Abstract
Pescadillo, which has been found to be involved in the process of ribosomal biogenesis, has been demonstrated to play a role in embryonic development, DNA replication, and gene transcription. While deregulation of ribosomal biogenesis was also found to contribute to carcinogenesis, and proteins that regulate ribosomal biogenesis are commonly overexpressed in primary tumors, little is known about the clinical significance and biological function of pescadillo in human breast cancer. In the current study, we found that the expression of pescadillo was markedly up‐regulated in human breast cancer cells and tissues at both mRNA and protein levels. Immunohistochemical analysis revealed that pescadillo expression in clinical stage I–IV primary breast cancer tissues was statistically significantly higher than that in normal breast tissues (P < 0.05). Furthermore, we demonstrated that knockdown pescadillo with RNAis inhibited cell proliferation and the colony‐forming ability of the cells. Anchorage‐independent growth ability assay indicated that ablation of pescadillo led to the reduction of breast cancer cells tumorigenicity in vitro. Moreover, depletion of endogenous pescadillo resulted in decreased expression of cell cycle protein cyclin D1 and up‐regulation of cyclin‐dependent kinase inhibitor p27Kip1, as well as attenuated protein kinase B (Akt)/glycogen synthase kinase 3 beta (GSK‐3β) signaling. Taken together, our results suggest that pescadillo might play a role in promoting the proliferation and carcinogenesis of human breast cancer, and thereby might be a potential target for human breast cancer treatment. (Cancer Sci 2009; 100: 2255–2260)
Ribosomal biogenesis, the process via which ribosomes are made, is essential for cell growth, proliferation, and animal development.( 1 , 2 ) Ribosome biogenesis represents a key metabolic requirement in a proliferating cell, and its tight regulation is crucial for a cell to grow and proliferate.( 3 , 4 ) Mounting evidence has shown that ribosomal proteins are frequently up‐regulated in primary tumors,( 5 , 6 , 7 ) and that mutations in the genes encoding for proteins that regulate rRNA synthesis are also associated with cancer and other human diseases.( 8 , 9 , 10 ) Thus, identification of proteins that regulate ribosomal biogenesis and are involved in the molecular events leading to abnormal homeostasis and carcinogenesis may represent an approach to identifying new therapeutic targets and development of effective therapeutic strategies against cancers.
Pescadillo, originally identified in the embryo of zebrafish, has been reported to be involved in ribosome biogenesis and regulation of DNA replication.( 11 , 12 ) The pescadillo gene codes for a nuclear protein, namely, the pescadillo protein, which contains a breast cancer associated gene 1 (BRCA1) C‐terminal (BRCT) protein–protein interaction domain, numerous nuclear localization signals, and a putative SMT3 suppressor of mif two 3 homolog 1 (SUMO‐1) binding site.( 13 , 14 , 15 ) It has been reported that pescadillo plays a role in the processing of pre‐rRNA molecules during assembly of 60S ribosomal subunits, through formation of the PeBoW complex via combining with Bop1 and WDR12 proteins.( 16 , 17 , 18 , 19 ) Truncation of the BRCT domain of pescadillo protein generates the dominant‐negative mutant which blocks the assembly of the PeBoW complex and processing of the 32S pre‐rRNA into mature 28S rRNA, suggesting that the BRCT domain of pescadillo is crucial for its function in ribosome biogenesis.( 13 , 19 ) Furthermore, pescadillo has been shown to interact with the insulin receptor substrate‐1 (IRS‐1) that acts as an intracellular substrate for various cytokine receptors, including insulin and insulin‐like growth factor‐I (IGF‐I), and serves to send mitogenic or metabolic signals from the cell surface to the nucleus.( 20 ) Moreover, pescadillo has been demonstrated to bind DNA directly and to regulate gene transcription,( 21 ) demonstrating that pescadillo is a multifunctional protein contributing to multiple biological processes in addition to embryo development. Recently, deregulation of pescadillo expression has been found to be associated with cancer initiation and development.( 15 , 22 , 23 , 24 , 25 ) Maiorana and colleagues demonstrated that up‐regulation of pescadillo in AR5 cells, a human fibroblast cell line expressing SV40 T antigen, significantly enhanced the ability of the cells to form colonies in soft agar. Furthermore, they revealed that pescadillo could interact with both IRS‐1 and the SV40 T antigen, and markedly decreased the interaction of T antigen with p53.( 22 ) Consistent with the suggested oncogenic property of pescadillo, it was found that the pescadillo protein was up‐regulated in malignant human astrocytomas as compared with differentiated astrocytes.( 15 ) In addition, Killian and colleagues demonstrated that pescadillo could directly cause chromosomal instability and contribute to carcinogenesis.( 24 ) All these published studies have suggested the possibility that pescadillo is involved in cancer development or progression.
In the current study, we found that the expression of pescadillo was markedly up‐regulated in human breast cancer cells and tissues. Knockdown of peccadillo with RNAi inhibited the cell proliferation and colony‐forming ability of the cells on soft agar. Furthermore, we demonstrated that silenced expression of pescadillo could induce the expression of p27Kip1, reduce the expression of cyclin D1, and lead to inactivation of protein kinase B (Akt)/glycogen synthase kinase 3 beta (GSK‐3β) pathway. Our findings suggest that pescadillo has an important function in the proliferation and carcinogenesis of human breast cancer, indicating that pescadillo might be a potential target for human breast cancer treatment.
Materials and Methods
Cell lines. Primary normal breast epithelial cells (NBEC) were obtained from mammoplasty specimens from a 30‐year‐old woman at the Department of Plastic Surgery, the First Affiliated Hospital of Sun Yat‐sen University, China, in accordance with rules and regulations concerning ethical issues on research use of human subjects, and prior patient consent and approval from the Institutional Research Ethics Committee were obtained. NBEC were cultured in keratinocyte serum‐free medium (Invitrogen, Carlsbad, CA, USA).( 26 ) Breast cancer cell lines, including MDA‐MB‐435, T47D, MDA‐MB‐453, MDA‐MB‐231, MCF‐7, ZR‐75‐30, and SK‐BR‐3, were grown in the DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA).
Patient information and tissue specimens. A total of 92 breast cancer paraffin‐embedded specimens from female patients, which had been histopathologically and clinically diagnosed as breast cancer at the First Affiliated Hospital of Sun Yat‐sen University from 2000 to 2002, were used in the present study. Clinical staging was determined according to the American Joint Committee on Cancer (AJCC) criteria.( 27 ) The median age at the time of surgery was 46 years (range 27–69 years). For the use of these clinical materials for research purposes, prior patient consent and approval from the Institutional Research Ethics Committee were obtained.
Vectors and gene transduction. To introduce RNAis to MDA‐MB‐435 and ZR‐75‐30 cells, we used the pSuper‐retroviral vector to clone and express the following RNAi oligonucleotides to knock down pescadillo expression: pescadillo‐RNAi‐1, CCAGAGGACCTAAGTGT GA; and pescadillo‐RNAi‐2, ACACAAGAAGAAGGTTAAC. Recombinant retroviral vectors were produced by transient co‐transfection as described previously.( 28 ) Viral infection was performed serially, and stable cell lines expressing pescadillo‐RNAis were selected with 0.5 μg/mL puromycin 48 h after infection. After a 10‐day selection, whole cell lysates were fractionated on SDS‐PAGE to examine the level of pescadillo protein.
RNA extraction and real‐time RT‐PCR. Total RNA from cultured cells and surgically obtained tumor tissues was extracted using Trizol reagent according to the manufacturer’s instruction. The extracted RNA was pretreated with RNAase‐free DNase, and 2 μg RNA from each sample was used for cDNA synthesis primed with random hexamers. cDNAs were amplified and quantified in ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Real‐time RT‐PCR primers and probes were designed with the assistance of the Primer Express version 2.0 software (Applied BioSystems). Sequences of the primers were: pescadillo, 5′‐ATCCGAGAGGCCAACAAGCT‐3′ (forward); pescadillo, 5′‐TCAGACCTCACCGCCTCA TC‐3′ (reverse); pescadillo probe, 5′‐(FAM)AGAAG CGGAAAGCC (TAMRA)‐3′; GAPDH, 5′‐GACTCATGACCACAGTC CATGC‐3′ (forward); GAPDH, 5′‐AGAGGCAGGGATGATGTTCTG‐3′ (reverse); GAPDH probe, 5′‐(FAM)CATCAC TGCCACCCAGAAGACTGT G(TAMRA)‐3′. Expression data were normalized to the geometric mean of housekeeping gene GAPDH to control the variability in expression levels and calculated as
where CT represents the threshold cycle for each transcript.
Western blotting. Western blotting was performed as described previously,( 26 ) using anti‐pescadillo (Bethyl Laboratories, Montgomery, TX, USA); anti‐p‐Akt (ser473), anti‐Akt, anti‐p‐GSK‐3β (ser9), anti‐GSK‐3β, anti‐p‐Rb(ser608), anti‐p27 (Cell Signaling Technology, Danvers, MA, USA); anti‐cyclin D1 (Becton Dickinson, Franklin Lakes, NJ, USA); anti‐cyclin A2, anti‐B1 (Epitomics, Burlingame, CA, USA); and anti‐Rb (Santa Cruz Biotechnology, Santa Cruz, CA, USA) as primary antibodies. The membranes were stripped and re‐probed with an anti‐α‐tubulin monoclonal antibody (Sigma, St Louis, MO, USA) as a loading control.
Immunohistochemical (IHC) analysis. IHC analysis was performed to study altered protein expression in five normal breast tissues and 92 human breast cancer tissues using rabbit anti‐pescadillo antibody (1:250; Bethyl Laboratories). The IHC procedure was performed using previously described methods.( 26 ) For negative controls, the rabbit anti‐pescadillo antibody was replaced with normal goat serum, or the anti‐pescadillo antibody was blocked with a recombinant pescadillo polypeptide by co‐incubation at 4°C overnight preceding the immunohistochemical staining procedure.
IHC staining for protein expression in tumor lesions and normal tissues was quantitatively analyzed with the AxioVision Rel.4.6 computerized image analysis system assisted with an automatic measurement program (Carl Zeiss, Oberkochen, Germany). Specifically, the stained sections were evaluated at ×200 magnification, and 10 representative staining fields of each section were analyzed to verify the mean optical density (MOD), which represents the strength of staining signals as measured per positive pixels.
MTT assay. Cells were seeded on 96‐well plates at initial density of (0.2 × 104/well). At each time point, cells were stained with 100 μL sterile MTT dye (0.5 mg/mL) for 4 h at 37°C, followed by removal of the culture medium and addition of 150 μL of dimethyl sulfoxide (DMSO). The absorbance was measured using a Synergy 2 multi‐mode microplate reader (BioTek Instruments, Winooski, VT, USA) at wave‐length of 490 nm. All experiments were performed in triplicate.
Colony formation assays. Cells were plated on 100‐mm plates (1 × 103 cells per plate) and cultured for 10 days. The colonies were stained with 1% crystal violet for 30 s after fixation with 10% formaldehyde for 5 min.
Anchorage‐independent growth ability assay. Five hundred cells were trypsinized and suspended in 2 mL complete medium plus 0.3% agar. The agar–cell mixture was plated on top of a bottom layer with 1% complete medium agar mixture. After 10 days, viable colonies that contained more than 50 cells or were larger than 0.1 mm were counted. All experiments were performed in triplicate.
Bromodeoxyuridine labeling and immunofluorescence. Cells grown on coverslips were incubated with 5‐bromodeoxyuridine (BrdUrd) for 1 h and stained with anti‐BrdU antibody according to the manufacturer’s instructions. Gray level images were acquired under a laser scanning microscope (Axioskop 2 plus; Carl Zeiss, Jena, Germany).
Statistical analysis. All statistical analyses were carried out using the SPSS 10.0 statistical software package (SPSS, Chicago, IL, USA). Means ± SD were calculated, and the two‐tailed Student’s t‐test was performed for paired samples using the data analysis tools provided by the software. In all cases, P < 0.05 was considered statistically significant.
Results
Pescadillo expression was elevated in human breast cancer. Western blotting analysis and real‐time RT‐PCR analysis demonstrated that pescadillo protein and mRNA were highly expressed in all breast cancer cell lines, including T47D, MCF‐7, MDA‐MB‐231, MDA‐MB‐453, SK‐BR‐3, ZR‐75‐30, and MDA‐MB‐435, compared with in human NBEC (Fig. 1A,B). Furthermore, comparative analysis of pescadillo expression was conducted on four cases of paired primary breast cancer tissue and adjacent‐noncancerous tissue. Consistent with the aforementioned results, the expression of pescadillo was also found to be differentially up‐regulated in all four human primary breast cancer tissues compared with their matched adjacent‐noncancerous tissues, using Western blotting and real‐time RT‐PCR (Fig. 1C,D).
Figure 1.
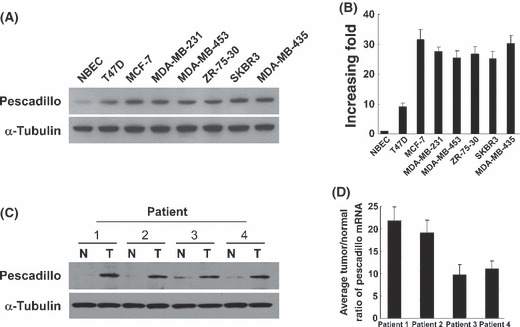
Overexpression of pescadillo in human primary breast cancer. (A) Western blotting analysis of pescadillo protein in normal breast epithelial cells (NBEC) and indicated breast cancer cell lines. α‐Tubulin was probed for loading control. (B) Expression of pescadillo mRNA in the NBEC and cultured breast cancer cell lines. Expression levels were normalized for GAPDH. (C) Western blotting analysis of pescadillo protein from human primary breast cancer (T) and paired tumor‐adjacent non‐cancerous breast tissues (n), with each pair taken from a same patient. (D) Real time RT‐PCR analysis of pescadillo mRNA from the same four pairs of breast cancer and adjacent non‐cancerous tissues. Error bars represent SDs calculated from three parallel experiments.
To further examine the prevalence of pescadillo up‐regulation in breast cancer, the following samples were subjected to immunohistochemical staining with a human pescadillo antibody: five paraffin‐embedded, archived non‐cancerous human breast tissues; and 92 paraffin‐embedded, archived breast cancer tissue samples, including 24 cases of stage I, 25 cases of stage II, 31 cases of stage III, and 12 cases of stage IV tumors. Pescadillo protein was detected in 88 of 92 (95.7%) cases. As shown in Figure 2(A), pescadillo was undetectable or only marginally detectable in the normal breast tissues. High levels of pescadillo expression were present in areas containing tumor cells of the primary breast cancer cells. Quantitative analysis indicated that the average MODs of pescadillo staining in clinical stage I–IV primary tumors were statistically significantly higher than that in normal breast tissues (P < 0.05, Fig. 2B). Taken together, these results clearly demonstrated that pescadillo expression was elevated in human breast cancer.
Figure 2.
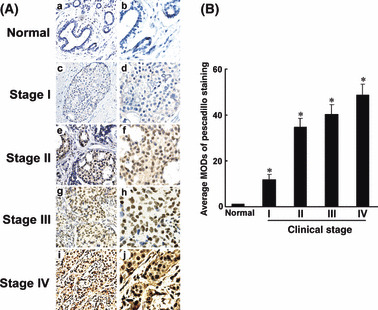
Increased expression of pescadillo protein in histopathological sections of breast cancer as shown by immunohistochemical analysis. (A) Representative examples of IHC staining for pescadillo expression in normal breast tissues and breast cancer of American Joint Committee on Cancer (AJCC) stages I–IV. (a,b) Normal breast tissue (c,d) clinical stage I; (e,f) clinical stage II; (g,h) clinical stage III; and (i,j) clinical stage IV. Magnifications: ×200 (a, c, e, g, and i); ×400 (b, d, f, h, and j). (B) Average mean optical densities (MODs) of pescadillo staining in all stages of breast cancers (randomly chosen 12 cases per stage) were statistically higher than that in normal breast tissues (five cases). *Statistical significance (P < 0.05).
Down‐regulation of pescadillo inhibited proliferation and tumorigenicity of breast cancer cells. To further investigate the biological function of pescadillo in the pathogenesis of breast cancer, we knocked down endogenous pescadillo by two specific RNAis in MDA‐MB‐435 and ZR‐75‐30 cells. As shown in Figure 3(A), both RNAis effectively knocked down the expression of endogenous pescadillo protein in both MDA‐MB‐435 and ZR‐75‐30 cells. MTT assay and colony formation assay revealed that depletion of endogenous pescadillo in either MDA‐MB‐435 or ZR‐75‐30 cells caused significant inhibition of cell growth (Fig. 3B,C, P < 0.05). Moreover, the effect of pescadillo on the tumorigenic activity of breast cancer cells was examined using anchorage‐independent growth ability assay, and Figure 3(D) shows that down‐regulation of pescadillo in MDA‐MB‐435 and ZR‐75‐30 cells significantly reduced the anchorage‐independent growth abilities of both breast cancer cell lines, as indicated by the reduction in colony size and number on soft agar (P < 0.05). These results suggest that down‐regulation of pescadillo in breast cancer cells could result in inhibition of tumorigenicity in in vitro models.
Figure 3.
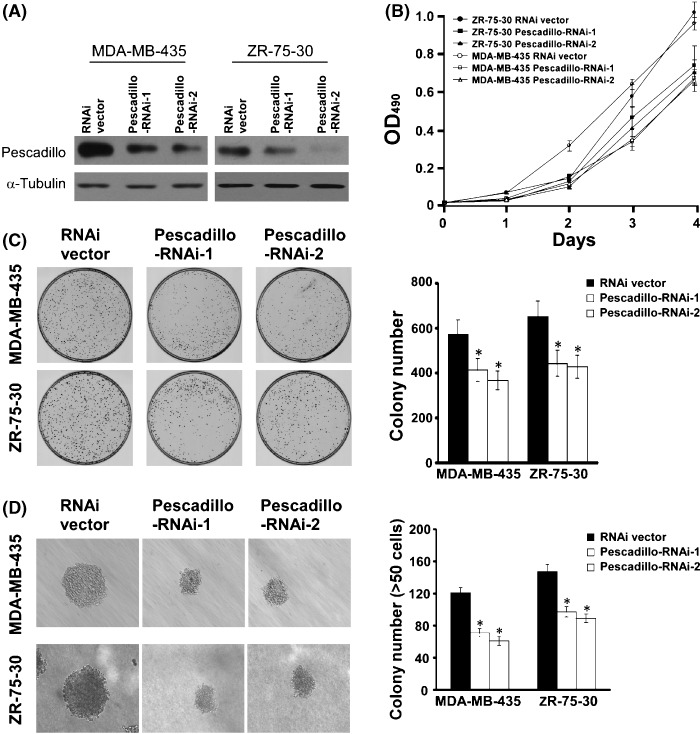
Suppression of pescadillo inhibits breast cancer cell proliferation and tumorigenicity. (A) Western blotting analysis for pescadillo protein in MDA‐MB‐435 and ZR‐75‐30 cells transduced with RNAi vector control and pescadillo RNAi constructs (pescadillo‐RNAi‐1 and pescadillo‐RNAi‐2), respectively. (B) Growth curves of MDA‐MB‐435 and ZR‐75‐30 cells transduced with pescadillo RNAis as examined by MTT assay. Values presented represent means ± SD from three independent experiments. (C) The photos demonstrate results of colony‐formation assay of cells in the plate (left panel) and the mean (±SD) (right panel) colony numbers obtained from three independent experiments. (D) Silencing endogenous pescadillo abrogated the ability of anchorage‐independent growth of breast cancer cells. Colonies that contained more than 50 cells or were larger than 0.1 mm were scored (right panel). Each bar represents the mean ± SD of three independent experiments. *Statistical significance (P < 0.05).
Pescadillo regulated cell cycle modulator cyclin D1 and p27Kip1 in breast cancer cells. The above observations indicated that pescadillo might play a role in the proliferative phenotype of breast cancer cells. The expression and phosphorylation of several protein factors, known to be involved in cell cycle regulation, were further examined. Western blotting analysis revealed that knockdown of endogenous pescadillo did not alter the expression of cyclin A2 and cyclin B1. In contrast, the expression of cyclin D1 was dramatically down‐regulated, and cyclin‐dependent kinase inhibitor p27Kip1 was significantly up‐regulated in the pescadillo‐knocked down cells as compared with vector control cells (P < 0.05, Fig. 4A). In addition, we found that the phosphorylation of Akt and GSK‐3β were decreased in pescadillo RNAis‐transduced cells, as compared with the transduction control cells (P < 0.05), suggesting a possible role of pescadillo in modulating the Akt/GSK3‐β pathway.
Figure 4.
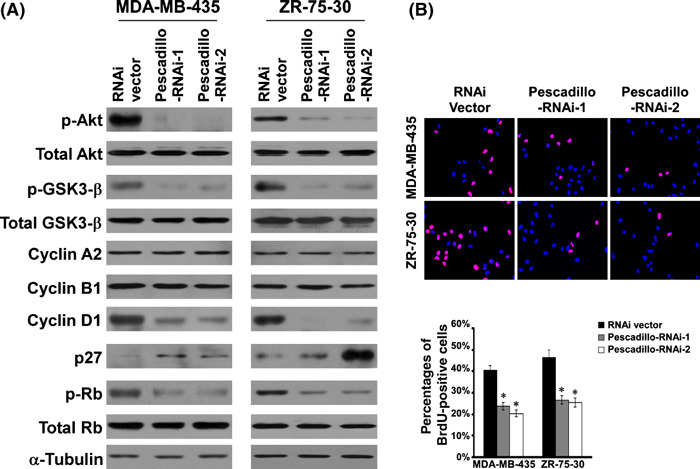
Down‐regulation of pescadillo regulates cell cycle proteins cyclin D1 and p27Kip1 and deactivates protein kinase B (Akt)/glycogen synthase kinase 3 beta (GSK‐3β) signaling in breast cancer cells. (A) Western blotting analysis of expression of p‐Akt (ser473), total Akt, p‐GSK3‐β (ser9), total GSK3‐β, cyclin A2, cyclin B1, cyclin D1, p27, p‐Rb, and total Rb in vector control‐infected and pescadillo RNAi(s)‐infected cells. α‐Tubulin was used as loading control. (B) The figures shown are representative images of cells processed for BrdU incorporation and the mean ± SD for the quantities of BrdU incorporation assessed by three independent experiments. *Statistical significance (P < 0.05).
It is well known that cyclin D1 and p27Kip1 play important roles in the regulation of the progression from the G1 to S phase in mammalian cells.( 29 , 30 , 31 ) To further understand the effect of pescadillo on cell cycle progression in the G1/S transition, BrdU incorporation assay was performed. As shown in Figure 4(B), 39.8% and 46.2% of control MDA‐MB‐435 and ZR‐75‐30 cells, whereas only 19.9% and 24.9% of pescadillo RNAi(s)‐infected MDA‐MB‐435 and ZR‐75‐30 cells, respectively, showed BrdU‐positive signals, supporting the notion that the expression level of pescadillo in breast cancer cells might impact on G1/S transitional phase entry.
Discussion
Our data presented in the current study provide, for the first time, evidence that pescadillo is up‐regulated in breast cancer cell lines and clinical tumors, at both mRNA and protein levels, in comparison to in normal breast epithelial cells and normal breast tissues. Furthermore, we have demonstrated that down‐regulation of endogenous pescadillo could inhibit the proliferation and tumorigenicity of breast cancer cells. These phenomena are associated with down‐regulation of cell cycle regulator cyclin D1 and up‐regulation of cyclin‐dependent kinase inhibitor p27Kip1. Moreover, we have found that the silencing of endogenous pescadillo could deactivate the Akt/GSK‐3β signaling pathway. These findings suggested that deregulation of pescadillo might play an important role in inhibiting carcinogenesis and progression of breast cancer.
Identification of mutations in zebrafish pescadillo gene, which resulted in reduced organ sizes, suggested that pescadillo might be involved in regulating cell proliferation during embryo development.( 12 ) Interestingly, the primary structure of pescadillo contains a BRCT domain that is known to be involved in DNA repair.( 13 , 14 ) Several conserved motifs for covalent attachment of SUMO‐1 were also identified in pescadillo, and previous studies have demonstrated that proteins covalently modified by SUMO‐1 are frequently involved in cell cycle control.( 15 , 32 ) Indeed, pescadillo mutants have been shown to display growth arrest in the G1 or G2 phase of the cell cycle in cells of species ranging from yeast to mammals.( 13 , 14 , 15 , 18 , 19 , 23 , 33 , 34 ) In our current study, we found that inhibition of pescadillo slowed down the growth of human breast cancer cells, as determined by three different methods, namely, MTT assay, BrdU incorporation and colony formation. In conjunction with our observation that RNAis‐induced pescadillo suppression, and decreased the number and size of cell colonies formed in soft agar, the finding supports the notion that pescadillo might be functionally relevant to the carcinogenesis of breast cancer.
The mechanism underlying the cell cycle arrest induced by inhibition of pescadillo remains to be determined. In the current study, we have shown attenuation in the Akt/GSK‐3β/cyclin D1/p27Kip1 signaling in pescadillo‐depleted cells. Akt family proteins have been previously shown to activate a signaling network that promotes G1/S progression through inactivation of GSK‐3β, leading to increased cyclin D1 and reduction of p27Kip1, and may also play a role key in the G2/M transition.( 29 , 30 , 31 , 35 , 36 , 37 ) Thus, one may speculate that the observed G1‐ or G2‐growth arrest in the cell cycle in pescadillo‐depleted cells was, at least in part, mediated through the diminishment of activated Akt and subsequently cyclin D1 protein. Alternatively, pescadillo has recently been shown to interact with the IRS‐1, an important mediator of insulin activities, which can also activate the Akt pathway in the cytoplasm.( 20 , 22 , 23 , 38 , 39 ) Whether pescadillo activates Akt through a mechanism associated with IRS‐1 activation remains to be clarified. On the other hand, it has been reported that depletion of pescadillo inhibits ribosome biogenesis, leading to release of ribosomal proteins L5, L11, and L23, which subsequently interact with and inactivate Mdm2, an E3 ubiquitin ligase that targets p53 for proteasomal degradation, resulting in p53 accumulation and consequent cell cycle arrest.( 40 , 41 , 42 ) It is intriguing that the reported biochemical functions of pescadillo, including modulation of cell cycle and apoptosis, are very much dependent on the accumulation of p53.( 15 , 34 , 43 ) To address this question, we knocked down the expression of pescadillo in two human breast cancer cell lines, MDA‐MB‐435 (p53 mutant, 266G‐E) and ZR‐75‐30 (p53, wild type),( 44 ) and found that the depletion of pescadillo impaired cell proliferation, indicating that the observed cell cycle control mediated by pescadillo is p53‐independent. Indeed, recent studies also provided evidence that pescadillo could perform its biological functions independent of p53. Sikorski et al. showed that pescadillo could directly bind DNA and regulate gene transcription.( 21 ) Moreover, the levels of pescadillo increased in various p53‐/‐ carcinoma cell lines.( 15 , 22 ) Thus, existing evidence appears to support the notion that pescadillo modulation of the cell cycle may be through p53‐independent mechanisms.
The induction of pescadillo expression has been demonstrated to be mediated by two mechanisms. Kinoshita et al. reported that pescadillo was up‐regulated in malignant mouse astrocytes following the loss of p53, using a culture model of glial tumorigenesis.( 15 ) On the other hand, Charpentier et al. have found that pescadillo expression was increased in 190 000 mRNA transcripts in the breast cancer cell line MCF‐7 after exposure to estrogen.( 45 ) In our study, we found that the expression of pescadillo was markedly up‐regulated in human breast cancer cells and tissues at both mRNA and protein levels. As determined by immunohistochemical analysis, 88 of 92 (95.7%) paraffin‐embedded archival breast cancer biopsies displayed moderate to strong staining of pescadillo in tumor cells, whereas no significant staining of pescadillo was detected in the adjacent noncancerous epithelial cells, supporting the notion that pescadillo might play a role in the development and progression of breast cancer. Whether the up‐regulation of pescadillo in the breast cancer specimens examined in the current cohort correlates with the status of p53 and estrogen receptor needs to be further investigated.
A recent study showed that pescadillo could substitute for the transforming function of the SV40 T antigen in mouse embryonic fibroblasts (MEFs),( 22 ) and interestingly, another study suggested that inactivation of the RRB1–pescadillo pathway induced chromosomal instability.( 24 ) These seemingly contradictory data further complicate the investigation into the precise function of pescadillo in tumorigenesis, and whether pescadillo displays differential biological functions in different tissue types or when interacting with different cooperative molecules remains to be determined. Nonetheless, the observation that pescadillo was substantially up‐regulated upon estrogen treatment of breast cancer cells raises the possibility that pescadillo could be an appealing therapeutic target and biomarker for breast cancer and warrant further investigation.( 45 )
Acknowledgments
This study was supported by grants from the Ministry of Science and Technology of China ([973]2005CB724605); the Foundation of the Ministry of Science and Technology of China (30771110, 30870963, 30872930, 30831160517); the Science and Technology Department of Guangdong Province, Zhuhai City, China (PC20071076); the Science and Technology Department of Guangdong Province, China, (07001503, 8251008901000006); the Science and Technology Department of Guangdong Province, China (2006Z3‐E4081); and Foundation of Ministry of Education [No. (2008)890 and No. 200805580047]. The study was also supported by a key grant from the 985‐II project, the Science and Technology Department of Guangdong Province, China, (2008B030301071, 0811220600155).
References
- 1. Pardee AB. G1 events and regulation of cell proliferation. Science 1989; 246: 603–8. [DOI] [PubMed] [Google Scholar]
- 2. Pyronnet S, Sonenberg N. Cell‐cycle‐dependent translational control. Curr Opin Genet Dev 2001; 11: 13–8. [DOI] [PubMed] [Google Scholar]
- 3. Spence J, Gali RR, Dittmar G et al. Cell cycle‐regulated modification of the ribosome by a variant multiubiquitin chain. Cell 2000; 102: 67–76. [DOI] [PubMed] [Google Scholar]
- 4. Klein J, Grummt I. Cell cycle‐dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci U S A 1999; 96: 6096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Draptchinskaia N, Gustavsson P, Andersson B et al. The gene encoding ribosomal protein S19 is mutated in Diamond‐Blackfan anaemia. Nat Genet 1999; 21: 169–75. [DOI] [PubMed] [Google Scholar]
- 6. Kondoh N, Shuda M, Tanaka K et al. Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res 2001; 21: 2429–33. [PubMed] [Google Scholar]
- 7. Ruggero D, Grisendi S, Piazza F et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 2003; 299: 259–62. [DOI] [PubMed] [Google Scholar]
- 8. Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site‐specific synthesis of pseudouridine in ribosomal RNA. Cell 1997; 89: 565–73. [DOI] [PubMed] [Google Scholar]
- 9. Iritani BM, Delrow J, Grandori C et al. Modulation of T‐lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J 2002; 21: 4820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol 2000; 20: 5516–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Du YC, Stillman B. Yph1p, an ORC‐interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 2002; 109: 835–48. [DOI] [PubMed] [Google Scholar]
- 12. Allende ML, Amsterdam A, Becker T et al. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev 1996; 10: 3141–55. [DOI] [PubMed] [Google Scholar]
- 13. Holzel M, Grimm T, Rohrmoser M et al. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res 2007; 35: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haque J, Boger S, Li J et al. The murine Pes1 gene encodes a nuclear protein containing a BRCT domain. Genomics 2000; 70: 201–10. [DOI] [PubMed] [Google Scholar]
- 15. Kinoshita Y, Jarell AD, Flaman JM et al. Pescadillo, a novel cell cycle regulatory protein abnormally expressed in malignant cells. J Biol Chem 2001; 276: 6656–65. [DOI] [PubMed] [Google Scholar]
- 16. Oeffinger M, Leung A, Lamond A et al. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA 2002; 8: 626–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adams CC, Jakovljevic J, Roman J et al. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 2002; 8: 150–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lapik YR, Fernandes CJ, Lau LF et al. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell 2004; 15: 17–29. [DOI] [PubMed] [Google Scholar]
- 19. Grimm T, Holzel M, Rohrmoser M et al. Dominant‐negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW‐complex. Nucleic Acids Res 2006; 34: 3030–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka S, Mohr L, Schmidt EV et al. Biological effects of human insulin receptor substrate‐1 overexpression in hepatocytes. Hepatology 1997; 26: 598–604. [DOI] [PubMed] [Google Scholar]
- 21. Sikorski EM, Uo T, Morrison RS et al. Pescadillo interacts with the cadmium response element of the human heme oxygenase‐1 promoter in renal epithelial cells. J Biol Chem 2006; 281: 24423–30. [DOI] [PubMed] [Google Scholar]
- 22. Maiorana A, Tu X, Cheng G et al. Role of pescadillo in the transformation and immortalization of mammalian cells. Oncogene 2004; 23: 7116–24. [DOI] [PubMed] [Google Scholar]
- 23. Prisco M, Maiorana A, Guerzoni C et al. Role of pescadillo and upstream binding factor in the proliferation and differentiation of murine myeloid cells. Mol Cell Biol 2004; 24: 5421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Killian A, Le Meur N, Sesboue R et al. Inactivation of the RRB1‐Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene 2004; 23: 8597–602. [DOI] [PubMed] [Google Scholar]
- 25. Zhang H, Fang Y, Huang C et al. Human pescadillo induces large‐scale chromatin unfolding. Sci China C Life Sci 2005; 48: 270–6. [DOI] [PubMed] [Google Scholar]
- 26. Li J, Zhang N, Song LB et al. Astrocyte elevated gene‐1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res 2008; 14: 3319–26. [DOI] [PubMed] [Google Scholar]
- 27. Singletary SE, Connolly JL. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J Clin 2006; 56: 37–47; quiz 50‐1. [DOI] [PubMed] [Google Scholar]
- 28. Hahn WC, Dessain SK, Brooks MW et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol 2002; 22: 2111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakayama KI, Hatakeyama S, Nakayama K. Regulation of the cell cycle at the G1‐S transition by proteolysis of cyclin E and p27Kip1. Biochem Biophys Res Commun 2001; 282: 853–60. [DOI] [PubMed] [Google Scholar]
- 30. Slingerland J, Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J Cell Physiol 2000; 183: 10–7. [DOI] [PubMed] [Google Scholar]
- 31. Sherr CJ. D‐type cyclins. Trends Biochem Sci 1995; 20: 187–90. [DOI] [PubMed] [Google Scholar]
- 32. Johnson ES, Blobel G. Cell cycle‐regulated attachment of the ubiquitin‐related protein SUMO to the yeast septins. J Cell Biol 1999; 147: 981–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rohrmoser M, Holzel M, Grimm T et al. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol 2007; 27: 3682–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lerch‐Gaggl A, Haque J, Li J et al. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J Biol Chem 2002; 277: 45347–55. [DOI] [PubMed] [Google Scholar]
- 35. Diehl JA, Cheng M, Roussel MF et al. Glycogen synthase kinase‐3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 1998; 12: 3499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995; 81: 323–30. [DOI] [PubMed] [Google Scholar]
- 37. Shtivelman E, Sussman J, Stokoe D. A role for PI 3‐kinase and PKB activity in the G2/M phase of the cell cycle. Curr Biol 2002; 12: 919–24. [DOI] [PubMed] [Google Scholar]
- 38. Baserga R. The insulin receptor substrate‐1: a biomarker for cancer? Exp Cell Res 2008; 315: 727–32. [DOI] [PubMed] [Google Scholar]
- 39. Easton JB, Kurmasheva RT, Houghton PJ. IRS‐1: auditing the effectiveness of mTOR inhibitors. Cancer Cell 2006; 9: 153–5. [DOI] [PubMed] [Google Scholar]
- 40. Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 2003; 22: 6068–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pestov DG, Strezoska Z, Lau LF. Evidence of p53‐dependent cross‐talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol 2001; 21: 4246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai MS, Zeng SX, Jin Y et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 2004; 24: 7654–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gessert S, Maurus D, Rossner A et al. Pescadillo is required for Xenopus laevis eye development and neural crest migration. Dev Biol 2007; 310: 99–112. [DOI] [PubMed] [Google Scholar]
- 44. Neve RM, Chin K, Fridlyand J et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Charpentier AH, Bednarek AK, Daniel RL et al. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res 2000; 60: 5977–83. [PubMed] [Google Scholar]


