Abstract
Evidence exists that sex steroids such as estrogens affect epithelial ovarian cancer. The expression profiles of the estrogen receptors (ER) and ERβ in particular have not been fully described. Therefore, in our present study, we examined the methylation status of the promoters 0K and 0N, and the expression of ERβ isoforms in human epithelial ovarian carcinoma. We then correlated methylation status with ER expression status. Twelve ovarian carcinoma cell lines, six primary cultures of ovarian surface epithelial cells (OSE), and 64 cases of ovarian carcinoma tissues were examined. Bisulfite sequencing and quantitative reverse transcription–polymerase chain reaction were used to evaluate methylation status and expression of ERβ isoforms. The relative abundance of exon 0N, ERβ1, ERβ2, and ERβ4 mRNA was significantly lower in ovarian cancer cell lines and tissues than in their corresponding normal counterparts. However, ERβ5 mRNA level was relatively higher in the cancers, in clear cell adenocarcinoma in particular, than in the normal ovary. Bisulfite sequencing analysis demonstrated that the two promoters of the ERβ gene exhibited distinct methylation patterns. Promoter 0N was unmethylated in OSE, rarely methylated in normal ovarian tissues, and extensively methylated in ovarian cancer cell lines and tissues (11/15 cell lines and 18/32 cancer tissues were extensively methylated). The promoter 0K was, however, unmethylated in both normal and malignant ovarian cells and tissues. A significant correlation between promoter 0N hypermethylation and the loss of exon 0N, ERβ1, ERβ2, and ERβ4 mRNA expression was detected in ovarian carcinoma cells and tissues. Treatment of ovarian carcinoma cells with 5‐aza‐2′ deoxycytidine resulted in reexpression of the ERβ gene. The results of our present study suggest that ERβ is inactivated mainly through aberrant DNA methylation. This process may play an important role in the pathogenesis of epithelial ovarian cancer. (Cancer Sci 2008; 99: 2365–2372)
Of the gynecological malignancies, epithelial ovarian cancer is the leading cause of death in the great majority of developed countries.( 1 ) Sex steroid hormones have been implicated in the etiology and progression of some epithelial ovarian cancers. Human epithelial ovarian carcinomas can express both estrogen receptors (ER) and progesterone receptors (PR).( 2 , 3 ) In endometrioid endometrial and breast carcinomas, steroid hormone receptor status is correlated well with response to hormonal manipulation and prognosis.( 4 , 5 , 6 ) However, in epithelial ovarian carcinoma, the prognostic significance of tumor ER and PR status remains controversial.( 2 , 7 , 8 ) Forty to 60% of ovarian cancers express ERα, but only a small proportion of patients (7–18%) respond clinically to anti‐estrogen treatment.( 9 , 10 )
Most estrogen actions are mediated through its binding to two ER proteins, ERα and ERβ, which belong to a large family of transcription factors, the nuclear receptor family.( 11 ) Wild‐type ERβ (ERβ1) encodes the full‐length, 530 amino‐acid receptor protein, whereas ERβ2 (ERβcx) to ERβ5, which utilize alternative exons, encode variant receptors with different C‐ and N‐termini.( 7 , 12 ) Two novel ERβ mRNA isoforms containing distinct 5′‐untranslated regions, exons 0K and 0N, have recently been demonstrated to be generated by alternative splicing of exon 1.( 13 ) These results indicate that the transcription of the human ERβ gene occurs from at least two different promoters, those of 0N and 0K.
DNA methylation has an essential regulatory function in mammalian development, suppressing gene activity by changing chromatin structure.( 14 , 15 ) Aberrant DNA methylation of promoter region CpG islands may therefore serve as an alternate mechanism that leads to the inactivation of tumor‐suppressor genes in human cancers.( 16 ) Therefore, the identification of genes regulated by methylation‐associated silencing could lead to the characterization of novel regulators of the initiation and progression of human neoplasia.
In the mammary gland, ERβ1 mRNA is downregulated during breast tumorigenesis.( 17 , 18 , 19 ) Decreases in the mRNA expression levels of ERβ1and ERβ2 in breast carcinoma are significantly correlated with the status of promoter 0N hypermethylation.( 19 ) The expression of ERβ1 mRNA is also significantly decreased in localized prostatic adenocarcinoma compared with normal tissues, and this is correlated with aberrant DNA methylation of promoter 0N.( 20 ) In ovarian carcinoma, several studies have demonstrated that the expression of ERβ1 is decreased in ovarian cancer tissues compared with normal ovarian tissue.( 21 , 22 , 23 ) ERβ1 functions as an important regulator of cell proliferation and has a proapoptotic role in ovarian cancer cells. This suggests that ERβ1 is a potential tumor‐suppressor gene in this tumor.( 24 ) However, the expression of the various ERβ isoforms in epithelial ovarian cancer has not been described and the mechanism of their action is unclear. Therefore, in the present study, we measured the mRNA expression levels of the ERβ isoforms and correlated expression levels with the DNA methylation status of the 0K and 0N promoters.
Materials and Methods
Surgical specimens and clinical data. Sixty‐four specimens of histologically confirmed primary ovarian carcinomas were obtained from patients who underwent surgical therapy from 1998 to 2004 at Tohoku University Hospital. Six specimens of normal ovary were also obtained from patients with benign non‐ovarian disease. The samples were stored at –80°C and embedded immediately in OCT compound (Sakura Finetechnical, Tokyo, Japan) until further use. Only sections containing a minimum of 80% carcinoma by examination with hematoxylin–eosin staining were used for total RNA preparation. Clinicopathological variables for the patients in this study, including International Federation of Gynecology and Obstetrics (FigO) stage, histological subtype, and grade, are summarized in Table 1. Histological subtypes and stage were determined according to FigO criteria. Grade was evaluated by one of the authors (J.A.) using a universal grading system for epithelial ovarian cancer.( 25 ) Overall survival was calculated from the time of initial surgery to death, or the date of last contact. The survival times of patients still alive or lost to follow up in September 2007 were censored. The research protocol was approved by the ethics committee of Tohoku University Graduate School of Medicine.
Table 1.
Characteristics of epithelial ovarian cancer patients in the present study
| Characteristic | n | % |
|---|---|---|
| Age | ||
| <50 years | 22 | 34.4 |
| >50 years | 42 | 65.6 |
| Stage | ||
| I/II | 24 | 37.5 |
| III/IV | 40 | 62.5 |
| Histology | ||
| Serous | 31 | 48.4 |
| Endometrioid | 8 | 12.5 |
| Mucinous | 7 | 10.9 |
| Clear cell | 17 | 26.6 |
| Grade | ||
| 1 | 29 | 45.3 |
| 2 | 25 | 39.1 |
| 3 | 10 | 15.6 |
Ovarian cancer cell lines and primary culture of surface epithelial cells. OVCAR3, Caov3, SKOV3, TOV112D, TOV21G, OV90, and ES2 (adenocarcinoma, OVCAR3, SKOV3; serous adenocarcinoma, Caov3, OV90; clear cell adenocarcinoma, TOV21G, ES2; endometrioid adenocarcinoma, TOV112D) cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). JHOS2, JHOS3, JHOS4, HTOA, OMC3, JHOC5, JHOC7, and JHOC8 (serous adenocarcinoma, JHOS2, JHOS3, JHOS4, HTOA; mucinous adenocarcinoma, OMC3; clear cell adenocarcinoma, JHOC5, JHOC7, JHOC8) cell lines were purchased from Riken Cell Bank (Tsukuba, Japan). Cell lines derived from prostate cancer (PC3), breast cancer (MCF‐7), and endometrial cancer (Ishikawa cells) were also analyzed.
Primary cultures of normal human ovarian surface epithelium (OSE) cells were initiated from surface scrapings of normal ovaries removed from eight women undergoing abdominal total hysterectomy or radical hysterectomy for non‐ovarian disorders at Tohoku University Hospital. The purity of OSE cells was confirmed by immunostaining for cytokeratin and vimentin.( 26 )
RNA preparation and real‐time quantitative reverse transcription–polymerase chain reaction. Total RNA was isolated from frozen tissues, ovarian cancer cell lines, and OSE cells with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). A reverse transcription–polymerase chain reaction (RT‐PCR) kit (Superscript First‐Strand Synthesis System; Invitrogen, Carlsbad, CA, US) was used and complementary DNA (cDNA) synthesis was carried out according to the kit instructions.
Real‐time quantitative polymerase chain reaction was carried out using the Light Cycler system (Roche Diagnostics, Mannheim, Germany). The primers used were: exon 0K, 5′‐TACTGAGTCCGATGAATGTG‐3′ and 5′‐TAAGGCTAGATGG‐TGAGTTT‐3′; exon 0N, 5′‐GTCCGCATTTTAGAGAAGGC‐3′ and 5′‐GGAGGAAGGAGAATTAAGGCT‐3′; ERβ1, 5′‐CCTGGCTAACCTCCTGATGC‐3′ and 5′‐ACCCCGTGATGGAGGACTT‐3′; ERβ2, 5′‐GATCTTGTTCTGGACAGGGATG‐3′ and 5′‐AGGCCTTTTCTGCCCTC‐3′; ERβ3, 5′‐ATGCTTTGGTTTGGGTGAT‐3′ and 5′‐CTTCCCTCAGCATAAACAATC‐3′; ERβ4, 5′‐CTTTGGTTTGGGTGATTG‐3′ and 5′‐CACATGACTCTTGGCACTA‐3′; ERβ5, 5′‐CTTTGGTTTGGGTGATTG‐3′ and 5′‐TTCCTTAACTTGCAGACACT‐3′; ERα, 5′‐AGACACTTTGATCCACCTGA‐3′ and 5′‐CAAGGAATGCGATGAAGTAG‐3′; and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH), 5′‐GCACCGTCAAGGCTGAGAAC‐3′ and 5′‐ATGGTGGTGAAGACGCCAGT‐3′. GAPDH cDNA fragments were amplified as positive controls. Negative controls without RNA and without reverse transcriptase were also done. Two independent RT‐PCR reactions were carried out.
DNA preparation and bisulfite sequencing. Genomic DNA from frozen tissues, ovarian cancer cell lines, and OSE cells was extracted using the AquaPure Gnomic DNA kit (Bio‐Rad, Hercules, CA, USA). Genomic DNA (1 µg) was treated with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer's instructions. Sequence analysis of genomic DNA templates treated with bisulfite was carried out as described previously.( 27 ) The primers used were: exon 0N, 5′‐AGATTTTTTAAATTTGAGATTGGGGTTG‐3′ and 5′‐CTTACCTTACAAATAAACACACC‐3′; and exon 0K, 5′‐GTTGGGGTTATTTYGGGGTTGTT‐3′ and 5′‐CCTCCAACAAACACATTCA‐3′. The schematics of the primer sequences are depicted in Figure 1. Hot‐start polymerase chain reaction was carried out at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 55°C (62°C for exon 0K) for 30 s, and elongation at 72°C for 1 min. The correct‐sized band was isolated and its DNA was extracted using the QIAquick Gel Extraction Kit (Qiagen). The purified DNA was sequenced with the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Figure 1.
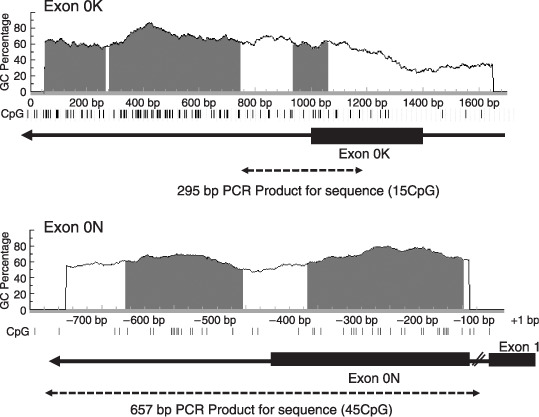
Schematics of the estrogen receptor‐β 5′‐flanking region structure, the CpG islands around the promoters of exons 0K and 0N, and the primer sequences for bisulfite sequencing. Vertical lines represent each CpG site. CpG islands were defined as stretches of DNA that had both a >50% GC content and an observed/overexpected frequency of CpG dinucleotides of >0.6.( 40 ) The polymerase chain reaction (PCR)‐amplified region for bisulfite sequencing is indicated by a dashed arrow. The approximately 10‐kbp intron between exon 0N and exon 1 is indicated by the diagonal line.
Statistical analysis. Statistical analysis was carried out using Stat View 5.0 (SAS Institute Inc., Cary, NC, US) software. The statistical significance among the ERβ isoforms and patient characteristics was evaluated using the Mann–Whitney U‐test, Kruskal–Wallis analysis of variance, and Scheffe analysis. Univariate analysis of the prognostic significance for prognostic factors was carried out using a log‐rank test, after each survival curve was obtained by the Kaplan–Meier method. All patients who could be assessed were included in the intention‐to‐treat analysis. A result was considered significant when the P‐value was less than 0.05.
Results
Expression of ERβ isoforms in ovarian cancer cells. The gene expression levels of the ERβ isoforms in OSE cells and ovarian cancer cell lines are summarized in Figure 2a. Quantitative RT‐PCR was carried out and the ratios of the ERβ isoforms to GAPDH were calculated to allow for comparison between the cell lines. In OSE cells, the expression pattern for each isoform was relatively homogeneous compared with the heterogeneous pattern in the ovarian cancer cell lines. The median values of exon 0N, ERβ1, and ERβ2 gene expression levels in the cancer cell lines were significantly lower than those in OSE cells (P < 0.001) (Fig. 2b). In contrast, the expression of the ERβ5 gene in the cancer cell lines, particularly in the clear cell adenocarcinomas, was significantly higher than in the OSE cells (P < 0.001) (Fig. 2a,b). The expression of exon 0K was very weak in both ovarian carcinoma cell lines and OSE cells (Fig. 2b). The expression patterns in the ovarian cancer cell lines resembled those in cell lines derived from other organs (PC3, MCF7, Ishikawa cells).
Figure 2.
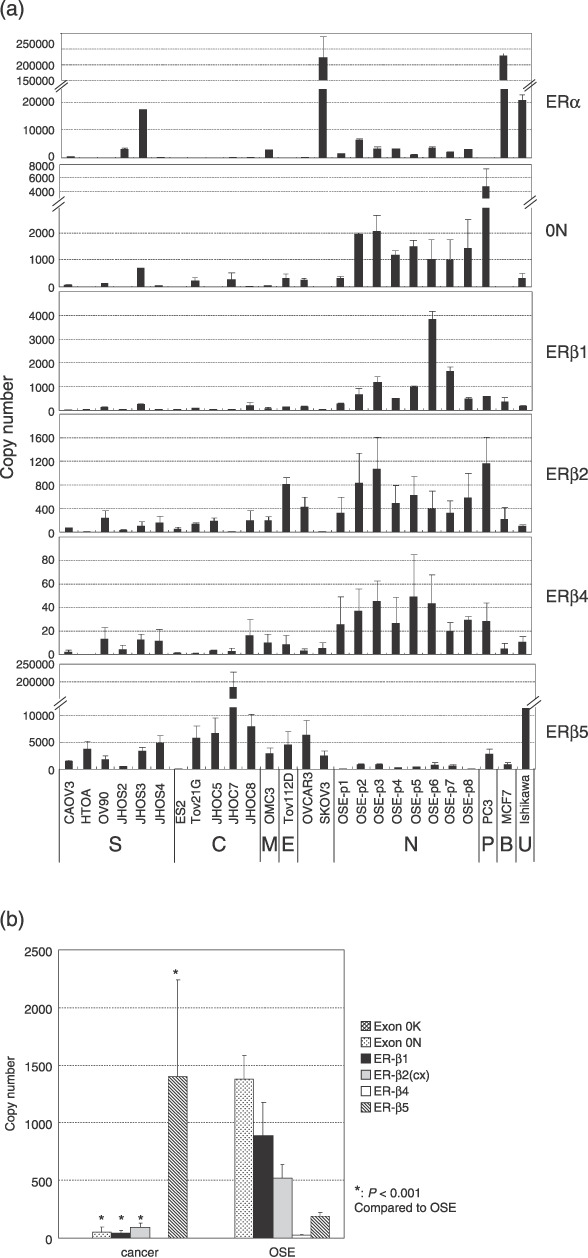
Expression of estrogen receptor (ER)‐β isoform mRNA in ovarian cancer cell lines, normal ovarian surface epithelial cells in primary culture (OSE), PC3, MCF3, and Ishikawa cells. (a) Each bar is the mean ± SD of two independent quantitative real‐time reverse transcription–polymerase chain reaction experiments, standardized against an internal positive control (glyceraldehyde 3‐phosphate dehydrogenase). The capital letters at the bottom of the figure refer to the origin of the cell lines (B, breast carcinoma; C, clear cell adenocarcinoma; E, endometrioid adenocarcinoma; M, mucinous adenocarcinoma; N, normal ovarian surface epithelial cells; P, prostatic carcinoma; S, serous adenocarcinoma; U, uterine carcinoma). Summary of the expression levels of ERβ isoform mRNA in ovarian cancer cell lines and OSE. (b) Each bar is the median ± SE of the expression levels of ERβ isoforms as shown in (a). Asterisks denote significant differences compared with OSE.
Expression of ERβ isoforms in ovarian cancer tissues. Gene expression levels of the ERβ isoforms in the ovarian cancer tissues are summarized in Figure 3. Median values of all of the ERβ isoforms including exon 0N were lower in the ovarian carcinoma tissues than in normal ovarian tissues (P < 0.001) (Fig. 3a). In all histological subtypes, the relative amounts of exon 0N and ERβ4 expression were significantly lower in ovarian cancer tissues than in normal ovarian tissues. In serous adenocarcinoma, the expression levels of all ERβ isoforms including exon 0N were significantly lower than in normal ovarian tissues (P < 0.01) (Fig. 3b). In mucinous and clear cell adenocarcinoma, the expression levels of ERβ1 and ERβ2 were relatively high in comparison to the other hisotological subtypes, although the differences did not reach statistical significance (Fig. 3b). ERβ5 expression was significantly higher in clear cell adenocarcinoma than in the other histological subtypes, and was comparable to the normal ovarian tissues (Fig. 3b). There were no statistically significant correlations between the expression levels of any of the ERβ isoforms or any clinicopathological parameters including prognosis.
Figure 3.
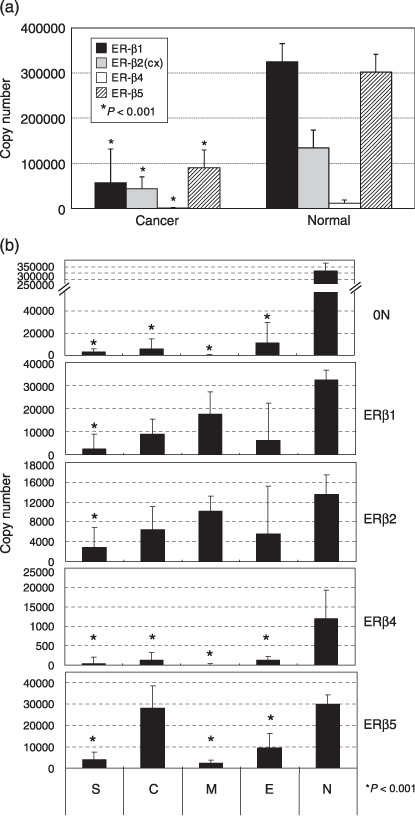
(a,b) Summary of the expression of estrogen receptor (ER)‐β isoform mRNA in normal ovarian and whole ovarian cancer tissues (a), and in each histological subtype (b). Each bar is the median ± SE of two independent quantitative real‐time reverse transcription–polymerase chain reaction experiments for ERβ isoforms, standardized against an internal positive control (glyceraldehyde 3‐phosphate dehydrogenase; GAPDH). Asterisks denote significant differences (P < 0.001) from the normal ovarian tissues. C, clear cell adenocarcinoma; E, endometrioid adenocarcinoma; M, mucinous adenocarcinoma; N, normal ovary; S, serous adenocarcinoma.
Methylation status of the 5′‐untranslated region of ERβ in ovarian cancer cell lines and tissues. We hypothesized that downregulation of the ERβ gene in ovarian cancer occurred through promoter methylation. The comprehensive analysis of the percentage of methylated CpG islands was carried out by bisulfite genomic sequencing (Fig. 4). A 295‐bp region of the promoter 0K, containing 15 CpG sites (Fig. 1), was amplified by polymerase chain reaction from bisulfite‐modified DNA and the methylation status for the 15 CpG sites was clearly determined by direct sequencing. These results demonstrate that all of the CpG sites were unmethylated in all OSE, ovarian cancer cell lines, and tissues examined in the present study. A 675‐bp region of the promoter 0N, containing 45 CpG sites (Fig. 1), was also analyzed. None of five cases and one of six cases were detected to be methylated in the promoter 0N in the OSE and normal ovarian tissues, respectively. Extensive methylation in this region was also detected in most ovarian cancer cell lines and tissues examined in the present study (11 of 15 cases and 18 of 32 cases, respectively) (Fig. 5). In ovarian carcinoma tissues, the frequency of methylation was higher in clear cell and endometrioid adenocarcinomas (seven of eight cases and four of five cases, respectively) than in serous and mucinous adenocarcinoma (7 of 17 cases and none of two cases, respectively), although the differences were not statistically significant (Fig. 5b).
Figure 4.
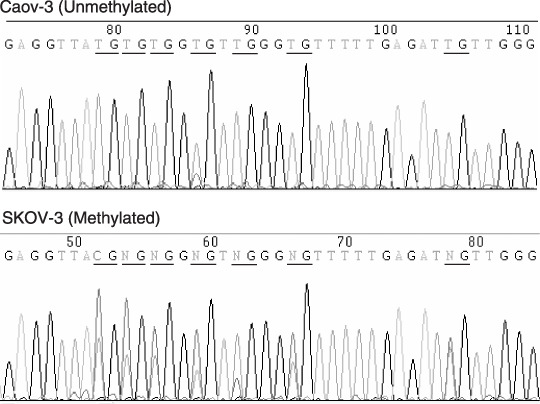
Representative DNA sequences of bisulfite‐treated DNA from ovarian cancer cell lines. CpG sites are underlined. Genomic DNA was modified with sodium bisulfite. This treatment resulted in the conversion of unmethylated cytosines to thymines, whereas the methylated cytosines remained unchanged. The degree of methylation at each CpG site was expressed as the peak height of the methylated cytosine relative to the peak height of total cytosine. No methylation of the promoter 0N region was present in the Caov‐3 cell line and partial methylation was present in the SKOV‐3 cell line.
Figure 5.
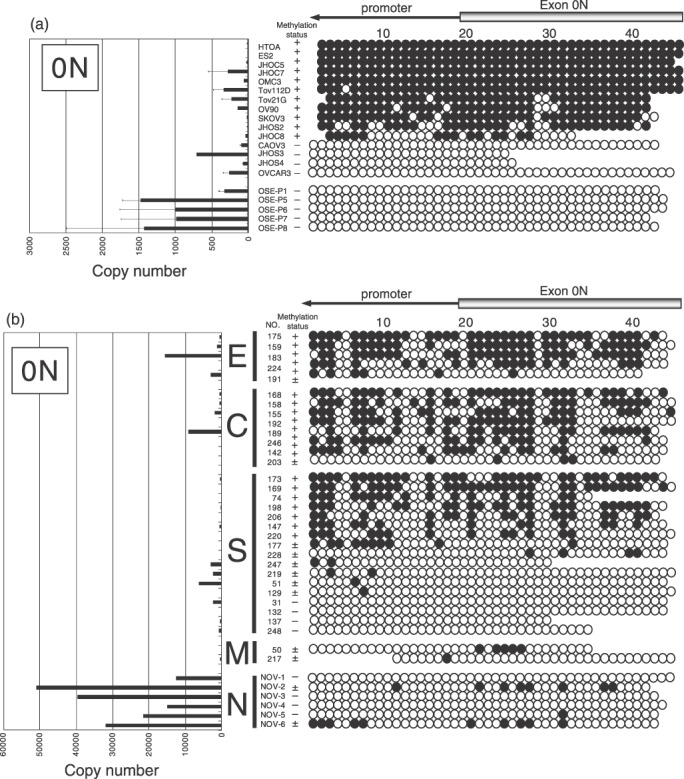
Methylation status of each CpG site in the 0N promoter and expression of 0N mRNA in cells and tissues. Genomic DNA was extracted from 15 ovarian cancer cell lines, five normal ovarian surface epithelial cells in primary culture (OSE), 32 primary ovarian cancer tissues, and six normal ovarian tissues. (a) Ovarian cancer cell lines and OSE. (b) Ovarian cancer and normal ovarian tissues. After bisulfite treatment, direct sequencing was carried out for 45 CpG sites at the exon 0N promoter. Each circle represents a CpG site, a filled circle indicates full or partial methylation, and an open circle indicates no methylation. Methylation status indicated extensive methylation (30–100% methylation of promoter 0N CpG sites, +), marginal methylation (1–29% methylation of promoter 0N CpG sites, ±), and unmethylation (0% methylation of promoter 0N CpG sites, –). Relative expression levels of exon 0N mRNA are shown on the left side. C, clear cell adenocarcinoma; E, endometrioid adenocarcinoma; M, mucinous adenocarcinoma; N, normal ovary; S, serous adenocarcinoma.
We further evaluated whether there was any correlation between the mRNA expression levels of the ERβ isoforms and the methylation status of promoter 0N. As demonstrated in Figure 5, all of the OSE cells and normal ovarian tissues expressed abundant 0N mRNA, whereas most of the ovarian cancer cell lines showed a loss of, or a very low level of, exon 0N mRNA expression (Fig. 5a). In serous carcinoma tissues, the methylation status of promoter 0N was significantly associated with the loss of exon 0N mRNA expression (P < 0.05) (data not shown).
Treatment with 5‐aza‐2′‐deoxycitidine. To further confirm that aberrant DNA methylation contributed to loss of expression of the ERβ gene, we assessed the effect of 5‐aza‐2′‐deoxycitidine (5‐aza‐dC), a demethylating agent, on exon 0N mRNA expression by quantitative RT‐PCR. 5‐Aza‐dC treatment of the ovarian cancer cell lines ES2 and JHOC5, in which the 0N promoter was completely methylated, resulted in marked re‐expression of the exon 0N gene on days 3 and 5 (Fig. 6). The 0N mRNA expression after treatment was significantly higher than before treatment (P < 0.05) (Fig. 6). In the Caov3 cell line, in which the 0N promoter was completely unmethylated, treatment did not restore expression of 0N mRNA. These results suggest that hypermethylation of the 0N promoter plays a critical role in silencing expression of the ERβ isoforms.
Figure 6.
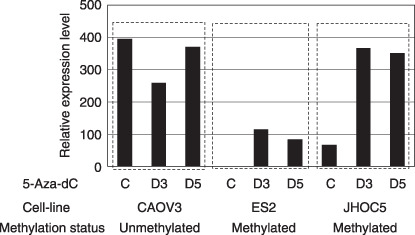
Reexpression of estrogen receptor (ER)‐β 0N mRNA by 5‐aza‐2′‐deoxycitidine (5‐aza‐dC) treatment. Methylation status and mRNA expression of exon 0N were assessed by bisulfite sequencing and quantitative reverse transcription–polymerase chain reaction of D0, D3, and D5 with or without 5‐aza‐dC (1.0 µmol/L) treatment. Methylation status showed methylation of 0N promoter CpG sites of each cell line before 5‐aza‐dC treatment.
Discussion
Aberrant methylation of the ERβ promoter 0N was associated with the loss of mRNA expression, which was restored by treatment with the demethylating agent 5‐aza‐dC. This demonstrates that aberrant DNA methylation is the main pathway of transcriptional silencing of the ERβ (0N) gene in ovarian carcinoma cells. In addition, decreased expression of the ERβ1, ERβ2, and ERβ4 genes occurs in a substantial proportion of ovarian cancer cells and tissues. This suggests that the reduced expression levels of these genes are associated with reduced expression of the ERβ (0N) gene. However, ERβ5 was increased in ovarian cancer cells. These results suggest that ERβ isoforms may be involved in the development and progression of epithelial ovarian cancer. To the best of our knowledge, this is the first report describing epigenetic silencing of ERβ isoforms in human ovarian cancer.
Changes in the mRNA and protein levels of the ERβ isoforms have been documented in various cancerous tissues. Campbell‐Thompson et al. reported that ERβ is the predominant ER subtype in the human colon and that decreased levels of ERβ1 and ERβ2 mRNA is associated with colonic tumorigenesis in women.( 28 ) Leygue et al. reported that the expression levels of ERβ2 and ERβ5 mRNA are higher than that of ERβ1 mRNA in cancer cell lines and breast tumors, and that changes in the relative expression of ERβ1, ERβ2, and ERβ5 mRNA occur during breast tumorigenesis and tumor progression.( 29 ) Zhao et al. demonstrated that both ERβ1 and ERβ2 mRNA levels are significantly lower in tumor cell lines than in normal breast epithelial cells, although ERβ2 mRNA expression is more pronounced than that of ERβ1.( 19 )
In prostate cancer, higher expression of ERβ2 is associated with poor survival.( 30 ) Leung et al. reported that levels of ERβ1 expression are low when compared with those of other ERβ isoforms in various cell lines and tissues.( 31 ) Skrzypczak et al. noted that some adenocarcinomas show a slight decrease in ERβ2 mRNA and a significant increase in ERβ5 mRNA.( 32 ) In epithelial ovarian cancer, several previous studies have demonstrated that ERβ mRNA levels are decreased in ovarian cancer samples compared to normal ovaries, whereas the level of ERα mRNA is similar or slightly higher in cancer samples compared to normal ovaries.( 21 , 22 , 23 , 24 ) ERβ2 (originally referred to as ERβcx) is expressed in several ovarian epithelial tumor types, whereas its splice variant (originally referred to as ERβ2) and the exon 5/6 deletion variant are expressed only at very low levels.( 33 ) However, detailed descriptions of the expression levels of the different ERβ isoforms in ovarian cancer have not been published, likely because previous studies have had small sample sizes with few normal samples for comparison.
The present study utilized substantial numbers of ovarian cancer cell lines and tissues in order to make valid correlations between the various ERβ isoforms and clinicopathological parameters including the histological subtype. Additionally, primary cultures of OSE were used as a normal control to reduce the contamination from stromal components. We studied the expression of those ERβ isoforms with a truncation, insertion, or exchange of sequences in the C‐terminal of ligand‐binding domains (ERβ1, ERβ2, ERβ4, ERβ5) as they were the most frequently observed ERβ isoforms in the breast, endometrial, prostate, and colon cancers described above. We did not investigate the expression of ERβ3 because ERβ3 expression appears to be restricted to the testis.( 7 )
The expression levels of ERβ1, ERβ2, and ERβ4 mRNA were decreased in both ovarian cancer cell lines and cancer tissues in comparison to normal ovarian tissues. The results, especially those for ERβ1 and Erβ2, are consistent with previous studies in that their expression levels are decreased in ovarian cancer. Bardin et al. reported that ERβ expression is decreased in cysts and ovarian carcinomas in comparison to normal ovaries. They demonstrated that ERβ strongly inhibits PEO14‐ and BG1‐dependent cell proliferation and cell motility in a ligand‐independent manner, and induces apoptosis when transfected into cells.( 24 ) Omoto et al. demonstrated that MCF‐7 cells stably expressing ERβ1 or ERβ2 display reduced cell proliferation and colony formation in anchorage‐independent conditions.( 34 ) Additionally, expression of ERβ in ERα‐positive breast cancer cells inhibits their growth.( 35 ) Interestingly, ERβ knockout analysis has shown that ERβ is critical for the regulation of epithelial growth, and its absence results in hyperplasia of the prostatic epithelium.( 36 ) Taken together, these data clearly demonstrate that ERβ is an inhibitor of proliferation and that the loss of ERβ expression could be an important event in the pathogenesis of estrogen‐dependent cancers. It is possible that ERβ acts as a tumor‐suppressor gene. If this is the case, our data suggest that ERβ1 is more important as a tumor suppressor in ovarian cancer because ERβ1 is more comprehensively repressed in ovarian cancers compared with other ERβ isoforms. The identification of ERβ‐regulated specific genes involved in epithelial proliferation and apoptosis may advance our understanding of the progression of ovarian cancer and aid in the design of new targeted therapies.
In contrast to ERβ1, ERβ2, and ERβ4, the expression of ERβ5 mRNA was increased in ovarian cancer cell lines compared to OSE. Remarkably, ERβ5 was strongly expressed preferentially in clear cell adenocarcinoma cell lines and tissues. Clear cell adenocarcinoma has a poor prognosis, is relatively resistant to conventional chemotherapy, and has a characteristic molecular profile.( 3 , 37 ) The ERβ5 mRNA expression level in human breast tumors and endometrial tumors exceeds that of ERβ1. Functionally, ERβ5 heterodimerizes with ERβ1 and significantly enhances its overall activity in a ligand‐dependent manner.( 31 ) Even though little is currently known about ERβ5, we suspect that the effects of either estrogens or antiestrogens may be mediated through dimerization of ERβ1 and ERβ5. This isoform may influence cellular proliferation in ovarian cancer, especially in the clear cell subtype.
With regard to the mechanism behind ERβ gene repression, no mutations or loss of heterozygosity at this gene locus have been reported in ovarian cancer.( 38 ) Recent studies have, however, demonstrated methylation of the ERβ promoter 0N and reduced expression of ERβ isoforms in primary prostate cancer and breast cancer tissues as well as in cancer cell lines.( 19 , 20 ) In the present study, a clear correlation between aberrant methylation and silencing of ERβ 0N mRNA was observed in ovarian cancer cell lines and tissues. Expression was restored by treatment with the demethylating agent 5‐aza‐dC. These results indicate that hypermethylation of the 0N promoter plays a role in silencing ERβ gene expression. We also observed that the two promoter regions of the ERβ gene, 0N and 0K, exhibited distinct methylation and expression patterns. Th 0K promoter was fully unmethylated and repressed in ovarian cancers and normal ovaries. Zhao et al. reported that the 0N promoter was unmethylated in normal breast epithelial cells, but extensively methylated in breast cancer cell lines. In contrast, the 0K promoter was unmethylated in both normal and malignant breast epithelial cells.( 19 ) Our results are consistent with observations in breast cancer. There is little data regarding the relative contributions of the 0N and 0K promoters to the final ERβ1 and ERβ2 transcripts. The ERβ1 cDNA sequence does contain exon 0N, whereas ERβ2 cDNA sequences contain either exon 0N or exon 0K.( 7 , 12 ) Leung et al. reported that ERβ isoforms including ERβ1, ERβ2, and ERβ4, but not ERβ5, were found to be under the control of both the 0K and 0N promoters, whereas ERβ5 was regulated exclusively by the 0K promoter in breast cancer.( 39 ) Further studies characterizing the 0K and 0N promoters as well as putative promoters in other locations are needed to completely elucidate the entire mechanism regulating human ERβ gene expression. Aberrant CpG island methylation is an epigenetic change that is largely responsible for the silencing of ERβ isoforms. Further studies are needed to clarify the potential role of ERβ as a tumor suppressor in the pathogenesis of ovarian cancer.
Acknowledgments
This work was supported in part by a Grant‐in‐Aid for Scientific Research from the Ministry of Health and Welfare, a Grant‐in‐Aid from the Ministry of Education, Science, and Culture, a Grant‐in‐Aid from the Kurokawa Cancer Research Foundation, and a Grant‐in‐Aid from the Japan Society of Gynecologic Oncology.
References
- 1. Akahira J, Suzuki T, Ito K et al . Expression of 5α‐reductases in human epithelial ovarian cancer: its correlation with androgen receptor status. Jpn J Cancer Res 2001; 92: 926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rao BR, Slotman BJ. Endocrine factors in common epithelial ovarian cancer. Endocrine Rev 1991; 12: 14–26. [DOI] [PubMed] [Google Scholar]
- 3. Akahira J, Inoue T, Suzuki T et al . Progesterone receptor isoforms A and B in human epithelial ovarian carcinoma: immunohistochemical and RT‐PCR studies. Br J Cancer 2000; 83: 1488–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGuire WL. Steroid receptors in human breast cancer. Cancer Res 1978; 38: 4289–91. [PubMed] [Google Scholar]
- 5. Ehrlich CE, Young PLM, Cleary RE. Cytoplasmic progesterone and estradiol receptors in normal, hyperplastic and carcinomatous endometria: Therapeutic implications. Am J Obstet Gynecol 1981; 141: 539–46. [DOI] [PubMed] [Google Scholar]
- 6. Saito S, Ito K, Nagase S, Suzuki T et al . Progesterone receptor isoforms as a prognostic marker in human endometrial carcinoma. Cancer Sci 2006; 97: 1308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moore JT, McKee DD, Slentz‐Kesler K et al . Cloning and characterization of human estrogen receptor β isoforms. Biochem Biophys Res Commun 1998; 247: 75–8. [DOI] [PubMed] [Google Scholar]
- 8. Akahira J, Yoshikawa H, Shimizu Y et al . Prognostic factors of stage IV epithelial ovarian cancer: a multicenter retrospective study. Gynecol Oncol 2001; 81: 398–403. [DOI] [PubMed] [Google Scholar]
- 9. Hatch KD, Beecham JB, Blessing JA et al . Responsiveness of patients with advanced ovarian carcinoma to tamoxifen. A Gynecologic Oncology Group study of second‐line therapy in 105 patients. Cancer 1991; 68: 269–71. [DOI] [PubMed] [Google Scholar]
- 10. Scambia G, Benedetti‐Panici P, Ferrandina G et al . Epidermal growth factor, oestrogen and progesterone receptor expression in primary ovarian cancer: correlation with clinical outcome and response to chemotherapy. Br J Cancer 1995; 72: 361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mangelsdorf DJ, Thumnel C, Beato M et al . The nuclear receptor superfamily: the second decade. Cell 1995; 83: 835–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogawa S, Inoue S, Watanabe T et al . Molecular cloning and characterization of human estrogen receptor beta cx: a potential inhibitor of estrogen action in human. Nucl Acids Res 1998; 26: 3505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirata S, Shoda T, Kato J et al . The multiple untranslated first exons system of the human estrogen receptor β (ER β) gene. J Steroid Biochem Mol Biol 2001; 78: 33–40. [DOI] [PubMed] [Google Scholar]
- 14. Ushijima T, Okochi‐Takada E. Aberrant methylations in cancer cells: where do they come from? Cancer Sci 2005; 96: 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Razin A, Ceder H. DNA methylation and gene expression. Microbiol Rev 1991; 55: 451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Esteller M, Corn PG, Baylin SB et al . A gene hypermethylation profile of human cancer. Cancer Res 2001; 61: 3225–9. [PubMed] [Google Scholar]
- 17. Dotzlaw H, Leygue E, Watson PH et al . Estrogen receptor‐β messenger RNA expression in human breast tumor biopsies: relationship to steroid receptor status and regulation by progestins. Cancer Res 1999; 59: 529–32. [PubMed] [Google Scholar]
- 18. Tong D, Schuster E, Seifert M et al . Expression of estrogen receptor beta isoforms in human breast cancer tissues and cell lines. Breast Cancer Res Treat 2002; 71: 249–55. [DOI] [PubMed] [Google Scholar]
- 19. Zhao C, Lam EW, Sunters A et al . Expression of estrogen receptor beta isoforms in normal breast epithelial cells and breast cancer: regulation by methylation. Oncogene 2003; 22: 7600–6. [DOI] [PubMed] [Google Scholar]
- 20. Zhu X, Leav I, Leung Y et al . Dynamic regulation of estrogen receptor‐β expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol 2004; 164: 2003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pujol P, Rey JM, Nirde P et al . Differential expression of estrogen receptor‐α and ‐β messenger RNAs as a potential marker of ovarian carcinogenesis. Cancer Res 1998; 58: 5367–73. [PubMed] [Google Scholar]
- 22. Brandenberger AW, Tee MK, Jaffe RB. Estrogen receptor alpha (ER‐α) and beta (ER‐β) mRNAs in normal ovary, ovarian serous cystadenocarcinoma and ovarian cancer cell lines: down‐regulation of ER‐β in neoplastic tissues. J Clin Endocrinol Metab 1998; 83: 1025–8. [DOI] [PubMed] [Google Scholar]
- 23. Rutherford T, Brown WD, Sapi E et al . Absence of estrogen receptor‐β expression in metastatic ovarian cancer. Obstet Gynecol 2000; 96: 417–21. [DOI] [PubMed] [Google Scholar]
- 24. Bardin A, Hoffmann P, Boulle N et al . Involvement of estrogen receptor β in ovarian carcinogenesis. Cancer Res 2004; 64: 5861–9. [DOI] [PubMed] [Google Scholar]
- 25. Shimizu Y, Kamoi S, Amada S et al . Toward the development of a universal grading system for ovarian epithelial carcinoma. Prognostic significance of histopathologic features – problems involved in the architectural grading system. Gynecol Oncol 1998; 70: 2–12. [DOI] [PubMed] [Google Scholar]
- 26. Nitta M, Katabuchi H, Ohtake H et al . Characterization and tumorigenicity of human ovarian surface epithelial cells immortalized by SV40 large T antigen. Gynecol Oncol 2001; 81: 10–17. [DOI] [PubMed] [Google Scholar]
- 27. Wong KK, Tsang YT, Chang YM et al . Genome‐wide allelic imbalance analysis of pediatric gliomas by single nucleotide polymorphic allele array. Cancer Res 2006; 66: 11 172–8. [DOI] [PubMed] [Google Scholar]
- 28. Campbell‐Thompson M, Lynch IJ, Bhardwaj B. Expression of estrogen receptor (ER) subtypes and ERβ isoforms in colon cancer. Cancer Res 2001; 61: 632–40. [PubMed] [Google Scholar]
- 29. Leygue E, Dotzlaw H, Watson PH et al . Expression of estrogen receptor β1, β2, and β5 messenger RNAs in human breast tissue. Cancer Res 1999; 59: 1175–9. [PubMed] [Google Scholar]
- 30. Fujimura T, Takahashi S, Urano T et al . Differential expression of estrogen receptor β (ERβ) and its C‐terminal truncated splice variant ERβcx as prognostic predictors in human prostatic cancer. Biochem Biophys Res Commun 2001; 289: 692–9. [DOI] [PubMed] [Google Scholar]
- 31. Leung YK, Mak P, Hassan S, Ho SM. Estrogen receptor (ER)‐β isoforms: a key to understanding ER‐β signaling. Proc Natl Acad Sci USA 2006; 103: 13 162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skrzypczak M, Bieche I, Szymczak S et al . Evaluation of mRNA expression of estrogen receptor beta and its isoforms in human normal and neoplastic endometrium. Int J Cancer 2004; 110: 783–7. [DOI] [PubMed] [Google Scholar]
- 33. Chu S, Mamers P, Burger HG et al . Estrogen receptor isoform gene expression in ovarian stromal and epithelial tumors. Clin Endocrinol Metab 2000; 85: 1200–5. [DOI] [PubMed] [Google Scholar]
- 34. Omoto Y, Eguchi H, Yamamoto‐Yamaguchi Y et al . Estrogen receptor (ER) β1 and ERβcx/β2 inhibit ERα function differently in breast cancer cell line MCF7. Oncogene 2003; 22: 5011–20. [DOI] [PubMed] [Google Scholar]
- 35. Paruthiyil S, Parmar H, Kerekatte V et al . Estrogen receptor β inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res 2004; 64: 423–8. [DOI] [PubMed] [Google Scholar]
- 36. Weihua Z, Makela S, Andersson LC et al . A role for estrogen receptor β in the regulation of growth of the ventral prostate. Proc Natl Acad Sci USA 2001; 98: 6330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tan DSP, Kaye S. Ovarian clear cell adenocarcinoma: a continuing enigma. J Clin Pathol 2007; 60: 355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Herynk MH, Fuqua SAW. Estrogen receptor mutations in human disease. Endocr Rev 2004; 25: 869–98. [DOI] [PubMed] [Google Scholar]
- 39. Leung YK, Mak P, Singh L et al . Transcriptional regulation of novel and know estrogen receptor‐β isoforms during prostate carcinogenesis. Proc Amer Assoc Cancer Res 2005; 46: 749. [Google Scholar]
- 40. Gardiner‐Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol 1987; 196: 261–82. [DOI] [PubMed] [Google Scholar]


