Abstract
Induction of the viral BZLF1 gene has previously been shown to be one of the first steps in the reactivation of Epstein-Barr virus (EBV). Using an EBV oriP episomal vector system, we have reconstituted the regulation of the promoter for BZLF1 on stably transfected episomes, mapped promoter elements required for that regulation, and investigated mechanisms that may control the switch between latency and the lytic cycle. Changes in histone acetylation at the promoter for the BZLF1 gene appear to be a key part of the reactivation mechanism of this herpesvirus.
Herpesviruses characteristically display latent persistence in their hosts and intermittent reactivation of replication; the reactivation of some human herpesviruses is associated with specific human diseases. The mechanism by which the viral genome is retained in the latent state is thus a key determinant of the pathogenesis of herpesviruses (30). Epstein-Barr virus (EBV; human herpesvirus 4) is thought to persist in a resting B lymphoid cell population since the viral genome can be detected selectively in these cells by using PCR on DNA extracted from normal virus carriers (2, 24). Reverse transcriptase PCR analysis of EBV gene expression in peripheral blood suggests that the viral gene expression is limited to EBNA-1, LMP-2, BamHI-A rightward transcripts, and the EBER RNAs (5, 29, 43); the humoral immune response is also consistent with this pattern of gene expression in persistence since antibodies to EBNA-1 are always present in the normal virus carrier state (30). It is difficult to study the mechanism of EBV reactivation in vivo because of the low abundance of the cells carrying latent virus, but a broadly similar pattern of EBV gene expression is observed in group I Burkitt's lymphoma (BL) cell lines in culture (32). These cell lines provide the best model available to study the switch between latency and the lytic replication of EBV. Cross-linking the surface immunoglobulin on BL cells so as to mimic the binding of cognate antigen (41, 42) or treatment of the BL cells with transforming growth factor β both activate the lytic cycle of EBV efficiently in some BL cell lines. Both of these systems are likely to reflect physiologically relevant mechanisms of reactivation from latency in vivo.
The key step in the switch from latency to the productive replicative cycle of EBV (6) is the expression of the viral immediate-early gene BZLF1 (also known as ZEBRA, EB1, and Zta). This b-zip transcription factor (9) binds a specific target sequence in the promoters of the early genes of the virus and cooperates with the EBV BRLF1 protein to activate early genes. The promoters for BRLF1 and BZLF1 are also targets for activation by BZLF1, and this autoactivation of expression of BZLF1 appears to be the basis for the efficient switch into the lytic cycle once it begins to be activated (reviewed in reference 39). The kinetics of these processes have previously been studied in detail in the BL cell line Akata treated with anti-immunoglobulin (anti-Ig) (37). Transcription promoter elements have been mapped in the promoter for BZLF1 (Zp) by using transient-transfection assays with induction of Zp expression either by tetradecanoyl phorbol acetate (TPA) in the presence of theophylline (11) or by anti-Ig treatment of the cells (35). The current model involves promoter elements designated ZIA, ZIB, ZIC, ZID, ZII, ZIIIA, and ZIIIB; mutation of either ZIIIA or ZIIIB results in a loss of activation of Zp by BZLF1 (11, 21, 39), and both sites can bind BZLF1 in footprinting assays (11, 18). The ZIIIB element has the higher affinity in footprinting and has therefore been proposed as the major target for BZLF1 autoactivation of the Zp promoter (11). Factors that are candidates for binding to the other sites have also been identified (39). However, it is difficult in the transient assays to relate the efficiency of gene expression to that of the intact EBV genome and thus to know whether the assays quantitatively reflect the true regulation. The physiological relevance of activation of Zp by TPA is also open to question (19).
There is a further important difference between the transient assays and the bona fide latent EBV genome regulation; transfection of an expression vector for BZLF1 into cells containing latent EBV activates the EBV lytic cycle but does not activate the endogenous BZLF1 promoter (17, 20). In contrast, transfected BZLF1 promoter reporter plasmids are transactivated by a cotransfected BZLF1 expression vector (11). This has led to the concept that latency is maintained in the EBV genome in latently infected cells by an unknown mechanism that keeps the Zp promoter switched off until the proper activation signal arrives, for example from anti-Ig signal transduction. This arrangement would prevent accidental activation of the virus replication and thus be a key determinant of latency. A variety of mechanisms have been proposed for negative regulation of EBV transcription in latency, including antisense transcription (28), repression by a viral gene product, and methylation of the EBV genome (31).
In EBV-immortalized lymphoblastoid cell lines (LCLs), the viral LMP-2 proteins can block activation of the promoter for BZLF1 but in BL cell lines such as Akata LMP-2 proteins are either absent or only expressed at a very low level, a finding consistent with the efficient activation of the lytic cycle in these cells. We have previously made recombinant EBV strains with site-directed mutations of regulatory elements to study the control of other aspects of viral gene expression (8), but the conventional genetic approaches for EBV result in recombinant virus in LCLs which express LMP-2. We have therefore sought an alternative (and more rapid) way to mimic accurately the regulation of the BZLF1 promoter by using BL cells as the host. In this study we reconstitute the regulation of the promoter for BZLF1 on stably transfected episomes, map promoter elements required for that regulation, and correlate the changes in histone acetylation at the Zp promoter with activation. The results are consistent with a model in which a repressive chromatin structure at the Zp promoter that can be overcome by histone acetylation is a key part of the mechanism by which the latency of EBV is maintained.
MATERIALS AND METHODS
Plasmid construction.
Plasmid p294 (also named CMVpEBNA-1 in reference 40) was a gift from Bill Sugden. The BZLF1 sequences were derived initially from pUCIE, which contains the EBV sequence of the B95-8 strain from 102114 to 106435. It was made by cloning the NcoI-NruI fragment of B95-8 EBV derived from the E23 cosmid (13) into the SmaI site of pUC12 (having filled the NcoI overhang with Klenow DNA polymerase), with the insert oriented so that the former NcoI end is close to the BamHI and SalI sites in the pUC12 polylinker.
p294:Zp-3241 was made by excising the EBV content of pUCIE by using XbaI and EcoRI sites in the polylinker of pUC, filling the overhangs with Klenow DNA polymerase, and then cloning into the SalI site of p294, which had been similarly blunted with Klenow DNA polymerase. Inserts were obtained in both orientations (p294:Zp-3241 and p294:Zp-3241rev).
p294:Zp-4263 was constructed by first cloning the BamHI R fragment of B95-8 EBV (sequence 103816 to 107402) into the BamHI site of p294, choosing a clone with the insert orientated similarly to p294:Zp-3241. This plasmid was then cut with SalI, and the larger fragment was ligated to the SalI fragment from pUCIE that contained the BZLF1 gene. This resulted in reconstruction of the EBV region from positions 102114 to 107402 in an orientation similar to that of Zp-3241.
p294:Zp-552 was prepared by cloning the BamHI fragment from pUCIE (containing B95-8 EBV sequence 102114 to 103746) into the BamHI site of p294, choosing the orientation similar to that of Zp-3241.
p294:Zp-226 was constructed by cloning the small SphI fragment from p294:Zp-552 into the SphI site of p294, choosing the orientation similar to that of Zp-3241.
In p294:Zp-552X, a XhoI linker was introduced to facilitate cloning of further site directed mutants of the Zp region. The equivalent of the BamHI Z fragment of EBV was subcloned from pUCIE into the BamHI site of pGEM4Z. This construct was cut at the unique NaeI site, an XhoI linker (CCTCGAGG) was inserted, and the plasmid was recircularized. The modified BamHI Z equivalent fragment was then excised and cloned into the BamHI-digested vector segment of p294:Zp-4263. This is thus analogous to p294:Zp-552 but contains the XhoI linker and has only one SphI site.
Wild-type and site-directed mutant sequences of Zp were then transferred into p294:Zp-552X from plasmids (kindly supplied by S. Speck) containing the mutated sites. The Zp region from the wild-type promoter and each site-directed mutant was PCR amplified by using primers W9668 (CCGCTCGAGTGCAATGTTTAGTGAGTT) and W9669 (ACATGCATGCCATGCATATTTCAACTGG). The PCR products were cut with XhoI and SphI and cloned between the XhoI and SphI sites of p294:Zp-552X. Plasmids in this series were called p294:Zp-552Xwt, p294:Zp-552XMIA/B, p294:Zp-552XMIC, p294:Zp-552XMID, p294:Zp-552XMII, p294:Zp-552XMIIIA, and p294:Zp-552XMIIIB, according to the mutation they carried. These plasmids were all sequenced through the Zp region to confirm their identity and to ensure that no mutations had been introduced during the PCR amplification. The PCR primers were designed so that the extra sequence of the linker was eliminated but six nucleotides were substituted to create the XhoI site with all the promoter elements in their normal spacing relative to the BZLF1 coding sequence.
Cell culture, transfection, and induction of BZLF1.
The Akata cell line was maintained in RPMI 1640 medium supplemented with penicillin, streptomycin, and 10% heat-inactivated fetal calf serum. Single-cell cloning of the parental Akata stock was performed according to the method reported by Shimizu et al. (36). Cells were transfected by electroporation at 250 V and 960 μF by using a Bio-Rad GenePulser as described previously (38). Stable transfectants were selected by the addition of 0.2 to 0.3 mg of hygromycin B per ml. Transfected cell lines were named systematically with the Akata cell clone number followed by the plasmid name; for example, AK31/p294:Zp-552Xwt would be Akata clone 31 cells containing plasmid p294:Zp-552Xwt. For each cell line, multiple independent pools were analyzed from two to four transfections so that reproducibility of the BZLF1 response could be ensured. In all cases, results from representative lines are shown.
Induction of the BZLF1 gene was performed by treating exponentially growing cultures with rabbit anti-human IgG 0.5% (vol/vol) (Dako). Unless otherwise indicated, cells were harvested 48 h after this treatment. To prevent spontaneous lytic viral replication prior to micrococcal nuclease digestion, cultures were pretreated with phosphonoacetic acid (0.2 to 0.4 mM for 10 days). Where indicated, cells were pretreated with trichostatin A (50 ng/ml) for 12 h prior to the addition of anti-Ig. B95-8 cells induced into the lytic cycle with the phorbol ester TPA (50 ng/ml for 3 days) were used as a positive control in Western blots.
RNA isolation and RPA.
To prepare cytoplasmic RNA, cells were washed three times with cold phosphate-buffered saline (PBS) and lysed in 150 mM NaCl–10 mM Tris-Cl (pH 7.5)–1 mM MgCl2–0.1% Triton X-100, and the nuclei were removed by centrifugation. The cytoplasmic extract was adjusted to 2% sodium dodecyl sulfate (SDS) and 20 mM EDTA, and proteinase K was added to 0.5 mg/ml. After 5 min at room temperature, the mixture was extracted twice with phenol-chloroform and once with chloroform. RNA was ethanol precipitated and redissolved in water.
An RNase protection assay (RPA) was performed with a probe synthesized from plasmid SP64-Z44, which contains the B95-8 EBV sequence from 102984 to 103650 cloned between the BamHI and EcoRI sites of SP64 (37). Prior to transcription, the plasmid was linearized by digestion with SphI. The plasmid was transcribed by using SP6 polymerase in the presence of [32P]UTP to yield a 477-nucleotide antisense probe containing EBV sequences 102984 to 103419. Overnight hybridization of the riboprobe with 30 μg of cytoplasmic RNA was performed at 42°C by using reagents supplied in the RPA II kit (Ambion). After RNase digestion, protected fragments were analyzed by electrophoresis on a 6% polyacrylamide gel containing 8 M urea. Hybridization of the riboprobe to correctly initiated BZLF1 mRNA generated a protected species of 219 nucleotides.
Western blotting.
Cells were collected by centrifugation, lysed in SDS-gel sample buffer, and sonicated to disperse the DNA (8). The equivalent of 106 cells was loaded into each lane of SDS–10 or 12.5% polyacrylamide gels. After electrophoresis, proteins were transferred to nitrocellulose membranes by electroblotting. After being blocked with milk (8), membranes were incubated either with the anti-BZLF1 monoclonal BZ1 (44) or the anti-LMP1 monoclonal S12 (23). Rabbit anti-mouse horseradish peroxidase conjugate (Amersham) was used at a dilution of 1:2,000 as a secondary antibody, and complexes were visualized by using enhanced chemiluminescence (Amersham).
DNA preparation and Southern blotting.
To prepare genomic DNA, cells were collected by centrifugation and washed in cold PBS. Cells were then resuspended in 100 mM NaCl–5 mM EDTA–10 mM Tris-Cl (pH 7.5) and lysed by bringing the solution to 1% SDS. Proteinase K was added to a concentration of 0.3 mg/ml, and the mixture was allowed to digest at room temperature for 12 h. DNA was extracted twice with phenol-chloroform and once with chloroform prior to ethanol precipitation. The resulting pellet was washed briefly with 70% ethanol and then redissolved in 10 mM Tris-Cl (pH 7.5)–1 mM EDTA.
Genomic DNA from 1.6 × 106 cells (10 μg) was digested with the restriction enzymes indicated, fractionated by electrophoresis on agarose gels, and Southern blotted to nitrocellulose filters (22). Filters were prehybridized in 65°C for 3 h, hybridized with probe overnight at 65°C, and then washed successively with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then 0.1× SSC at 65°C. The probes for hybridization were pBamWLd (B95-8 EBV BamHI fragments W, L, and d, all ligated into the BamHI site of pBR322) and the JH region probe M13 C76R51B (10). All the other probes used were generated by PCR from B95-8 EBV plasmids, and their corresponding positions on the viral genetic map (3) are indicated in the figure legends. Probes were labelled with [32P]dCTP (Amersham) by a random priming reaction.
Bisulfite sequencing.
Methylation of cytosine residues was assayed by the bisulfite sequencing method, in which unmethylated cytosine residues are converted to uracil, which is then fixed as thymidine in a PCR (12). Briefly, DNA from AK31/p294:Zp-3241 cells was denatured with alkali, neutralized, and then treated overnight with 3.1 M sodium bisulfite. After recovery of the DNA and PCR, clones of the PCR product were analyzed by nucleotide sequencing. The oligonucleotides used for PCR were D8097 (CCTCCTCCTCTTTTA) and D8098 (CTAACATCTCCCCTT) with an annealing temperature of 37°C.
Microccocal nuclease digestion.
The protocol used to prepare cell nuclei and micrococcal nuclease digestion was a modification of that described by Dyson and Farrell (7). First, 2 × 107 cells were collected by centrifugation, resuspended in 1.3 ml of 10 mM NaCl–5 mM MgCl2–10 mM Tris-Cl (pH 7.5), and left on ice for 30 min. Cells were then lysed by the addition of 0.7 ml of 5% NP-40 and disrupted with 20 strokes of a Dounce homogenizer (B pestle). Nuclei were purified by sedimentation through 0.8 M sucrose in the same buffer at 700 × g for 5 min and then resuspended in 60 mM KCl–15 mM NaCl–3 mM MgCl2–15 mM Tris-Cl (pH 7.5)–0.25 M sucrose–0.5 mM dithiothreitol–50 μM CaCl2. For digestion, the CaCl2 was supplemented to 2.2 mM, and 11.4 U of micrococcal nuclease (Pharmacia) was added in a final volume of 250 μl. Aliquots of 50 μl were removed at the specified time points into 200 μl of 0.4 M Tris-Cl (pH 7.5)–0.1 M EDTA–1% SDS to terminate the reaction. DNA was purified by digestion with proteinase K–phenol-chloroform extraction and ethanol precipitation.
Chromatin immunoprecipitation.
The method of Braunstein et al. (4) as modified by Alberts et al. (1) was used for chromatin immunoprecipitation. Briefly, aliquots of 106 cells were left untreated or were treated with 0.5% anti-Ig for 3 h. Cells were then fixed by the addition of formaldehyde to a final concentration of 1% for a further 10 min. After the cells were washed, crude nuclei were prepared (4) and resuspended in 1% SDS–10 mM EDTA–50 mM Tris-Cl (pH 8.0). The resulting solution was sonicated such that the modal DNA fragment length was reduced to under 1 kb. This solution was diluted 10-fold with immunoprecipitation buffer (4) and precleared for 2 h with protein A-Sepharose beads preabsorbed with sonicated single-stranded DNA. Immunoprecipitation was performed by incubation for 12 h at 4°C with polyclonal antibody to acetylated histone H4 (Upstate Biotechnology; catalog number 06-598) or a control rabbit serum. Immune complexes were collected by a further incubation with protein A-Sepharose beads and washed extensively prior to elution from the beads. Histone DNA cross-links were reversed by heating to 65°C for 4 h. DNA fragments were purified by phenol-chloroform extraction, ethanol precipitated, and dissolved in 50 μl of water.
Next, 2 μl of the solution of immunoprecipitated DNA fragments was used for each 50-μl PCR. The progress of the reaction was monitored such that the amplification remained in the exponential phase. The primers used were AK1A and AK1B (26), which span the Zp region (B95-8 EBV positions 103180 to 103751); 257V (AGGGCAGTGATAGCGAC) and 982T (TTTCCATCATGTGTTTA, positions 111360 to 111737) in the BKRF4 early gene; 543V (ATTTTATTCTGGGGGCG) and 641M (GGGAAACACTGTTTCGG, positions 8748 to 9560), which span the promoter of the late gene BCRF1; and 018R (TACTTTGGCTTGCCGGG) and 019R (ACGCAGAGGCCTGCACC, positions 149061 to 149637) in the BdRF1 late gene.
RESULTS
Isolation and characterization of Akata cell clones.
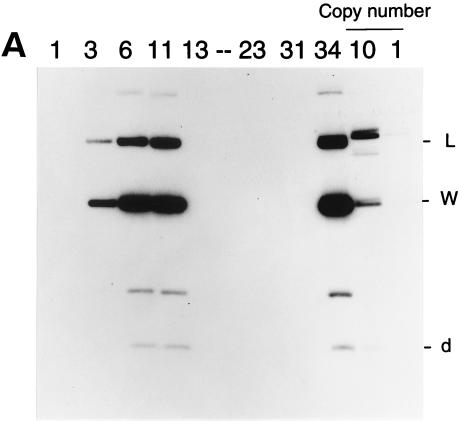
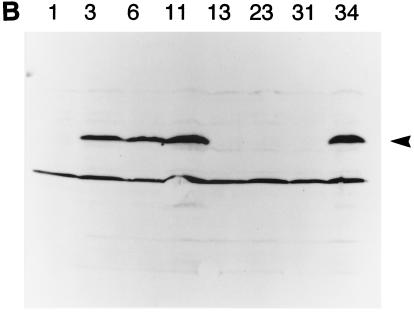
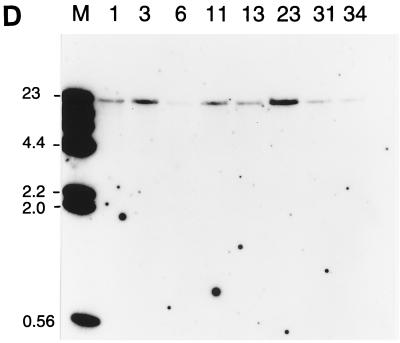
To determine whether BZLF1 EBV gene expression can be properly reconstituted on transfected oriP plasmids, it would be necessary to have comparable human cell lines either containing or lacking EBV so that expression of genes transfected into the EBV-negative cell line could be compared with that of genes in the virus in the EBV-positive cell line. A convenient system became available when it was found (36) that the EBV genome could be lost spontaneously at a low rate from the Akata BL cell line so that EBV-negative variants of this line could be isolated by single-cell cloning. We repeated the cloning of Akata cells and isolated several EBV-negative Akata cell lines, some of which were used in the work described here. Several EBV-positive Akata clones were also isolated during this procedure. The EBV status of the clones was determined by Southern blotting with a probe that could detect EBV DNA (Fig. 1A); overexposures of this autoradiograph in which the single-copy plasmid control was strongly detectable also gave no signal in the EBV-negative Akata clone DNA, and the multiple copies of BamHI-W in EBV gave a sensitivity of 0.1 copies of EBV per cell (not shown). Western blotting for the EBV EBNA-1 (Fig. 1B) protein was also consistent with the presence of EBV in clones 3, 6, 11, and 34. The EBV-positive clones retained the characteristic pattern of expression of EBNA-1 but no expression of LMP-1 (Fig. 1C). Clones 1, 13, 23, and 31 were EBV negative by all criteria. One of the EBV-negative Akata clones (AK31) was particularly appropriate for the subsequent experiments since it grew well in culture and sustained transfection and drug selection reliably. All of the clones retained the appearance of Akata cells under the microscope, and Southern blotting the DNA with a probe for the rearranged JH region (Fig. 1D) showed that all of the Akata clones had the same, unique Ig heavy-chain rearrangement, confirming their common origin.
FIG. 1.
(A) Southern blot for EBV DNA in Akata cell clones. Genomic DNA extracted from Akata cell clones 1, 3, 6, 11, 13, 23, 31, and 34 was digested with BamHI and electrophoresed on an agarose gel. A Southern blot of the gel was probed with EBV BamHI L, W, and d fragments. Dilutions of a plasmid containing EBV BamHI L, W, and d fragments cut with BamHI were coelectrophoresed to give hybridization signals equivalent to 10 and 1 copies per cell. The positions of the BamHI L, W, and d fragments are indicated. The extra band in the Akata tracks between BamHI-d and -W is presumed to result from an extra BamHI restriction site in Akata, which reduces the size of the BamHI L fragment relative to the B95-8 standard. (B) Western blot for EBNA-1. Extracts of Akata cell clones 1, 3, 6, 11, 13, 23, 31, and 34 in an SDS-gel sample buffer were electrophoresed on a polyacrylamide gel and analyzed by Western immunoblotting by using a human serum to detect the EBNA-1 protein (arrowhead). (C) Western blot for LMP-1. Extracts were analyzed as in panel B but with the S12 monoclonal antibody for LMP-1. The LMP-1 signal in the positive control (B95-8 cell extract) is marked with an arrowhead. (D) Southern blot for JH rearrangement. DNA from the Akata cell clones was digested with EcoRI and analyzed by Southern blotting by using a fragment of the JH region clone M13 C76R51B (10) as a probe. Markers were a HindIII digest of phage lambda DNA; the sizes are indicated in kilobases.
Reconstitution of anti-Ig induction of BZLF1.
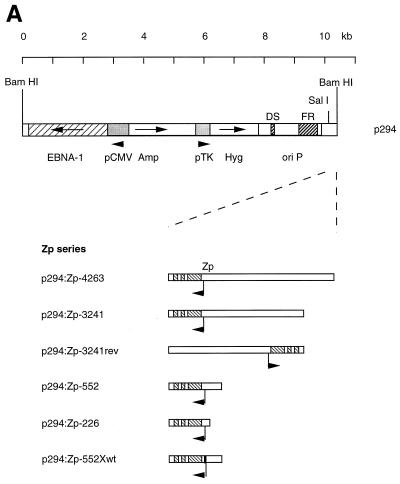
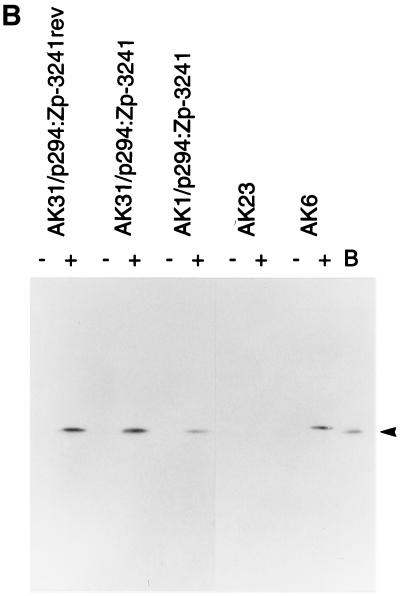
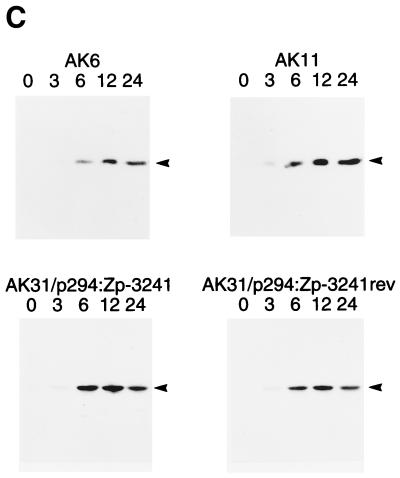
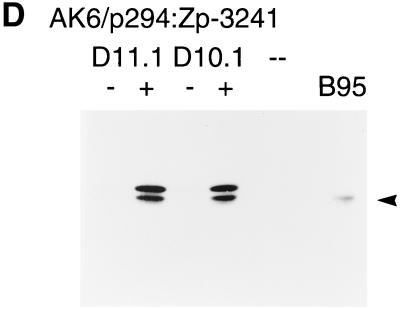
The BZLF1 gene is induced as the first part of the switch from EBV latency into the lytic cycle. This switch can be induced efficiently by treating the Akata cells with antibodies (anti-Ig) that cross-link the surface Ig present on the cells (41, 42). Various oriP/EBNA-1 plasmids containing the BZLF1 gene region were transfected into AK31 cells, and cells maintaining the plasmids were selected by using hygromycin. The structures of the plasmids used are shown in Fig. 2A. When AK31 cells containing the p294:Zp-3241 BZLF1 plasmid (AK31/p294:Zp-3241) were treated with anti-Ig, BZLF1 expression (measured by Western blotting) was induced to a level similar to that of the endogenous viral gene in EBV-positive AK6 cells (Fig. 2B). The induction worked equally well in transfected cells in which the orientation of the BZLF1 gene region was reversed relative to the plasmid vector sequences (plasmid AK31/p294:Zp-3241rev [Fig. 2B]). Inducibility was also not confined to the AK31 cell line as host since line AK1/p294:Zp-3241 contains the same BZLF1 plasmid in EBV-negative AK1 cells and also induced well (Fig. 2B). The kinetics of the induction of BZLF1 were very similar (Fig. 2C) for the transfected BZLF1 gene (AK31/p294:Zp-3241 and AK31/p294:Zp-3241rev) compared with EBV-positive AK6 and AK11 cells. Furthermore, when the p294:Zp-3241 plasmid was transfected into EBV-positive Akata 6 cells, both the endogenous and plasmid BZLF1 proteins were induced (Fig. 2D); the Akata and B95-8 BZLF1 proteins differ in a few amino acids (26) and can be distinguished by SDS-gel electrophoresis.
FIG. 2.
(A) Structures of p294 BZLF1 plasmids. The structures of the plasmids used here are illustrated beneath a kilobase scale. (B to D) Cell lines were cultured with (+) or without (−) anti-Ig, and extracts were analyzed by Western immunoblotting for BZLF1 expression by using the BZ-1 antibody. The BZLF1 signal is indicated with an arrowhead in each panel. (B) Western blot of BZLF1 induction in AK31/p294:Zp-3241rev (line H4), AK31/p294:Zp-3241 (line L4), and AK1/p294:Zp-3241 (line K1) cells after 48 h of anti-Ig treatment. An extract of B95-8 cells treated with TPA was used as a marker for B95-8 BZLF1 (track B), EBV-negative AK23 cells were the negative control, and AK6 cells were the positive control for anti-Ig induction. Two irrelevant tracks have been excised from the center of this blot during photography, but the samples shown were all electrophoresed and blotted together on the same filter and come from the same photographic print. (C) Time course of BZLF1 induction 0, 3, 6, 12, and 24 h after addition of anti-Ig in EBV-positive AK6 and AK11 cells and in transfected AK31/p294:Zp-3241 (line L4) and AK31/p294:Zp-3241rev (line H4) cells. (D) Induction of plasmids and endogenous BZLF1 in two isolates of AK6/p294:Zp-3241 cells (lines D11.1 and D10.1) after 48 h of anti-Ig treatment.
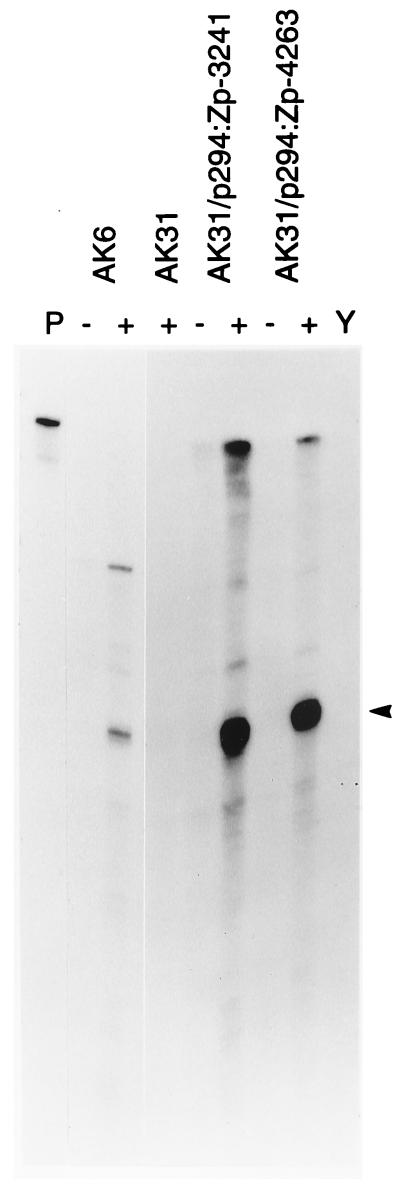
To ensure that the induction of BZLF1 expression was a result of transcription of correctly initiated BZLF1 mRNA from the plasmid, RNA was extracted from some of the cell lines and analyzed by using an RPA. The results (Fig. 3) with AK31/p294:Zp-3241 and AK31/p294:Zp-4263 cells demonstrate a strong induction in response to anti-Ig of RNA corresponding to transcription initiated at the point mapped previously for BZLF1 RNA in the B95-8 EBV genome and subsequently confirmed in Akata cells (26). The copy number of the episomes in the EBV-negative cell lines transfected with the BZLF1 plasmid was shown by Southern blotting Hirt supernatant extracts (15) of the transfected cell lines to be about 30, similar to that of EBV (data not shown).
FIG. 3.
RPA of transcription at Zp promoter in plasmids. RNA was extracted from cell lines AK31/p294:Zp-3241 (line L4) and AK31/p294:Zp-4263 (line C10.1) with (+) or without (−) 6 h of anti-Ig treatment and then analyzed by RPA by using probe SP64-Z44/SphI. The protected fragment of 210 nucleotides (arrowhead) indicates the correct initiation at position 103194 in the EBV sequence (3). RNA from AK6 cells served as a positive control, and AK31 cells treated with anti-Ig and yeast tRNA (Y) were the negative controls. The undigested probe (1/100 of the amount used in the RPA) is shown in lane P. The second, larger band in the AK6 lane corresponds to a sequence difference between Akata EBV and B95-8 EBV, which was used for the probe and oriP plasmids, and is not another transcription start in Akata EBV. It represents the upstream transcription of, for example, the RNA encoding BRLF1.
Regulation of Zp promoter.
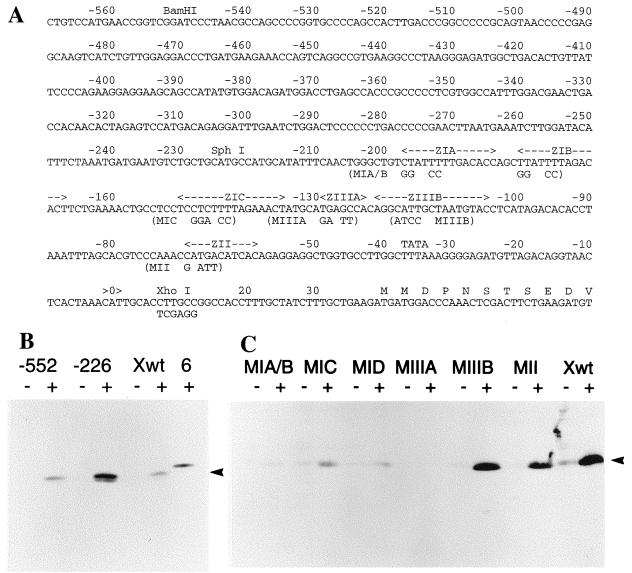
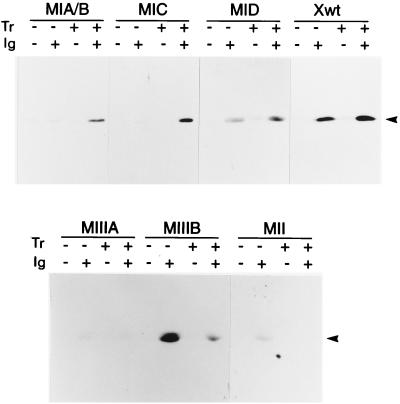
Many transient-transfection and footprinting experiments have resulted in the current model of regulation of Zp, the promoter for BZLF1. In this model (39), most of the sequences governing Zp activity are located within 200 bp upstream of the transcription start with some evidence for further negative elements in the region 200 to 550 bp upstream of the transcription start. The BZLF1 plasmids shown in Fig. 2 and 3 contained at least 3,241 bp of EBV DNA upstream of the Zp transcription start, so plasmids were made containing shorter regions upstream of Zp (Fig. 4A). These were transfected into AK31 cells and lines containing the plasmids were established. The BZLF1 response to anti-Ig was retained in constructs containing only 226 bp upstream of the transcription start (Fig. 4B), but the response of p294:Zp-226 was a little stronger than that of p294:Zp−552, supporting the previous data from transient assays that weak negative elements may lie in the region between positions −226 and −552 (25, 33). These elements have not yet been investigated further since they did not appear to determine the anti-Ig response, but subsequent experiments were performed in the context of the −552 construct so that their effect would be included in the analysis. Several elements have been identified within the −226 region as being crucial for Zp regulation in transient assays and have also been shown to bind specific cell and viral proteins (Fig. 4A). A series of plasmids was therefore produced in which these elements were mutated in the context of the whole −552 promoter (Fig. 4A). Up to this point in the analysis, the sequences of the transfected plasmids were exactly the same as those for B95-8 EBV but, to clone the site-directed mutant constructs, it was necessary to introduce a restriction site which substituted six nucleotides downstream of the transcription start (Fig. 4A). This change made no difference to the anti-Ig response of the wild-type −552 sequence (Fig. 4B). However, disruption of the ZIA/B, ZIC, ZID, or ZIIIA elements in this context resulted in a substantial reduction in anti-Ig responsiveness (Fig. 4C), demonstrating the importance of all of these elements for Zp function. Mutation of ZII caused a partial reduction in anti-Ig responsiveness, which was variable between clones. Mutation of the ZIIIB element made surprisingly little difference to the anti-Ig response in this system (Fig. 4C). The ZIIIB element had the higher affinity of the two BZLF1 binding sites in footprinting assays and was found to be important in transient assays by using induction of the promoter with TPA and BZLF1, although it was less important in the response to TPA alone (11). Although the ZID site has been reported to bind proteins in footprinting analyses (11), its mutagenesis has not been reported in the transient system. To confirm the correct identity of all of these mutant plasmids in the cell lines, at the end of the experiments the Zp region was reisolated from the cell line DNA by PCR and characterized by nucleotide sequencing and restriction enzyme digestion by using EcoRI and BamHI, which produce characteristic cleavage patterns in the various mutants (data not shown).
FIG. 4.
(A) Sequence of B95-8 Zp region showing relevant restriction sites, boundaries of −552 and −226 truncations, and ZIA, ZIB, ZIC, ZIIIA, ZIIIB, and ZII promoter elements mapped in footprinting and transient-transfection experiments (39). Sequences altered in site-directed mutants (MIA, etc.) including the nucleotides substituted for the introduction of the XhoI site used in the cloning done in this study are also shown. The DNA sequence is reversed from the direction of the standard B95-8 map and numbered from the transcription start of the BZLF1 mRNA, shown as position 0 (position 103194 in the B95-8 sequence). The sequence of the start of the protein sequence of BZLF1 is shown in the one-letter amino acid code. The TATA box sequence TTTAAA is also marked. (B) Western blots of BZLF1 expression in cell lines treated with anti-Ig for 48 h (+) or left untreated (−). Lanes: −552, AK31/p294:Zp-552; −226, AK31/p294:Zp-226; Xwt, AK31/p294:Zp-552Xwt; 6, AK6. The BZLF1 protein is marked with an arrowhead. (C) Western blots of BZLF1 expression in cell lines containing Zp mutations treated with anti-Ig for 48 h (+) or left untreated (−). Lanes: MIA/B, AK31/p294:Zp-552XMIA/B; MIC, AK31/p294:Zp-552XMIC; MID, AK31/p294:Zp-552XMID; MIIIA, AK31/p294:Zp-552XMIIIA; MIIIB, AK31/p294:Zp-552XMIIIB; MII, AK31/p294:Zp-552XMII; Xwt, AK31/p294:Zp-552Xwt. The BZLF1 protein from the plasmids is marked with an arrowhead.
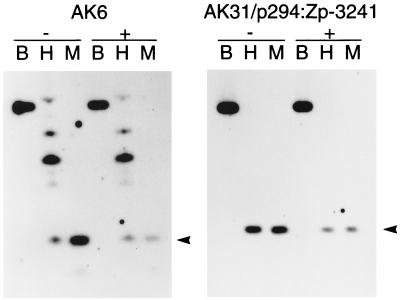
It has been suggested that latency of EBV might be maintained by antisense transcription of the EBNA-1 gene, interference by a latent viral gene product, or DNA methylation. Although they may play a role, these proposed mechanisms seem to be unnecessary at the level of Zp in our system, in which the Zp response to anti-Ig was apparently reconstituted on the stably maintained plasmids. Antisense RNA seems unlikely to be the controlling mechanism, since the regulation worked equally well for either orientation of Zp in the plasmids (Fig. 2B). There seems to be no requirement for another viral gene product apart from EBNA-1 in the maintenance of Zp latency since there are no other EBV genes in the cell lines shown in Fig. 2C and since Zp in these plasmids behaved similarly to EBV. The possibility of DNA methylation of the Zp promoter being required for latency was investigated directly by measuring CpG methylation on the plasmids and on the endogenous EBV in AK6 cells by using comparative sensitivity to restriction digestion by MspI and HpaII (Fig. 5). DNA is cleaved to completion by MspI, but cleavage by HpaII is prevented at CpG methylated restriction sites, resulting in slower-migrating bands in the HpaII tracks on the Southern blot that differ from the MspI bands, if there is methylation. Although there was some methylation of the EBV DNA, no methylation was detected by this method in the Zp promoter region of the plasmid, which regulated Zp properly. There is one CpG dinucleotide within the −226-bp promoter that is not within an MspI restriction site and thus cannot be monitored by restriction digestion. Methylation at this site (position −79 in Fig. 4A) was tested by using sequencing of bisulfite-modified DNA from the AK31/p294:Zp-3241 cells. This showed no methylation in the Zp plasmid at this position (six bisulfite-modified clones sequenced; data not shown), although the −226 deletion regulates properly (Fig. 4B). So, while there is likely to be a role for methylation in the control of gene expression in EBV latency, methylation of Zp is not required to keep Zp inactive prior to induction by anti-Ig in the plasmids, which seem to accurately mimic the induction of Zp in the whole virus. There was also no change in the DNA methylation revealed in the restriction digestion assays in response to anti-Ig treatment (Fig. 5).
FIG. 5.
DNA methylation analysis of Zp region. DNA from cell lines AK6 or AK31/p294:Zp-3241 treated for 48 h with anti-Ig (+) or left untreated (−) was digested with BamHI (B), HpaII (H), or MspI (M). After electrophoresis and Southern blotting, the filters were probed with a Zp PCR fragment (nucleotides 103180 to 103751 of B95-8 EBV). The 498-bp MspI restriction fragment from Zp is marked with an arrowhead; the EBV BamHI Z fragment detected by the probe is 1,794 bp in length.
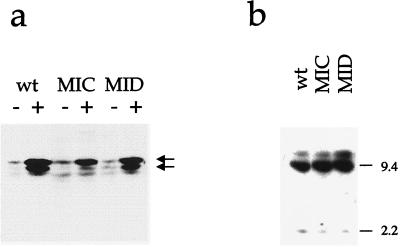
Our results are consistent with the model in which a primary signal from anti-Ig leads to some expression of BZLF1 which can then bind to its own promoter and strongly activate further transcription, this autoactivation being responsible for most of the measurable BZLF1 (11, 26, 39). Transfection of the BZLF1 gene under the control of a strong promoter is itself able to activate the lytic cycle but, interestingly, does not activate the endogenous BZLF1 gene through the Zp promoter (17, 20). The stably maintained plasmids in our system were tested for this level of regulation by comparing the response to anti-Ig of the wild-type Zp plasmid to the MIC mutant in EBV-positive AK6 cells (Fig. 6). The MIC mutant promoter only responded very weakly to direct anti-Ig activation (Fig. 4C), but in a transient-transfection assay it retains 70% of the wild-type inducibility in response to a cotransfected BZLF1 expression vector (11). We thus tested whether the BZLF1 produced from the endogenous Akata genome is able to activate the MIC mutant promoter in trans (Fig. 6). The Zp wild-type plasmid and another mutant (MID) responded directly to anti-Ig induction, but the MIC mutant again responded very weakly, even though BZLF1 was produced from the endogenous EBV (Fig. 6a). The lack of response with the MIC mutant was not due to a lack of template DNA in the cell line; a Southern blot showed a similar level of plasmid DNA in all three cell lines tested (Fig. 6b). The data suggest that the mechanism for protecting the promoter in latently infected cells from activation by BZLF1 in the absence of anti-Ig induction is present in our system. These results and our conclusions that antisense transcription, interference by a latent viral gene product, and DNA methylation of the Zp promoter are not required for correct regulation of Zp lead us to the notion that some form of chromatin structure at the Zp might be preventing Zp activation until the signal from anti-Ig released that repression so that BZLF1 autoactivation of transcription could occur.
FIG. 6.
(a) Western blot of BZLF1 protein expression in extracts from AK6/p294:Zp-552Xwt (wt), AK6/p294:Zp-552XMIC (MIC), and AK6/p294:Zp-552XMID (MID) cells either with (+) or without (−) anti-Ig treatment for 24 h. The two BZLF1 bands are shown by arrows (upper, Akata; lower, B95-8 plasmid). (b) DNA was extracted from the cell lines in panel a without anti-Ig treatment, digested with EcoRI, and analyzed by Southern blotting by using p294:Zp-552Xwt as a probe (sizes shown in kilobases). The 9.4-kb plasmid band hybridizes disproportionately strongly because it contains the oriP and EBNA-1 repeat sequences, so its intensity should not be compared with the larger cross-hybridizing 30-kb EcoRI B fragment derived from the endogenous EBV in the AK6 cells.
Chromatin structure of EBV and regulation of Zp.
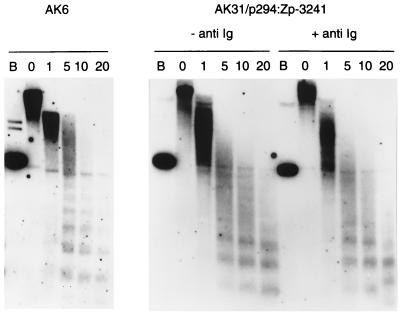
EBV DNA has been shown previously to be organized in nucleosomes in latently infected cells (7, 34). The standard nucleosome pattern was found in EBV by using a probe across the Zp region (Fig. 7). Nucleosome-associated chromatin structure was also detected in the stably maintained Zp plasmids with a Zp probe (Fig. 7), although this appeared to be less extensive than with the corresponding probe on EBV, perhaps because of technical aspects of the assay in view of the much smaller size of the plasmid or relative proximity of Zp in the plasmids to oriP. No difference in nucleosome pattern could be detected in response to anti-Ig induction, but only about 20% of the Akata cells induce into the lytic cycle in response to anti-Ig treatment, so it would be surprising if changes could be detected by examining the bulk DNA.
FIG. 7.
Nucleosome structure of Akata EBV in AK6 cells and BZLF1 plasmid in AK31/p294:Zp-3241 cells (with [+] or without [−] anti-Ig treatment) in the Zp region. Nuclei were treated with micrococcal nuclease, and the resulting DNA was analyzed by Southern blotting. Filters were probed with a Zp PCR fragment (nucleotides 103180 to 103751 of B95-8 EBV). Track B is a BamHI digest of the DNA without nuclease treatment and lanes 0 to 20 (min) are a time course of micrococcal nuclease treatment.
The presence of chromatin structure in the Zp promoter region of the plasmids (Fig. 7) encouraged us to investigate the possibility that some of the Zp elements might be related to recruitment of chromatin remodelling agents such as histone acetyltransferases. Histone acetylation is one mechanism by which chromatin conformation that regulates gene expression is maintained in cells (14, 16, 27). Trichostatin A is a specific inhibitor of histone deacetylases, so treatment of cells with trichostatin A results in an increase in histone acetylation and activation of genes that have been repressed by histone deacetylation. Treatment of AK6 cells with trichostatin A did not induce BZLF1 expression, and there was little evidence for synergy between trichostatin A and anti-Ig treatment on Zp in the wild-type Zp plasmid p294:Zp-552Xwt (Fig. 8). However, trichostatin A treatment rescued the anti-Ig inducibility of certain Zp mutant constructs, particularly the MIA/B and MIC mutants. These results suggest that Zp activation that results from signal transduction from anti-Ig through these elements involves histone acetylation. Factors binding to the ZIA/B and ZIC elements would be involved in the recruitment of histone acetylase activity to the promoter so that activation can proceed. When these elements were mutated, the promoter could not be activated unless it was artificially acetylated by trichostatin A treatment of the cells. There may also be additional steps involving histone acetylation or deacetylation in the regulation of Zp since with the MIIIB and MII mutants trichostatin reduced the inducibility by anti-Ig (Fig. 8).
FIG. 8.
Trichostatin A treatment of Akata cell clones. Western blots of BZLF1 expression in cell lines containing Zp mutations treated (+) or untreated (−) with anti-Ig and/or trichostatin A. Lanes: MIA/B, AK31/p294:Zp-552XMIA/B; MIC, AK31/p294:Zp-552XMIC; MID, AK31/p294:Zp-552XMID; MIIIA, AK31/p294:Zp-552XMIIIA; MIIIB, AK31/p294:Zp-552XMIIIB; MII, AK31/p294:Zp-552XMII; Xwt, AK31/p294:Zp-552Xwt. The BZLF1 protein is marked with an arrowhead.
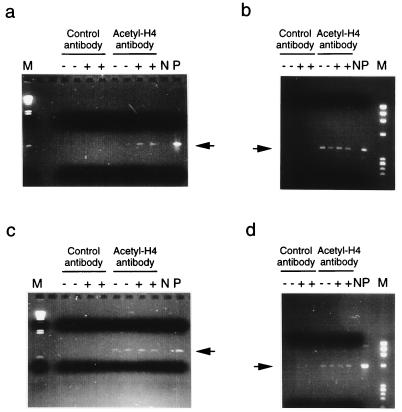
Although the immediate-early nature of Zp indicates that its activation does not require induction of intermediate gene expression, the data in Fig. 8 do not exclude the possibility of trichostatin A acting by inducing expression of other transcription factors that might transregulate Zp. To confirm that histone acetylation at Zp is involved in the activation of the Zp promoter, we directly studied the histone acetylation of the Zp region in response to anti-Ig treatment of the cells in the more physiologically relevant situation of induction of the endogenous Akata virus BZLF1 gene in Akata cells. After treatment with anti-Ig, the chromatin was temporarily stabilized by cross-linking with formaldehyde. The cross-linked chromatin was sonicated to reduce it to the size of a few nucleosomes and then immunoprecipitated with an antibody specific for acetylated histone H4. The precipitated fraction was isolated by using protein A-Sepharose beads, the cross-linking was reversed, and the presence of Zp DNA sequences in the acetylated fraction was determined by PCR with primers that amplify part of the Zp sequence. The amount of DNA template in the PCRs and the number of cycles were adjusted so that the amplification product was detected but not saturated. Anti-Ig treatment of the cells resulted in an increase in Zp sequences in the chromatin fraction that bound to the antibody specific for acetylated histone H4 (Fig. 9). In contrast, this short period of anti-Ig treatment made no difference to the level of DNA from various other regions of the EBV genome in the chromatin fraction that bound to the antibody specific for acetylated histone H4 (Fig. 9). The control regions tested were the promoter of a late gene (BCRF1) and sequences in the BKRF4 or BdRF1 genes. These data confirm that histone acetylation of the chromatin around Zp occurs during Zp activation in the EBV genome.
FIG. 9.
Immunoprecipitation of chromatin containing acetylated histone H4. Duplicate aliquots of Akata (AK6) cells were treated with anti-Ig (+) or were left untreated (−) for 3 h. Sonicated, cross-linked chromatin fragments were immunoprecipitated with antibody to acetylated histone H4 or a control rabbit serum containing an equivalent level of IgG. The immunoprecipitated DNA was analyzed by PCR for Zp promoter region sequences (nucleotides 103180 to 103751) (a) or, as controls, for sequences in the BKRF4 gene (nucleotides 111360 to 111737) (b), the promoter sequence of the EBV late gene BCRF1 (nucleotides 8748 to 9560) (c), and sequences in the BdRF1 gene (nucleotides 149061 to 149637) (d). Negative (N) and positive (P) controls for the PCR reactions, lacking template or with an Akata or B95-8 DNA template, respectively, were coelectrophoresed with the experimental samples. Size markers (M) were a HindIII digest of phage λ DNA (a and c) or a HaeIII digest of X174 DNA (b and d), and PCR products were detected by staining with ethidium bromide. The specific products of the PCR reactions are indicated by arrows.
DISCUSSION
Although much of the understanding of gene expression and regulation has come from transient-transfection assays with promoters linked to reporter genes, it is impossible to relate those results quantitatively to the real gene. The very low efficiency of transient transfection in lymphoid cells also complicates the interpretation of the results. We first addressed this problem in the analysis of EBV gene regulation by making recombinant viruses to study the Cp promoter for the EBNA genes (8). The site-directed mutants of Cp in EBV yielded valuable insight into the regulation of Cp but also illustrated several difficulties with the recombinant virus approach. Most approaches to EBV recombination require that the mutant progeny be competent in transformation of B lymphocytes, imposing considerable selection on the phenotypes of the progeny virus. Standard methods are also very lengthy and difficult to apply to large numbers of recombinant constructs and yield recombinant virus in LCLs, which express LMP-2 that suppresses activation of the lytic cycle, hindering the study of Zp. We have therefore developed an alternative strategy in which autonomously replicating episomes with oriP and EBNA-1 encoded within the plasmid are used as very small analogues of the EBV genome to reconstitute expression and regulation of the BZLF1 gene.
The regulation of the Zp promoter in response to anti-Ig was similar in the oriP plasmids described here to that observed in the EBV genome (Fig. 2). There appeared to be no requirement for sequences more than 226 bp upstream of the transcription start for the positive regulation by anti-Ig, but there was evidence for a weak negative regulatory effect on anti-Ig induction by the sequences between positions −226 and −552. Negative regulatory elements in this region have been investigated by using transient-transfection assays, focusing on YY1 sites (25) and sequences referred to as H1 elements (33), but it is not yet clear how that mapping, which was based on TPA induction or constitutive activity, relates to the regulation studied in our experiments. The activity of the negative elements between positions −226 and −552 thus deserves further investigation. All of our experiments so far have been performed in the context of the intact BZLF1 gene, permitting autoactivation of the Zp promoter by the BZLF1 protein, which is the normal biological situation. We cannot tell on the basis of our current results whether there might be regulatory elements downstream of the transcription start, within the BZLF1 coding sequence or introns that might be involved in the gene regulation, but this could also be investigated in future experiments.
The detailed footprinting and analysis of the effects of mutating Zp promoter elements in the response to TPA and theophylline in transient-transfection reporter assays (11, 18) formed a basis for our investigation. The anti-Ig response of the Zp promoter has also been studied directly by using Zp-CAT reporter plasmids (35); the induction by anti-Ig in those studies was lower than the induction of BZLF1 observed in EBV in Akata cells but nevertheless clearly directed attention to the ZIA/B element in the anti-Ig regulation of Zp (35). We were unable to obtain satisfactory anti-Ig regulation of Zp reporter constructs in transient-transfection experiments (G. Packham, P. Jenkins, and P. J. Farrell, unpublished results), and this led us to pursue the system described here. The effects of some of the site-directed mutants on BZLF1 expression in our system were similar to the transient assays, but there were some important differences.
One notable difference was that in the transient assay experiments, both of the ZIIIA and ZIIIB elements had been found to be essential for efficient autoactivation of the promoter by BZLF1 (11); since ZIIIB was found to have a slightly higher affinity for BZLF1 binding in a footprinting analysis, it was assumed that ZIIIB was the more important BZLF1 binding site (11). In our experiments with the stably transformed cells, which accurately mimic the response of the whole viral genome, ZIIIB can be mutated with only a modest reduction in the activation of BZLF1 expression. In contrast, mutation of the more canonical ZIIIA site greatly reduced activation of the promoter. The degree of inactivation in MIIIA suggests that this mutant may affect the direct response to anti-Ig as well as the autoactivation by BZLF1, perhaps consistent with a slight reduction in inducibility of MIIIA in response to TPA noted in an earlier transient assay (11). There was some variation in the reduction of activity caused by mutation of the ZII element between cell lines, but the result shown in Fig. 8 is typical of the reduction observed. Mutation of the ZIA/B or ZIC elements greatly reduced BZLF1 inducibility by anti-Ig. It is important to remember that we have not tested all promoter sequences within the −221 region systematically, so there may be further essential genetic elements that have not been investigated here. However, the site-directed mutants (devised originally by Flemington and Speck [11]) covered the regions of the promoter found to be protected in a footprinting assay, so they are likely to represent the major elements of importance.
The insensitivity of endogenous Zp in latent EBV in response to activation by BZLF1 compared to Zp in a transfected plasmid (17, 20) indicated that it was likely that Zp is protected from activation by BZLF1 protein in the EBV genomes in latently infected cells. We have argued against antisense transcription as a mechanism for the maintenance of EBV latency because the activation of Zp on the oriP plasmids in our experiments was equally effective when the orientation of the Zp sequences was reversed in the plasmid. There was a certain amount of readthrough transcription in the oriP plasmids (Fig. 3 and data not shown), particularly from the EBNA-1 gene in the plasmids, but there was no evidence that this was relevant to the control of Zp expression. DNA methylation within the minimal Zp region that we have tested also seemed to be unnecessary in the plasmids for apparently normal regulation of Zp. There is clearly DNA methylation of latent EBV in vivo, at least in the parts of the genome that have been examined (31), and the general underrepresentation of CpG residues is consistent with methylation of EBV DNA during evolution of the viral sequence. DNA methylation is likely to provide an additional level of repression of the viral genome in latency, or it might be that methylation outside the Zp promoter is important for the maintenance of latency, but our experiments suggest that the primary mechanism by which Zp is regulated between latency and lytic activation does not require DNA methylation of Zp DNA. We had anticipated that DNA methylation might be an explanation for why only a proportion of the cells in an Akata population (10 to 30% in our experiments) responds to anti-Ig, but the fraction of cells containing the transfected plasmids that responds is similar, indicating that DNA methylation of Zp is not the limiting factor.
Recently it has become clear that modification of histones in chromatin, particularly through acetylation, is a very widespread mechanism by which the activity of genes is regulated (14, 16, 27). The acetylation of the histones most likely results in the more open chromatin conformation which has long been known to be associated with transcriptionally active genes. Histone acetyltransferases associated with gene activation have been identified, and there are now several examples of transcriptional repressors which have been shown to act by recruiting histone deacetylase activity to the promoter. Based on the earlier transcription results (17, 20) and the data presented here, we suggest that a repressive chromatin conformation is present on the Zp DNA in latency that prevents activation of the promoter until the structure is released in response to signal transduction from anti-Ig. The MIA/B and MIC mutants of Zp showed the greatest restoration of anti-Ig inducibility in response to trichostatin A (Fig. 8), indicating that the contribution to promoter activity mediated by signal transduction through these elements in the wild-type Zp promoter results in changes to the promoter that are functionally equivalent to histone acetylation. Whether the factors that bind to these elements in fact contain histone acetylase or recruit a histone acetylase to the promoter remains to be determined, but it is clear that the acetylation of histone H4 occurs in the Zp region in response to anti-Ig signal transduction (Fig. 9). The reductions in anti-Ig-induced BZLF1 expression observed with the trichostatin treatment of MIIIB and MII (Fig. 8) imply that there may be multiple (possibly compensating) steps involving histone acetylation, but the simplest model that would explain the data so far would be that a transcriptionally repressive chromatin structure involving deacetylated histones at the Zp region causes the inactivity of Zp during latency. This repression would be overcome (so that virus reactivation can occur) in response to the signal transduction resulting from cross-linking surface Ig, a procedure designed to mimic the binding of antigen to the surface Ig. Multiple signals are involved in Zp activation (39), but the histone acetylation reported here appears to be a key early step in the process.
ACKNOWLEDGMENTS
We thank Sam Speck for supplying Zp mutant plasmids, Richard Ambinder for advice on bisulphite sequencing, Martine Hollyoake and Claudio Elgueta for excellent technical assistance, and Martin Allday and Graham Packham for critically reading the manuscript.
REFERENCES
- 1.Alberts A, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 2.Babcock G J, Decker L L, Volk M, Thorley-Lawson D A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 3.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P, Barrell B. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 4.Braunstein M, Rose A, Holmes S, Allis C, Broach J. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Smith P, Ambinder R F, Hayward S D. Expression of Epstein-Barr virus BamHI-A rightward transcripts in latently infected B cells from peripheral blood. Blood. 1999;93:3026–3032. [PubMed] [Google Scholar]
- 6.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyson P J, Farrell P J. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 8.Evans T J, Farrell P J, Swaminathan S. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J Virol. 1996;70:1695–1705. doi: 10.1128/jvi.70.3.1695-1705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan J G, Rabbitts T H. The sequence of a human immunoglobulin epsilon heavy chain constant region gene, and evidence for three non-allelic genes. EMBO J. 1982;1:655–660. doi: 10.1002/j.1460-2075.1982.tb01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frommer M, McDonald L, Millar D, Collis C, Watt F, Grigg G, Molloy P, Paul C. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin B E, Karran L. Immortalization of monkey epithelial cells by specific fragments of Epstein-Barr virus DNA. Nature. 1984;309:78–82. doi: 10.1038/309078a0. [DOI] [PubMed] [Google Scholar]
- 14.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 15.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 16.Kadonaga J. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 17.Kolman J L, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70:1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouzarides T, Packham G, Cook A, Farrell P J. The BZLF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene. 1991;6:195–204. [PubMed] [Google Scholar]
- 19.Laux G, Freese U K, Fischer R, Polack A, Kofler E, Bornkamm G W. TPA-inducible Epstein-Barr virus genes in Raji cells and their regulation. Virology. 1988;162:503–507. doi: 10.1016/0042-6822(88)90496-5. [DOI] [PubMed] [Google Scholar]
- 20.Le Roux F, Sergeant A, Corbo L. Epstein-Barr virus (EBV) EB1/Zta protein provided in trans and competent for the activation of productive cycle genes does not activate the BZLF1 gene in the EBV genome. J Gen Virol. 1996;77:501–509. doi: 10.1099/0022-1317-77-3-501. [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Liu S, Speck S. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene. J Virol. 1998;72:8230–8239. doi: 10.1128/jvi.72.10.8230-8239.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Mann K P, Staunton D, Thorley-Lawson D A. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 25.Montalvo E A, Cottam M, Hill S, Wang Y J. YY1 binds to and regulates cis-acting negative elements in the Epstein-Barr virus BZLF1 promoter. J Virol. 1995;69:4158–4165. doi: 10.1128/jvi.69.7.4158-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Packham G, Brimmell M, Cook D, Sinclair A, Farrell P. Strain variation in Epstein-Barr virus immediate early genes. Virology. 1993;192:541–550. doi: 10.1006/viro.1993.1070. [DOI] [PubMed] [Google Scholar]
- 27.Pazin M, Kadonaga J. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 28.Prang N, Wolf H, Schwarzmann F. Epstein-Barr virus lytic replication is controlled by posttranscriptional negative regulation of BZLF1. J Virol. 1995;69:2644–2648. doi: 10.1128/jvi.69.4.2644-2648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe P M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 31.Robertson K, Ambinder R. Methylation of the Epstein-Barr virus genome in normal lymphocytes. Blood. 1997;90:4480–4484. [PubMed] [Google Scholar]
- 32.Rowe M, Rowe D T, Gregory C D, Young L S, Farrell P J, Rupani H, Rickinson A B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarzmann F, Prang N, Reichelt B, Rinkes B, Haist S, Marschall M, Wolf H. Negatively cis-acting elements in the distal part of the promoter of Epstein-Barr virus trans-activator gene BZLF1. J Gen Virol. 1994;75:1999–2006. doi: 10.1099/0022-1317-75-8-1999. [DOI] [PubMed] [Google Scholar]
- 34.Shaw J E, Levinger L F, Carter C., Jr Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979;29:657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu N, Takada K. Analysis of the BZLF1 promoter of Epstein-Barr virus: identification of an anti-immunoglobulin response sequence. J Virol. 1993;67:3240–3245. doi: 10.1128/jvi.67.6.3240-3245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair A J, Brimmell M, Shanahan F, Farrell P J. Pathways of activation of the Epstein-Barr virus productive cycle. J Virol. 1991;65:2237–2244. doi: 10.1128/jvi.65.5.2237-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinclair A J, Jacquemin M G, Brooks L, Shanahan F, Brimmell M, Rowe M, Farrell P J. Reduced signal transduction through glucocorticoid receptor in Burkitt's lymphoma cell lines. Virology. 1994;199:339–353. doi: 10.1006/viro.1994.1132. [DOI] [PubMed] [Google Scholar]
- 39.Speck S, Chatila T, Flemington E. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 40.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 42.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young L S, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F, Rowe D T, Greenspan D, Greenspan J S, Rickinson A B, Farrell P. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol. 1991;65:2868–2874. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]