerous tissues.
When a cut‐off value was set at the mean + 2 SD for OLFM4 mRNA expression in non‐cancerous tissues, the positivity for OLFM4 mRNA expression was found to be 67.9% (19/28) and 50% (16/32) in colon and breast cancer tissues, respectively; in the lung cancer tissues, mRNA was detected in 61.5% (24/39) of the samples2, 3, 4). Comparison of the samples, when both the non‐cancerous and cancerous tissues were obtained from the same patient, revealed that OLFM4 mRNA levels were elevated in 90% (18/20), 68.8% (22/32) and 84.6% (11/13) of the colon, breast and lung cancer tissues, respectively.
In the colon and breast cancer tissues, analysis of the relationship between OLFM4 mRNA expressed by the cancer tissues and the clinical variables revealed no significant difference in the mean mRNA expression levels in groups categorized on the basis of age, sex or pathological findings (P > 0.05). However, with regard to the potential ability to detect cancer, OLFM4 mRNA was expressed abundantly even in the early stages of the disease (Fig. 5). Because limited cases of stage I colon and breast cancers were included, statistical comparisons between stage I and other stages could not be made. However, there was no significant difference in mRNA expression between the relatively early stages and late stages of these cancers (stage I + II vs III + IV in colon cancers; stage I + IIA vs IIB + III in breast cancers). This early expression was even more striking in lung cancers, where OLFM4 mRNA was detected in 61% (19/31) of stage I cases, and no statistically significant difference was observed between the expression in stage I and stage II + III + IV. A statistically significant difference was observed in different histological types of lung cancer tissues; higher mRNA expression was observed in squamous cell carcinoma than in adenocarcinoma (Fig. 6; P < 0.01).
Figure 5.
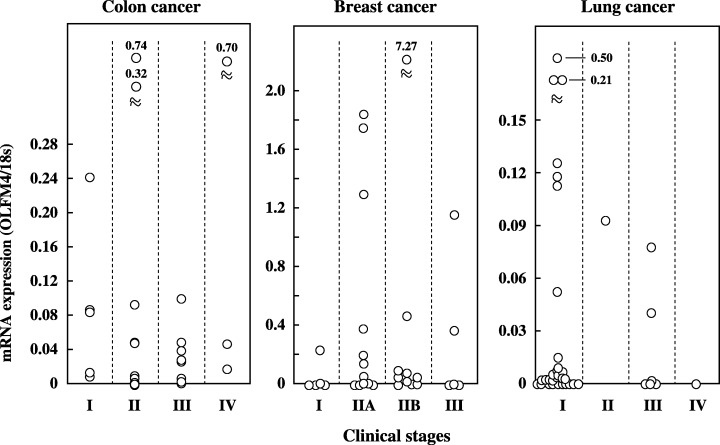
Expression of olfactomedin 4 (OLFM4) mRNA in different clinical stages of colon, breast and lung cancer patients. OLFM4 mRNA expression was quantitated as a value relative to 18S rRNA expression in each sample. Data are the mean values for three measurements in identical experiments.
Figure 6.
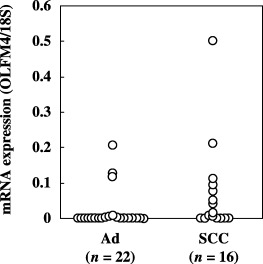
Expression of olfactomedin 4 (OLFM4) mRNA in different histological types in lung cancers. OLFM4 mRNA expression was quantitated as a value relative to 18S rRNA expression in each sample. One case with small cell carcinoma was excluded from this data. Ad, adenocarcinoma; SCC, squamous cell carcinoma.
To determine whether OLFM4 was related to other antiapoptotic molecules, we examined the correlation of its gene expression with that of survivin, a well‐known IAP overexpressed in various cancerous tissues.( 3 , 4 , 5 , 7 ) We found no correlation between the mRNA expression of these molecules in colon cancer (r = 0.006), breast cancer (r = −0.105) or lung cancer tissues (r = 0.001) (Fig. 7). This independency in the expression profile was also reflected by the combination assay carried out using OLFM4 and survivin, which resulted in higher positivity compared to the positivity shown when each molecule was used alone (Table 2).
Figure 7.
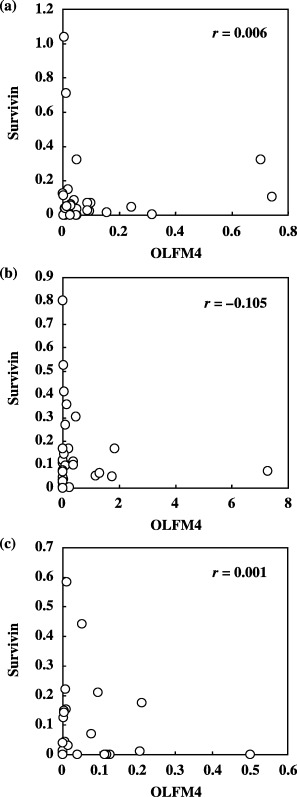
Correlation of mRNA expression between olfactomedin 4 (OLFM4) and survivin in various cancers. Survivin mRNA expression was measured using samples identical to those in which OLFM4 mRNA expression was assessed. Measured expression for both molecules in (a) colon, (b) breast and (c) lung cancers is plotted. Expression of mRNA was quantitated relative to 18S rRNA expression in each sample.
Table 2.
Positivity rate for the combination of olfactomedin 4 (OLFM4) with surviving
| Organ | OLFM4 | Survivin | Combination | |||
|---|---|---|---|---|---|---|
| % | n | % | n | % | n | |
| Colon (n = 28) | 67.9 | 19 | 71.4 | 20 | 92.9 | 26 |
| Breast (n = 32) | 50.0 | 16 | 65.6 | 21 | 78.1 | 25 |
| Lung (n = 39) | 61.5 | 24 | 41.0 | 16 | 71.8 | 28 |
Positivity rate was revealed by the percentage of the cases in which the measured value exceeded mean + 2 SD for expression in non‐cancerous tissues.
Effect of siRNA transduction on cell proliferation. To examine the importance of OLFM4 expression in the proliferation of cancer cells, we transduced OLFM4 siRNA into SBC‐1 lung cancer cells. SiRNA decreased OLFM4 mRNA expression to 4% of the base line after 3 days of transduction (Fig. 8a). We then examined how siRNA transduction affects cell numbers. Control cells, transduced with non‐silencing control RNA, increased approximately 20 times after 3 days, whereas 50% inhibition of this growth was noted in siRNA‐transduced cells (Fig. 8b).
Figure 8.
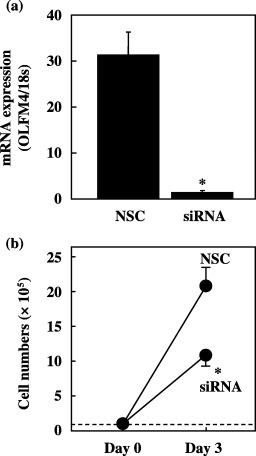
Silencing effect on olfactomedin 4 (OLFM4) mRNA expression due to small interfering RNA (siRNA) transduction in SBC‐1 cells. (a) Cells were transduced with non‐silencing control RNA or siRNA, and total RNA was extracted after 72 h in culture. NSC, non‐silencing control RNA. (b) Effect of OLFM4‐targeted siRNA transduction on proliferation by SBC‐1 cells. Cells were transduced with NSC or siRNA against OLFM4. Viable cell numbers were assessed after 3 days in culture. Mean values were obtained from three independent experiments at each data point. *P < 0.05 by Student's t‐test.
Discussion
OLFM4 is suspected to be highly expressed in cancer cells in view of its antiapoptotic function. However, there has been little investigation of OLFM4 expression in terms of cancer diagnosis. In the present study, we examined OLFM4 gene expression in various cancer tissues and corresponding non‐cancerous tissues. We detected a higher expression of OLFM4 mRNA in cancerous than in non‐cancerous tissues obtained from the colon, breast and lung. Using Northern blotting, Zhang et al. observed two mRNA species from the OLFM4 gene: the 2.9‐kb and 1.7‐kb species.( 8 ) Although only one blot for each organ was examined, different expression patterns were observed in their study (2.9 kb in colon, 1.7 kb in lung, and both in breast cancer tissue samples). Only the 2.9‐kb species has been reported in GenBank and the primer and probe used in our study was set based on the GenBank data. In our study, mRNA was detected in 96% (27/28) of colon cancers and the negative expression of the 2.9‐kb species in the study of Zhang et al. was obtained from only one lung cancer tissue. Therefore, it is considered that the 2.9‐kb species serves as a diagnostic marker. The 1.7‐kb species is possibly an alternative splicing variant as the probe used in the study of Zhang et al. could detect both species. However, it is still unclear whether our primer and probe can detect this variant because the exact position of the probe was not clarified in their study. The information regarding the splicing position of the 1.7‐kb species remains to be investigated.
In the present study, the siRNA experiment clarified that the significance of OLFM4 expression in cancer cells is to promote proliferation; this is consistent with the observation that overexpression of the OLFM4 gene promotes tumor growth.( 8 ) In contrast, siRNA transduction did not increase apoptosis in SBC‐1 cells (data not shown). In the original study by Zhang et al., induction of OLFM4 by gene transduction promoted cell resistance to apoptotic stimuli.( 8 ) Further, our preliminary observation was that OLFM4 gene expression is increased by apoptotic stimuli such as irradiation or anticancer drugs. Thus, it is speculated that OLFM4 functions as an inducible type of resistance factor, especially when apoptotic stimuli exist, whereas constitutively expressed OLFM4 promotes cell proliferation.
Gene expression of the IAP family member survivin has been reported to be a useful candidate marker for cancer diagnosis. In comparison with our previous studies on survivin, OLFM4 shows a similar extent of positivity in colon and breast cancers.( 4 , 5 , 7 ) In the present study, we found OLFM4 gene expression to be undetectable in all non‐cancerous lung samples. In contrast, approximately 62% of the lung cancer tissue samples were positive for OLFM4 expression, suggesting that OLFM4 could be a more useful genetic marker than survivin, which shows approximately 40% positivity in lung cancer tissue samples.( 6 ) Although the reason for higher expression of OLFM4 by squamous cell carcinoma than by adenocarcinoma is unclear, quantitative measurement of OLFM4 mRNA might be useful in the early diagnosis of lung cancer when only a limited number of cells can be obtained from the needle biopsy specimen, sputum or bronchoalveolar lavage fluid. Measurement of OLFM4 should be especially applied to tiny tumors in which the malignancy cannot be confirmed by cytological or histological examination.
We found no correlation between the expression of OLFM4 and survivin mRNAs in various cancers, indicating that expression of the OLFM4 marker is independent of survivin expression. Differences in the point of action in the apoptotic signaling pathway may underlie this independence in positivity between the two molecules.( 8 , 14 , 15 ) Another promising finding is that OLFM4 mRNA is highly expressed even in the early clinical stages, that is, before disease progression has been initiated. Because the combination assay of OLFM4 and survivin increases the positivity rate in cancers, measurement of OLFM4 may prove to be an important aid in detection of cancer.
References
- 1. Kaklamanis L, Savage A, Mortensen N et al. Early expression of bcl‐2 protein in the adenoma‐carcinoma sequence of colorectal neoplasia. J Pathol 1996; 179: 10–14. [DOI] [PubMed] [Google Scholar]
- 2. Rubio N, Espana L, Fernandez Y, Blanco J, Sierra A. Metastatic behavior of human breast carcinomas overexpressing the bcl‐x (L) gene: a role in dormancy and organospecificity. Laboratory Invest 2001; 81: 725–34. [DOI] [PubMed] [Google Scholar]
- 3. Nakamura M, Tsuji N, Asanuma K et al. Survivin as a predictor of cis‐diamminedichloroplatinum sensitivity in gastric cancer patients. Cancer Sci 2004; 95: 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nasu S, Yagihashi A, Izawa A et al. Survivin mRNA expression in patients with breast cancer. Anticancer Res 2002; 22: 1839–43. [PubMed] [Google Scholar]
- 5. Tsuji N, Furuse K, Asanuma K et al. Mutations of the p53 gene and loss of heterozygosity at chromosome 17p13.1 are associated with increased survivin expression in breast cancer. Breast Cancer Res Treat 2004; 87: 23–31. [DOI] [PubMed] [Google Scholar]
- 6. Tanabe H, Yagihashi A, Tsuji N, Shijubo Y, Abe S, Watanabe N. Expression of survivin mRNA and livin mRNA in non‐small‐cell lung cancer. Lung Cancer 2004; 46: 299–304. [DOI] [PubMed] [Google Scholar]
- 7. Endoh T, Tsuji N, Asanuma K, Yagihashi A, Watanabe N. Survivin enhances telomerase activity via up‐regulation of specificity protein 1‐ and c‐Myc‐mediated human telomerase reverse transcriptase gene transcription. Exp Cell Res 2005; 305: 300–11. [DOI] [PubMed] [Google Scholar]
- 8. Zhang X, Huang Q, Yang Z, Li Y, Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res 2004; 64: 2474–81. [DOI] [PubMed] [Google Scholar]
- 9. Zhang J, Liu WL, Tang DC et al. Identification and characterization of a novel member of olfactomedin‐related protein family, hGC‐1, expressed during myeloid lineage development. Gene 2002; 283: 83–93. [DOI] [PubMed] [Google Scholar]
- 10. Shinozaki S, Nakamura T, Iimura M et al. Upregulation of Reg 1α and GW112 in the epithelium of inflamed colonic mucosa. Gut 2001; 48: 623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snyder DA, Rivers AM, Yokoe H, Menco BP, Anholt RR. Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry 1991; 30: 9143–53. [DOI] [PubMed] [Google Scholar]
- 12. Moreno TA, Bronner‐Fraser M. Noelins modulate the timing of neuronal differentiation during development. Dev Biol 2005; 288: 434–47. [DOI] [PubMed] [Google Scholar]
- 13. Torrado M, Trivedi R, Zinovieva R, Karavanova I, Tomarev SI. Optimedin: a novel olfactomedin‐related protein that interacts with myocilin. Hum Mol Genet 2002; 11: 1291–301. [DOI] [PubMed] [Google Scholar]
- 14. Huang G, Lu H, Hao A et al. GRIM‐19, a cell death regulatory protein, is essential for assembly and function of mitochondrial complex I. Mol Cell Biol 2004; 24: 8447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J, Yang J, Roy SK et al. The cell death regulator GRIM‐19 is an inhibitor of signal transducer and activator of transcription 3. Proc Natl Acad Sci USA 2003; 100: 9342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monzo M, Rosell R, Felip E et al. A novel anti‐apoptosis gene: re‐expression of survivin messenger RNA as a prognosis marker in non‐small‐cell lung cancers. J Clin Oncol 1999; 17: 2100–4. [DOI] [PubMed] [Google Scholar]
- 17. Span PN, Sweep FC, Wiegerinck ET et al. Survivin is an independent prognostic marker for risk stratification of breast cancer patients. Clin Chem 2004; 50: 1986–93. [DOI] [PubMed] [Google Scholar]
- 18. Kami K, Doi R, Koizumi M et al. Survivin expression is a prognostic marker in pancreatic cancer patients. Surgery 2004; 136: 443–8. [DOI] [PubMed] [Google Scholar]
- 19. Tamm I, Wang Y, Sausville E et al. IAP‐family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), BAX, caspases, and anticancer drugs. Cancer Res 1998; 58: 5315–20. [PubMed] [Google Scholar]
- 20. Wang Z, Fukuda S, Pelus LM. Survivin regulates the p53 tumor suppressor gene family. Oncogene 2004; 23: 8146–53. [DOI] [PubMed] [Google Scholar]
- 21. Kotake K, Honjo S, Sugihara K et al. Changes in colorectal cancer during a 20‐year period: an extended report from the multi‐institutional registry of large bowel cancer. Japan Dis Colon Rect 2003; 46: S32–43. [DOI] [PubMed] [Google Scholar]
- 22. Saito K, Yagihashi A, Nasu S et al. Gene expression for suppressors of telomerase activity (telomeric‐repeat binding factors) in breast cancer. Jpn J Cancer Res 2002; 93: 253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


