Abstract
RUNX3 is a candidate tumor suppressor gene localized in 1p36, a region frequently inactivated through hypermethylation, histone modulation, and other processes in various human tumors. In this study, to elucidate a causal link between RUNX3 expression and biliary tract cancer, we investigated 17 human biliary cancer specimens. In addition, to examine roles of RUNX3 in biliary tract cancer, we restored silenced RUNX3 in the human biliary cancer cell line Mz‐ChA‐2 using a histone deacetylase inhibitor. Thirteen of 17 human cancer specimens exhibited suppressed RUNX3 expression compared with normal biliary ducts. Moreover, the decreased RUNX3 expression was related to a lower accumulation of acetylated histone H3 associated with RUNX3. In in vitro experiments, vorinostat, a member of a new class of highly potent histone deacetylase inhibitors, restored RUNX3 expression in Mz‐ChA‐2 cells. Furthermore, vorinostat‐induced RUNX3 significantly enhanced p21 expression and growth inhibition of Mz‐ChA‐2 cells through restoration of TGF‐β signaling. These data suggest the significance of histone deacetylation‐associated suppression of RUNX3 expression in biliary tract carcinogenesis. Furthermore, vorinostat might hold promise for treating biliary tract cancer through enhancement of TGF‐β signaling by restoration of RUNX3. (Cancer Sci 2011; 102: 776–783)
The incidence of biliary tract cancer is increasing, especially in Asia.( 1 ) The only potential cure for this disease is surgical resection. However, most cases are unresectable at the time of diagnosis due to the late clinical presentation,( 2 ) and the prognosis is extremely poor because of high resistance to chemotherapies and radiation therapies.( 2 , 3 ) Thus, elucidating the biological characteristics of biliary tract cancer cells is necessary to develop effective treatment strategies against the disease, thereby improving its prognosis.
The RUNX3 gene is located on chromosome 1 at 1p36.1, and encodes a protein belonging to the runt domain family of transcription factors that act as master regulators of gene expression in major developmental pathways.( 4 , 5 , 6 ) In transforming growth factor (TGF)‐beta signaling, RUNX3 is also identified as a tumor suppressor in terms of induction of cyclin‐dependent kinase inhibitor p21( 7 ) in cooperation with homologs of both the drosophila protein, mothers against decapentaplegic (MAD) and the Caenorhabditis elegans protein SMA (SMAD)( 8 ) and upregulation of a proapoptotic BH3‐only protein Bim.( 9 ) The RUNX3 gene is frequently inactivated through promoter hypermethylation, histone modification, hemizygous deletion, cytoplasmic mislocalization, or mutations( 10 ) in various carcinomas including gastric,( 6 ) pancreatic and biliary tract,( 11 ) lung,( 12 , 13 ) hepatocellular,( 14 ) breast,( 15 ) colon,( 16 ) prostate,( 17 ) and laryngeal.( 15 )
In biliary tract cancer, we previously showed that RUNX3 gene expression is silenced by hypermethylation of the promoter region in 70% of cell lines.( 11 ) Thus, RUNX3 appears to play a pivotal role in biliary cancers, although its expression has not been examined in patient specimens.( 11 ) Generally, hypermethylation in CpG islands in the promoter region of tumor suppressor genes is considered to silence gene expression through recruitment of histone deacetylase. Indeed, inhibition of histone deacetylases by histone deacetylase inhibitors (HDACIs) can reverse histone modifications and restore gene expression of which CpG island is hypermethylated. Recently, growth inhibition and induction of apoptosis by RUNX3 reactivation in gastric cancer cells following treatment with vorinostat, a member of a new class of highly potent HDACIs, has been reported.( 18 ) However, the contribution of histone acetylation to RUNX3 expression in biliary tract cancer cells remains unknown.
In this study, we first examined RUNX3 expression in resected specimens from biliary tract cancer patients. Second, we analyzed the status of histone acetylation associated with RUNX3 in both biliary tract cancer cell lines and resected specimens. Finally, we evaluated the effect of histone deacetylase inhibition on biliary tract cancer cells with special emphasis on RUNX3 expression, TGF‐β signaling, and growth activities.
Materials and Methods
Cell lines and biliary cancer patient specimens. The human biliary tract cancer cell line Mz‐ChA‐2 was a kind gift from Dr. A. Knuth (Krankenhaus Nordwest, Frankfurt, Germany). The human pancreatic cancer cell line MiaPaca2 was purchased from American Type Culture Collection (Rockville, MD, USA). All cell lines were cultured in DMEM (Gibco‐BRL, Tokyo, Japan) supplemented with 10% FBS and antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin) in a water‐saturated atmosphere of 5% CO2. The study population consisted of biliary tract cancer patients undergoing surgery at Kyoto University Hospital from 2002 to 2006. Written informed consent was obtained from each patient before tissue acquisition. Vorinostat (Cayman, Ann Arbor, MI, USA) was dissolved and diluted in DMSO, and TGF‐β1 (R&D Systems, Tokyo, Japan) was dissolved and diluted in 4 mM HCl containing 1 mg/mL BSA.
RNA isolation and real‐time quantitative RT‐PCR. Total RNA was isolated using an RNeasy Mini kit (Qiagen, Hilden, Germany). Equal amounts of total RNA were reverse‐transcribed using an Omniscript Reverse Transcriptase kit (Qiagen). The primers used for real‐time quantitative RT‐PCR analyses of RUNX3, p21, and GAPDH were Assays‐on‐Demand Gene Expression products (Applied Biosystems, Foster City, CA, USA). The primers for GAPDH served as an internal control for normalization of the results. Real‐time RT‐PCR amplification and data analysis were carried out using an ABI Prism 7700 (Life Technologies, Carlsbad, CA, USA).
Small interfering RNA. SiRNA targeting human RUNX3 and non‐targeting siRNA were purchased from Dharmacon (Lafayette, CO, USA). Cells were transfected with siRNA using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA).
Chromatin immunoprecipitation PCR. Chromatin immunoprecipitation for biliary cancer patient specimens was performed using the EpiQuik Tissue Acetyl‐Histone H3 ChiP kit (Epigentek, New York, NY, USA) according to the manufacturer’s instructions. Chromatin immunoprecipitation for Mz‐ChA2 cells, with or without 5 μM vorinostat for 48 h, was performed using the Acetyl‐Histone H3 and H4 Immunoprecipitation Assay kit (Upstate Biotechnology, Lake Placid, NY, USA), following the manufacturer’s instructions, as previously described.( 14 ) PCR was performed using three pairs of primers targeting RUNX3 sequences in the region from −282 bp of the promoter region to +2267 downstream of the transcription start site. Those pairs were: A (−1187 to −1047): 50‐tgt ttt tca aag agc cac agg ccg cc‐30 (sense), 50‐ggg agt ctc cta ggg acc cta agt ag‐30 (antisense); B (+450 to +567): 50‐gtt ccg ttt tgg atg cgc cct gca‐30 (sense), 50‐ca aaa ccc cat ccg ccc att tcc gca‐30 (antisense); C (+2137 to +2267): 50‐ggt ggt ggc att ggg gga cgt gcc gga‐30 (sense), 50‐gtc gtt gaa cct ggc cac ctg gtt ctt‐30 (antisense).
Western blot analysis. Cells were washed twice with ice‐cold PBS and lysed in 20 mM Tris–HCl buffer (pH 7.4) containing 150 mM NaCl, 2 mM EDTA, 1% Nonidet P‐40, 50 mM NaF, 1 mM Na3VO4, 1 mM Na2MoO4, 10 μM aprotinin, and 10 μM leupeptin. The solution was centrifuged at 15 000 g for 5 min, and the supernatant was removed. Equal amounts of cell lysate protein extracts were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a PVDF membrane. The membranes were incubated with anti‐p21 (Transduction Laboratories, Newington, NH, USA), anti‐human acetylated histone H3 and rabbit anti‐human acetylated histone H4 (Upstate Biotechnology), and β‐actin antibodies for 24 h at 4°C, and then with peroxidase‐conjugated secondary antibodies for 1 h at 37°C. The blotted proteins were detected using an ECL kit (Amersham Pharmacia Biotech, Buckinghamshire, UK).
Immunohistochemistry. Paraffin‐embedded tissue sections were routinely stained with hematoxylin‐eosin. Specimens and Mz‐ChA‐2 cells were stained immunohistochemically with a rabbit polyclonal antibody against human RUNX3 in a 1:200 dilution (Activemotif, Carlsbad, CA, USA). A positive reaction was indicated by the development of a reddish‐brown precipitate.
Luciferase assay. To measure TGF‐β signaling, the p3TP‐Lux reporter plasmid (a generous gift from Dr. J. Massague, Howard Hughes Medical Institute, Memorial Sloan‐Kettering Cancer Center, NY, USA) containing the plasminogen activator inhibitor‐1 promoter was used. Cells were seeded at a density of 3 × 105/well in six‐well plates for 20 h, then transiently transfected with 1.0 μg p3TP‐Lux reporter plasmids and, as an internal control, 0.2 μg pRL‐TK vectors (Promega, Madison, WI, USA) using Lipofectamine 2000. Four hours later, cells were treated with the appropriate concentration of vorinostat (0, 5 μM) in the presence or absence of 5 ng/mL TGF‐β1 in DMEM with 0.5% FBS for 48 h. The cell lysates were analyzed for luciferase activity using the Dual‐Luciferase Reporter Assay System (Promega) with or without siRNA targeting human RUNX3. Reporter activities are presented as means ± SD of at least three independent experiments.
Cell growth inhibition assays. Cell growth was monitored in a six‐well plate format. Cells were seeded in triplicate in six‐well dishes at a density of 3 × 105 cells/well, allowed to attach overnight, and the medium was replaced with medium containing the appropriate concentration of vorinostat (0, 2.5, 5 μM), TGF‐β1 (0, 5 ng/mL), and anti‐cancer drugs. Cell number was determined by counting using a hemocytometer at various times after initiation of vorinostat treatment, and cell viability was assessed using trypan blue dye exclusion.
Statistical analysis. All numerical data were expressed as means ± SD. Differences among the mean values were evaluated using Student’s t‐test. P‐values <0.05 were considered statistically significant.
Results
RUNX3 expression is decreased in biliary tract cancer. To examine RUNX3 expression in human biliary tract cancer specimens, immunohistochemical staining of RUNX3 was performed in 17 biliary tract cancer specimens that included both tumor and non‐tumor parts. In non‐tumor parts of all 17 specimens, RUNX3 protein was highly expressed in normal cytoplasm of biliary epithelial cells, but not in other lineages of cells, such as hepatocytes, vascular endothelial cells, and mesenchymal cells. Interestingly, in tumor parts, RUNX3 expression in biliary cancer cells was diminished in 13/17 specimens (76.5%) (Fig. 1). Thus, RUNX3 expression is decreased in biliary tract cancer cells.
Figure 1.
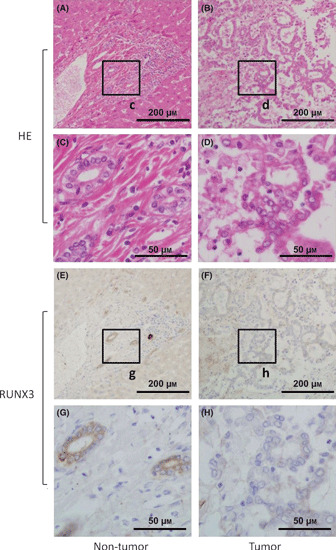
(A–D) Representative hematoxylin and eosin (HE) and (E–H) immunohistochemical stainings of RUNX3 in biliary tract cancer specimen, including non‐tumor tissue, are shown. The c, d, g, h boxes indicate the positions of the enlargements shown below. Scale bars in (A,B,E,F) panels: 200 μm; (C,D,G,H) panels: 50 μm.
Histone acetylation in chromatin associated with RUNX3 is decreased in biliary tract cancer specimens. To determine whether decreased RUNX3 expression in biliary tract cancer specimens is associated with the status of histone acetylation, in chromatin a ChIP‐PCR assay was performed using anti‐acetylated histone H3 antibody. Three pairs of primers complementary to RUNX3 from the upstream promoter region to downstream of the transcriptional start site were used. Three of six specimens in which RUNX3 expression was diminished in tumor parts by immunohistochemical staining could be prepared for ChIP‐PCR, and 2/3 (67%) specimens showed a decrease in PCR products in tumor parts compared with non‐tumor parts (Fig. 2), suggesting that decreased RUNX3 expression in biliary tract cancer cells is related to lower accumulation of acetylated histone H3 associated with RUNX3 in some cases.
Figure 2.
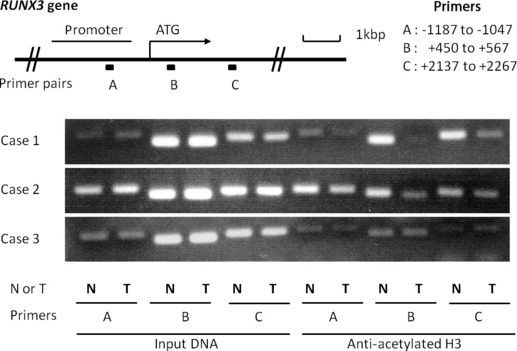
The top panel is a schematic presentation of the RUNX3 gene, indicating the location of three primer pairs used for PCR amplification in ChIP assays. Chromatin fragments were immunoprecipitated with an antibody against acetylated histone H3 in biliary tract cancer specimens. Input DNA, without immunoprecipitation by anti‐acetylated H3 antibody, was also subjected to PCR as a positive control. The right part of the bottom three panels shows representative ChIP assays prepared using anti‐deacetylated H3. Cases 1 and 2 show lower accumulations of acetylated histone H3 in chromatin associated with RUNX3 in tumors, especially in primer B. N, non‐tumor; T, Tumor.
Histone acetylation in chromatin associated with RUNX3 is decreased in biliary tract cancer cell lines. To confirm the suppressed accumulation of acetylated histone H3 associated with RUNX3 in biliary tract cancer, Western blot analysis of cell nuclear lysates and a ChIP‐PCR assay was performed in Mz‐ChA‐2 cells, a biliary tract cancer cell line whose RUNX3 expression was suppressed. Acetylated histone H3 and H4 were not detected by western blot analysis in a basal condition, whereas they were induced by treatment with an HDACI (vorinostat) in a dose‐dependent manner (Fig. 3a). ChIP‐PCR analysis also indicated almost no PCR products in Mz‐ChA‐2 cells (Fig. 3b); however, cells treated with an HDACI showed significantly increased levels of PCR products (Fig. 3b), suggesting that RUNX3 is a gene associated with histone deacetylation in chromatin in the biliary tract cancer cell line Mz‐ChA‐2.
Figure 3.
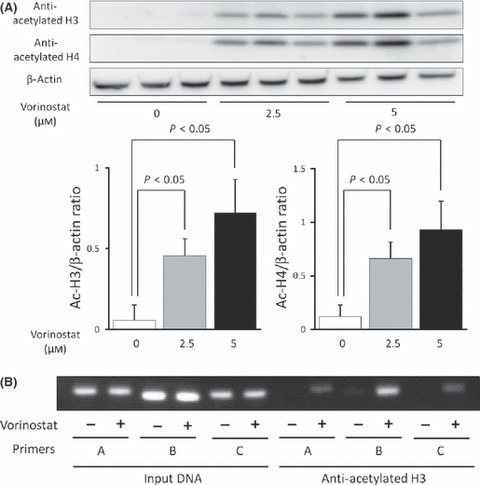
(A) Vorinostat stimulated histone acetylation in a dose‐dependent manner. Mz‐Ch‐A2 cells were treated with 0–5 μM vorinostat for 48 h. Cells were harvested and histones were prepared as described. Histone acetylation was detected by Western blot with antibodies against acetylated H3 and H4. Densitometric ratios of H3 and H4 are also shown. *P < 0.05 when compared with the value derived by non‐stimulation. (B) Vorinostat‐induced accumulation of acetylated histone H3 in chromatin associated with RUNX3. Three primer pairs described in Fig. 2 were used for PCR amplification in ChIP assays. Chromatin fragments from cells cultured with or without 5 μM vorinostat for 48 h were immunoprecipitated with an antibody against acetylated histone H3 in Mz‐Ch‐A2 cells. The panel shows ChIP assays prepared using anti‐deacetylated H3. Input DNA, without immunoprecipitation by anti‐acetylated H3 antibody, was also subjected to PCR as a positive control.
RUNX3 expression is restored by inhibition of histone deacetylase. To directly examine whether histone deacetylation affects silenced RUNX3 expression in biliary cancer cells, Mz‐Ch‐A2 and MiaPaca‐2, RUNX3‐negative biliary and pancreatic cancer cell lines, respectively, were treated with 0–10 μM vorinostat, and RUNX3 mRNA and protein expressions were assessed by quantitative real‐time PCR and immunohistochemistry, respectively. Vorinostat effectively induced RUNX3 mRNA expression in these cell lines in a dose‐ and time‐dependent manner (Fig. 4a). In the same manner, RUNX3 protein expression was also up‐regulated in cytoplasm of Mz‐ChA‐2 cells by 10 μM vorinostat in 48 h as assessed by immunohistochemical staining (Fig. 4b). These data suggest that RUNX3 gene expression is silenced by chromosomal deacetylation in biliary cancer cells.
Figure 4.
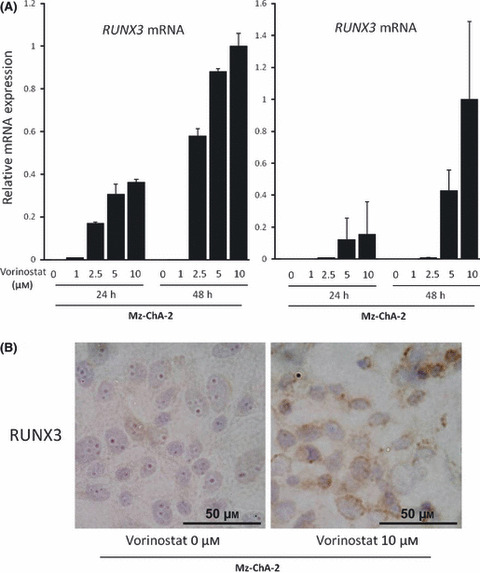
(A) Vorinostat restored RUNX3 in a dose‐ and time‐dependent manner. The Mz‐Ch‐A2 and MiaPaca‐2 cells, RUNX3‐non expressing biliary tract and pancreatic cancer cell lines, respectively, were treated with 0–10 μM vorinostat for 24 and 48 h, total RNA was isolated, and cDNA was synthesized as described. RUNX3 mRNA was measured by real‐time PCR. The data are expressed as means ± SD of three independent experiments. (B) Immunohistochemical staining of RUNX3 in Mz‐Ch‐A2 cells treated with 0 or 10 μM vorinostat for 48 h.
Vorinostat enhances TGF‐β‐induced p21 expression in biliary cancer cells. To investigate the effect of RUNX3 restoration by vorinostat on the TGF‐β‐signaling pathway, we further analyzed Mz‐ChA‐2 cells because these cells are known to have no genetic alteration in major molecules of the TGF‐β‐signaling pathway, such as the TGF‐β type II receptor or homologs of both the drosophila protein, mothers against decapentaplegic (MAD) and the Caenorhabditis elegans protein SMA (SMAD)4.( 12 ) As previously reported in other cells,( 19 , 20 ) TGF‐β1 alone induced a slight increase in p21 mRNA expression in Mz‐ChA‐2 cells. Vorinostat also dose‐dependently increased p21 mRNA expression; moreover, the TGF‐β action on p21 expression was greatly enhanced by vorinostat (Fig. 5a). As expected, vorinostat and TGF‐β1 synergically enhanced p21 protein expression (Fig. 5b,c). These data suggest a potentiating effect of RUNX3 restored by the addition of vorinostat on the TGF‐β signaling pathway.
Figure 5.
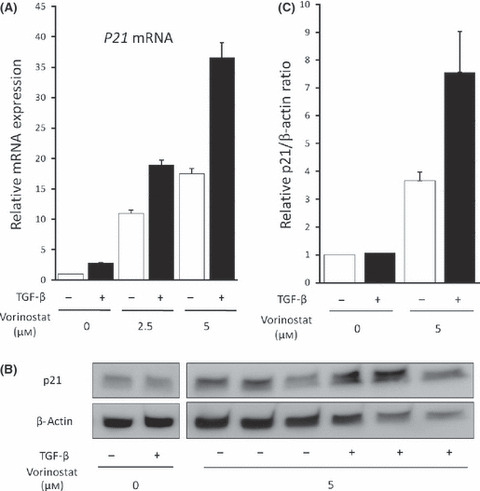
(A) p21 mRNA induction in Mz‐ChA‐2 cells by 0–5 μM vorinostat for 48 h with or without 5 ng/mL TGF‐β1 was measured by real‐time PCR. (B) p21 protein expression in Mz‐ChA‐2 cells induced by 0 or 5 μM vorinostat for 48 h with or without 5 ng/mL TGF‐β1 was detected by Western blot. (C) Relative densitometric ratios of p21 and β‐actin are also shown. The data are expressed as means ± SD of three independent experiments.
Enhancement of TGF‐β signaling by vorinostat is dependent on RUNX3 expression. To confirm whether restoration of RUNX3 is involved in the potentiating effect of vorinostat on the TGF‐β signaling pathway, RUNX3 was knocked down by siRNA in Mz‐ChA‐2 cells treated with vorinostat. Transfection of siRNA for RUNX3 significantly decreased the expression of RUNX3 mRNA to approximately the half level (Fig. 6a). The effect of RUNX3 knockdown was assessed by TGF‐β1‐mediated luciferase activity. The transcriptional activity of TGF‐β1 in the presence of vorinostat was significantly reduced by knockdown of RUNX3 (Fig. 6b). Also, TGF‐β1‐induced p21 mRNA expression was significantly reduced by RUNX3 knockdown. Of interest, knockdown of vorinostat‐induced RUNX3 did not affect the basal level of p21 mRNA expression without TGF‐β1 stimulation (Fig. 6c), suggesting that RUNX3 is involved only in p21 expression induced by TGF‐β, but not the base level. These results confirmed that vorinostat‐restored RUNX3 enhances TGF‐β signaling in biliary cancer cells.
Figure 6.
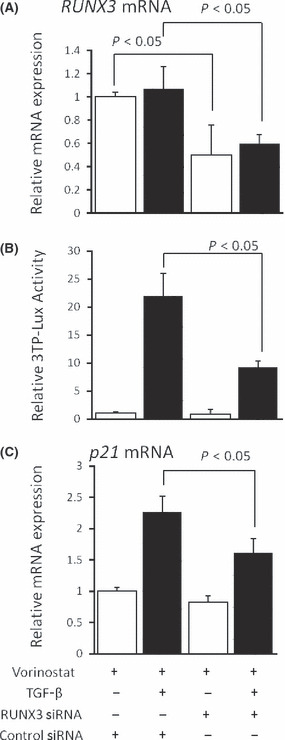
(A) Effect of RUNX3 siRNA on RUNX3 mRNA expression in Mz‐ChA‐2 cells by real‐time PCR. (B) Effect of RUNX3 siRNA on TGF‐β signaling in Mz‐ChA‐2 cells was assessed by a luciferase assay using the p3TP‐Lux reporter plasmid. (C) Effects of RUNX3 siRNA on TGF‐β1‐dependent p21mRNA expression was assessed by real‐time PCR. Cells were cultured with 5 μM vorinostat with or without 5 ng/mL TGF‐β1 for 48 h. The data are expressed as means ± SD of three independent experiments. *P < 0.05 when compared with the value derived by non‐RUNX3 siRNA stimulation.
Vorinostat enhances growth inhibition of cancer cells by TGF‐β. Vorinostat or TGF‐β alone inhibits cancer cell growth as previously reported.( 18 , 21 ) To examine whether vorinostat enhances the inhibitory effects of TGF‐β on biliary cancer cell growth, growth inhibition of Mz‐ChA‐2 cells by TGF‐β1 was assessed with or without treatment of vorinostat. The reduction rate of cell numbers by TGF‐β1 treatment was significantly greater in the presence of than in the absence of vorinostat (Fig. 7). Thus, vorinostat enhances biliary cancer cell growth inhibition by TGF‐β.
Figure 7.
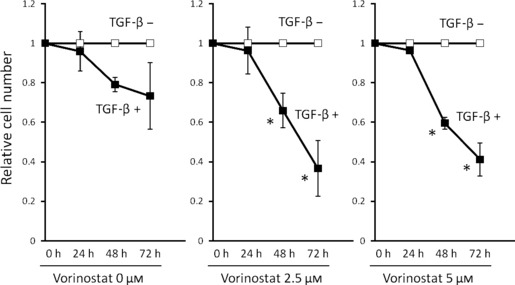
Effects of vorinostat on TGF‐β1‐induced growth inhibition of Mz‐ChA‐2 cells. Cells (1 × 105) were incubated with 0–5 μM vorinostat with or without 5 ng/mL TGF‐β1 for 0–72 h. Values are expressed as the growth rate of the cells without TGF‐β stimulation at each point. The data were calculated as the means ± SD of three experiments. *P < 0.05 when compared with the value derived by non‐TGF‐β1 stimulation.
Vorinostat/TGF‐β enhances growth inhibition of cancer cells by anti‐cancer drugs. As described above, vorinostat and TGF‐β synergistically induce p21 expression and inhibit biliary cancer cell growth through RUNX3 restoration. These results lead us to speculate that the combination of vorinostat and TGF‐β could effectively increase the sensitivity of biliary cancer cells to anti‐cancer drugs. In Mz‐ChA‐2 cells, p21 mRNA was not induced by 5‐Fluorouracil (5‐FU) treatment (Fig. 8a), but vorinostat/TGF‐β newly induced p21 mRNA expression in the presence of 5‐Fluorouracil. Furthermore, we analyzed growth inhibition of Mz‐ChA‐2 cells by 5‐FU with or without vorinostat/TGF‐β1. As expected by p21 mRNA data (Fig. 8a), treatment of vorinostat/TGF‐β enhanced the growth inhibitory effects of 5‐FU (Fig. 8b). These results suggest that vorinostat could be one of the therapeutic options against biliary cancers, especially those whose RUNX3 expression is decreased.
Figure 8.
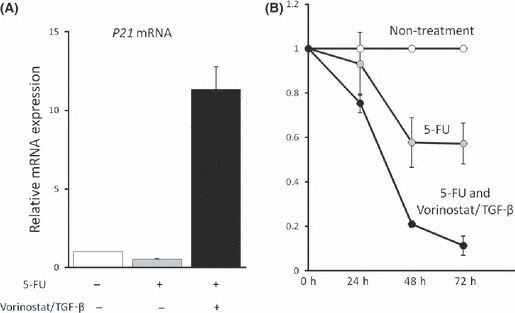
(A) Effects of anti‐cancer drug on p21 mRNA expression of Mz‐ChA‐2 cells. Cells (1 × 105) were incubated with 400 mM 5‐Fluorouracil (5‐FU) for 48 h with or without 5 μM vorinostat+ 5 ng/mL TGF‐β1. Values are expressed as relative mRNA expression of non‐treated cells. The data were calculated as the means ± SD of three experiments. (B) Effects of vorinostat + TGF‐β1 on 5‐Fluorouracil‐induced growth inhibition of Mz‐ChA‐2 cells. Cells (1 × 105) were incubated with 400 mM 5‐Fluorouracil with or without 5 μM vorinostat + 5 ng/mL TGF‐β1 for 0–72 h. Values are expressed as the rate of growth of non‐treated cells at each point. The data were calculated as the means ± SD of three experiments.
Discussion
RUNX3, a tumor suppressor gene with a runt domain, is frequently inactivated in various cancers, including gastric, colorectal, and pancreatic.( 6 , 11 , 12 , 13 , 14 , 15 , 16 , 17 ) However, the majority of the detailed expression levels and mechanisms underlying the regulation of RUNX3 expression in biliary tract cancer is unknown. In this study, we found that RUNX3 expression is decreased in most human biliary tract cancer tissues. We also demonstrated that, in some cases, this decrease is mediated by histone deacetylation associated with RUNX3. Furthermore, we showed that inhibition of histone deacetylation in biliary cancer cells restored RUNX3 expression, which in turn leads to enhancement of TGF‐β signal transduction and subsequent growth inhibition of the cells. These results suggest that inactivation of RUNX3 plays an important role in biliary tract carcinogenesis, and could be a therapeutic target for the disease.
Although RUNX3 expression has been extensively studied in gastrointestinal organs, especially in the stomach,( 6 ) its distribution in hepatobiliary organs has not been reported. In this study, we demonstrated by immunohistochemistry using human liver tissue samples that RUNX3 protein is expressed exclusively in biliary epithelial cells as opposed to being poorly expressed in hepatocytes. More interestingly, and consistent with our previous report showing frequent inactivation of RUNX3 expression in human biliary tract cancer cell lines,( 11 ) expression of RUNX3 protein was decreased in biliary tract cancer cells compared with normal biliary epithelial cells in most cases. To the best of our knowledge, this is the first report that examines the expression of RUNX3 protein in both normal and pathological states of biliary cells. We could not find any difference in clinico‐pathological data including prognosis between the thirteen RUNX3‐suppressed cases and the rest of four cases, probably because of small number of cases. Further analysis with larger number of cases might elucidate the significance of RUNX3 expression in clinical course and pathological findings of biliary tract cancers.
Previous reports have shown that RUNX3 is often inactivated through promoter hypermethylation in various cancers.( 10 ) Indeed, we previously reported that RUNX3 expression is silenced by hypermethylation of the promoter region in 70% of the human biliary tract cancer cell lines examined.( 11 ) In the current study, considering the close link between hypermethylation and histone deacetylation in tumor suppressor gene silencing,( 22 ) we focused on histone modulation in RUNX3 expression in biliary cancer cells. As expected, we observed accumulation of deacetylation of the histone associated with RUNX3 not only in the surgically resected biliary cancer tissues, but also in the human biliary cancer cell line Mz‐ChA‐2. Moreover, inhibition of histone deacetylation by HDACI significantly restored mRNA as well as protein expression of RUNX3 in Mz‐ChA‐2 cells. Thus, consistent with the results from gastric cancer cell lines,( 18 ) histone deacetylation appears to be involved in RUNX3 gene suppression in biliary cancer cells.
In this study, we found that RUNX3 restoration by the HDACI vorinostat induced enhancement of TGF‐β signaling and resulted in an increase in p21 expression and enhancement of TGF‐β‐induced inhibition of growth of a biliary cancer cell line. Moreover, RUNX3 knockdown by RUNX3 siRNA reduced not only TGF‐β signaling but also TGF‐β‐dependent p21 expression. These data are in agreement with several previous studies using other cancer cells demonstrating that some of the anti‐oncogenic functions of RUNX3 are exerted in the TGF‐β signal transduction pathway, at least in part through induction of cyclin‐dependent kinase inhibitor p21.( 7 , 12 ) Of note is that knockdown of vorinostat‐induced RUNX3 did not affect basal levels of p21 mRNA expression without TGF‐β stimulation. Although we do not know the reason why RUNX3 is present in cytoplasm in both normal biliary cells and biliary cancer cells, we propose that RUNX3 function is regulated by its expression level rather than by its localization in biliary cells. Taken together, these data confirm that RUNX3 is a tumor suppressor and suggest that RUNX3 exerts its anti‐oncogenic function mainly in the TGF‐β signaling pathway. This cytostatic function of RUNX3 was also confirmed by knockdown experiments in RUNX3‐positive biliary cancer cells such as TGBC24TKB cells or KMBC cells, showing that the expression levels of p21 were paralleled by those of RUNX3 in these cells (data not shown).
Very few treatment methods for advanced biliary tract cancer have been introduced, and their prognoses are generally very poor. In this study, we demonstrated that the combination of vorinostat and TGF‐β enhances growth inhibition of Mz‐ChA‐2 cell by 5‐FU. In this experiment, 5‐FU alone revealed growth inhibitory effect without affecting p21 expression. On the other hand, additional treatment of vorinostat/TGF‐β further inhibited cell growth with significant induction of p21. These findings suggest that cytostatic effect of 5‐FU was strongly enhanced by p21‐dependent growth inhibitory effect of vorinostat/TGF‐β. Our results account possible use of HDACIs as sensitizers for chemotherapies by induction of p21 expression that may improve the prognosis of advanced biliary tract cancer.
In conclusion, we demonstrated in this study that RUNX3 expression is suppressed in most biliary tract cancers, at least in part due to histone deacetylation, which appears to be involved in the pathophysiology of biliary tract cancers. Because an HDACI significantly enhanced TGF‐β signaling and inhibition of cell growth in association with recovery of RUNX3 expression in human biliary cancer cells, whether manipulation of histone modulation is a promising therapeutic strategy for biliary tract cancer needs further examination.
Disclosure Statement
None.
Acknowledgments
This work was supported by Grants‐in‐Aid for Scientific Research 18012029,18209027, and 19659181 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and Grant‐in‐Aid for Research on Measures for Intractable Diseases, and Research on Advanced Medical Technology from the Ministry of Health, Labor, and Welfare, Japan.
References
- 1. Shaib Y, El‐Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis 2004; 24: 115–25. [DOI] [PubMed] [Google Scholar]
- 2. Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology 2008; 48: 308–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Gron PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM. Biliary tract cancers. N Engl J Med 1999; 341: 1368–78. [DOI] [PubMed] [Google Scholar]
- 4. Ito Y. Molecular basis of tissue‐specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells 1999; 4: 685–96. [DOI] [PubMed] [Google Scholar]
- 5. Levanon D, Berstein Y, Negreanu V et al. A large variety of alternatively spliced and differentially expressed mPNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol 1996; 15: 175–85. [DOI] [PubMed] [Google Scholar]
- 6. Li QL, Ito K, Sakakura C et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002; 109: 113–24. [DOI] [PubMed] [Google Scholar]
- 7. Hasegawa K, Yazumi S, Wada M et al. Restoration of RUNX3 enhances transforming growth factor‐beta‐dependent p21 expression in a biliary tract cancer cell line. Cancer Sci 2007; 98: 838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chi XZ, Yang JO, Lee KY et al. RUNX suppresses gastric epithelial cell growth by inducing p21(WAF/Cip1) expression in cooperation with transforming growth factor beta –activated SMAD. Mol Cell Biol 2005; 25: 8097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yano T, Ito K, Fukamachi H et al. RUNX3 tumor suppressor upregulates Bim in gastric epitherial cells undergoing transforming growth factor beta‐inducede apoptosis. Mol Cell Biol 2006; 26: 4474–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene 2010; 29: 2605–15. [DOI] [PubMed] [Google Scholar]
- 11. Wada M, Yazumi S, Takaishi S et al. Frequent loss of RUNX3 geneexpression in human bile duct and pancreatic cancer cell lines. Oncogene 2004; 23: 2401–7. [DOI] [PubMed] [Google Scholar]
- 12. Li QL, Kim HR, Kim WJ et al. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochem Biophys Res Commun 2004; 314: 223–8. [DOI] [PubMed] [Google Scholar]
- 13. Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T. Promoter hypermethylation of tumor suppressor and tumor‐related genes in non‐small cell lung cancers. Cancer Sci 2003; 94: 589–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao WH, Liu WW. Hemizygous deletion and hypermethylation of RUNX3 gene in hepatocellular carcinoma. World J Gastroenterol 2004; 10: 376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim TY, Lee HJ, Hwang KS et al. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest 2004; 84: 479–84. [DOI] [PubMed] [Google Scholar]
- 16. Goel A, Arnold CN, Tassone P et al. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer 2004; 112: 754–9. [DOI] [PubMed] [Google Scholar]
- 17. Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol 2004; 202: 233–40. [DOI] [PubMed] [Google Scholar]
- 18. Huang C, Ida H, Ito K, Zhang H, Ito Y. Contribution of reactivated RUNX3 to inhibition of gastric cancer cell growth following suberoylanilide hydroxamic acid (vorinostat) treatment. Biochem Pharmacol 2007; 73: 990–1000. [DOI] [PubMed] [Google Scholar]
- 19. Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol 2007; 25: 84–90. [DOI] [PubMed] [Google Scholar]
- 20. Kumagai T, Wakimoto N, Yin D et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer 2007; 121: 656–65. [DOI] [PubMed] [Google Scholar]
- 21. Miyazaki M, Ohashi R, Tsuji T, Mihara K, Gohda E, Namba M. Transforming growth factor‐beta 1 stimulates or inhibits cell growth via down‐ or up‐regulation of p21/Waf1. Biochem Biophys Res Commun 1998; 246: 873–80. [DOI] [PubMed] [Google Scholar]
- 22. Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene 2001; 20: 3156–65. Review. [DOI] [PubMed] [Google Scholar]


