Abstract
Runt‐related transcription factor 3 (RUNX3) is a well known gene for its functions in gastric cancer suppression, but the effect of its genetic variations on the risk of gastric cancer remains unclear. In this study, ten tagging single nucleotide polymorphisms (tSNPs) of the RUNX3 gene were selected and genotyped in a hospital‐based case‐control study of 312 gastric cancer patients and 329 cancer‐free controls in a Chinese population. In the single‐locus analysis, three RUNX3 intronic tSNPs associated with significantly increased risk of gastric cancer were observed: the SNP3 rs11249206 CC genotype (adjusted odds ratio [OR] = 1.75, 95% confidence interval [CI] = 1.03–2.99), compared with the TT genotype; the SNP7 rs760805 AA genotype (adjusted OR = 1.82, 95% CI = 1.14–2.92), compared with the TT genotype; and the SNP8 rs2236852 GG genotype (adjusted OR = 1.69, 95% CI = 1.05–2.72), compared with the AA genotype. In the combined analyses of these three tSNPs, we found that the combined genotypes with four to six variant (risk) alleles (i.e. SNP3 C, SNP7 A, and SNP8 G alleles) were associated with an increased risk of gastric cancer compared with those with one to three variant (risk) alleles (adjusted OR = 2.00, 95% CI = 1.41–2.85), and this increased risk was more pronounced among subgroups of age ≥65 years, never smokers, and never drinkers. However, no significant association was observed in the clinicopathological features analyses. In conclusion, the RUNX3 genetic variants may modulate the risk of gastric cancer in a Chinese population. Further larger and functional studies are warranted to validate the findings. (Cancer Sci 2009; 100: 1688–1694)
Gastric cancer is the fourth most common cancer and second leading cause of cancer death worldwide with an estimated 934 000 new cases in 2002 and 700 000 deaths annually.( 1 ) Almost two‐thirds of the cases occur in developing countries and 42% in China alone.( 1 ) Despite some environmental agents such as Helicobacter pylori infection, salt‐preserved food consumption, and tobacco smoking, are found to be major risk factors for gastric cancer, only a fraction of individuals exposed to these factors develop gastric cancer during their lifetime, suggesting that genetic alteration is an important risk determinant in gastric carcinogenesis.
Runt‐related transcription factor 3 (RUNX3), also known as CFBA3/AML2/Pebp2αC, is a member of the runt‐related transcription factors (RUNXs). The RUNX3 gene is involved in neurogenesis of the dorsal root ganglia and T‐cell differentiation.( 2 , 3 ) Notably, the gene also functions as a tumor suppressor implicated in various types of cancers.( 4 , 5 , 6 ) As a key cytokine, RUNX3 participates in the tumor suppressor transforming growth factor (TGF)‐β signaling pathway that negatively regulates cell growth and promotes apoptosis of epithelial cells.( 7 ) Silencing of the RUNX3 gene may induce many epithelial malignancies,( 8 , 9 ) including gastric cancer.( 10 ) Both human and animal studies in vivo and in vitro had indicated that RUNX3 is closely related to gastric carcinoma. Moreover, RUNX3 is inactivated in more than 60% of human gastric cancers and various human gastric cancer cell lines have decreased RUNX3 expression.( 10 , 11 )
Recently, studies have reported that silencing a tumor suppressor gene may result in loss of its function in tumorigenesis.( 12 ) Inactivation mechanisms of the RUNX3 gene are believed to be promoter hypermethylation and homozygous deletion.( 13 , 14 ) Genetic variants of the RUNX3 gene may also play an inactivation role. For example, Li et al. found that the RUNX3 Arg122Cys mutation within the RUNX3 conserved Runt domain could affect the tumor‐suppressive activity of RUNX3.( 10 ) Also, Kim et al. examined mutations in the RUNX3 coding regions in 34 bladder tumors and found missense mutations and single nucleotide deletion in the conserved Runt domain that abolished the DNA‐binding ability of RUNX3 and resulted in truncation of the protein.( 15 ) This evidence generated the hypothesis that RUNX3 genetic variants may affect the functions of RUNX3, and consequently modulate the cell growth and apoptosis capacity of TGF‐β, and participate in the etiology of human cancers. Importantly, our previous molecular epidemiologic study had shown that genetic variants in RUNX3 may contribute to the risk of bladder cancer.( 16 ) Given the role of RUNX3 in carcinogenesis, we hypothesized that genetic variations in the RUNX3 gene may confer individual susceptibility to gastric cancer.
In the present study, 10 tagging single‐nucleotide polymorphisms (tSNPs) were selected to evaluate the association between these common genetic variants in RUNX3 and the risk of gastric cancer in our ongoing, hospital‐based, case‐control study in a Chinese population.
Materials and Methods
Study subjects. The study subjects consisted of 312 gastric cancer patients and 329 cancer‐free controls recruited from the Yangzhong and Yixin cities, two areas with high mortality of gastric cancer, in the central Jiangsu Province of China between March 2006 and May 2008. All cases were histologically confirmed gastric cancer patients. Those cases who had previous cancer or metastasized cancer from other origin were excluded. Only Han Chinese patients were included in this study and the participation rate was approximately 95%. In this study, all of the control subjects were frequency matched with the cases by age (±5 years) and sex. All of the 329 cancer‐free controls were genetically unrelated to the cases, had no individual history of cancers, and were recruited from the hospital who were seeking health care or doing routine health examinations. Informed consent was obtained from each of the eligible subjects before recruitment. A questionnaire was used to obtain demographic and risk factor information of the study subjects. The response rate of the eligible controls was approximately 85%. Those subjects who smoked daily for more than 1 year were defined as regular smokers. Individuals who consumed one or more alcoholic drinks per week for at least 1 year were considered regular drinkers. After interview, an approximately 5‐mL venous blood sample was collected from each participant. The research protocol was approved by the institutional review board of Nanjing Medical University.
For gastric cancer patients, the clinicopathological variables, including tumor site, tumor histotype, invasion, and lymph node status, were obtained from the medical records of patients. None of the patients had undergone radiotherapy or chemotherapy before surgery. Lauren's criteria were used to classify the tumors into intestinal or diffuse types.( 17 ) Tumor invasion was evaluated and classified as T1, T2, T3, or T4 according to the American Joint Commission for Cancer Staging in 2002 (sixth edition).( 18 ) Lymph nodes were examined and staged according to tumor‐node‐metastasis (TNM) classification (AJCCS, 6th).( 18 )
Single nucleotide polymorphism (SNP) selection and genotyping assays. The detailed method of tSNP selection for the present study has been described previously.( 16 ) Briefly, the tSNP had a minor allele frequency 0.10 in Han Chinese in Beijing (CHB) within a 67‐kb region spanning the RUNX3 gene. As a result, ten tSNPs were selected using a pairwise Tagger method( 18 ) with an r2 cut‐off value of 0.8 to capture all of the common single nucleotide polymorphisms (SNPs) in RUNX3 and the mean of r2 was 0.965. The rs number and relative position of selected 10 tSNPs are shown in Table 1 and their LD values were calculated and visually presented using the Haploview 4.0 software( 18 ) in Figure 1. The LD values among the ten tSNPs were weak with r2 less than 0.60.
Table 1.
Information on ten genotyped tagging single‐nucleotide polymorphisms (tSNPs) of the Runt‐related transcription factor 3 (RUNX3) gene
| SNP no. | NCBI reference SNP no. | Chromosome position † | Location | Base change | MAF | P * | P for HWE** | ||
|---|---|---|---|---|---|---|---|---|---|
| Database ‡ | Cases | Controls | |||||||
| 1 | rs6672420 | 25163597 | Exon 1 | T A | 0.289 | 0.314 | 0.288 | 0.329 | 0.268 |
| 2 | rs11249208 | 25155713 | Intron 1 | A G | 0.330 | 0.267 | 0.255 | 0.332 | 0.129 |
| 3 | rs11249206 | 25150569 | Intron 1 | T C | 0.378 | 0.466 | 0.387 | 0.005 | 0.388 |
| 4 | rs7551188 | 25145787 | Intron 1 | T C | 0.489 | 0.407 | 0.433 | 0.364 | 0.501 |
| 5 | rs1395621 | 25143159 | Intron 1 | C T | 0.411 | 0.420 | 0.426 | 0.865 | 0.147 |
| 6 | rs906296 | 25137245 | Intron 1 | G C | 0.256 | 0.250 | 0.263 | 0.608 | 0.152 |
| 7 | rs760805 | 25124510 | Intron 3 | T A | 0.464 | 0.393 | 0.467 | 0.008 | 0.306 |
| 8 | rs2236852 | 25116354 | Intron 4 | A G | 0.477 | 0.434 | 0.472 | 0.177 | 0.067 |
| 9 | rs9438876 | 25113703 | Intron 4 | A G | 0.244 | 0.285 | 0.289 | 0.902 | 0.514 |
| 10 | rs2282718 | 25113643 | Intron 4 | G A | 0.333 | 0.383 | 0.382 | 1.000 | 0.296 |
SNP position in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP).
Minor allele frequency (MAF) for Han Chinese in Beijing in the HapMap database (http://www.hapmap.org).
P‐value for the allele distribution difference between the cases and controls.
Hardy–Weinberg equilibrium (HWE) P‐value in the control group.
Figure 1.
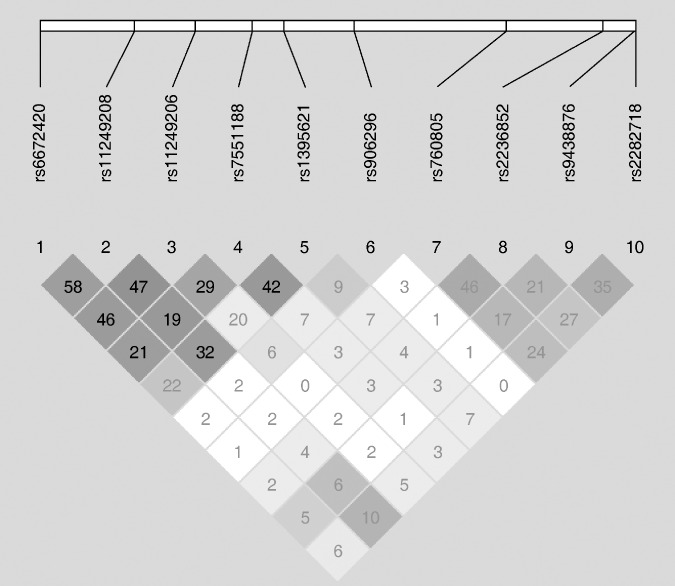
Reconstructed linkage disequilibrium (LD) plot among the Runt‐related transcription factor 3 (RUNX3) 10 tagging single‐nucleotide polymorphisms (tSNPs) in 329 control subjects.
The selected tSNPs were genotyped in all 641 subjects using the PCR–RFLP method. The tSNP information, primers, and restriction enzymes have been described previously.( 16 ) The genotype analysis was done by two persons independently in a blind fashion. Approximately 10% of the samples were randomly selected for repeated genotyping for confirmation, and the results were 100% concordant.
Statistical analysis. Chi‐square test was used to evaluate differences in frequency distributions of selected demographic variables, smoking status, alcohol use, as well as each allele and genotype of the RUNX3 polymorphisms between the cases and controls. Chi‐square test was also used to assess the difference between the RUNX3 polymorphisms and clinicopathological characteristics. The crude and adjusted odds ratios (ORs) and their 95% confidence intervals (CIs) were obtained to assess the association between the RUNX3 polymorphisms and gastric cancer risk using unconditional univariate and multivariate logistic regression models. The multivariate adjustment included age, sex, tobacco smoking, and alcohol use. Hardy–Weinberg equilibrium of the genotype distribution among control groups was tested by a goodness‐of‐fit chi‐square test. The combined genotypes were further stratified by subgroups of age, sex, smoking status, and alcohol use.
The statistical power was calculated by using power and sample size calculation (PS) software (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize). The computation of LD between SNPs was estimated using the normalized measure of allelic association D′ and r2 , and the characterization of these patterns was determined using Haploview 4.0 software.( 19 ) The dataset was randomly partitioned into two equal parts using random number generators, named dataset1 and dataset2 for assessing overfitting.( 20 )
All of the statistical analyses were carried out with SAS software (version 9.1.3; SAS Institute, Cary, NC, USA), unless indicated otherwise. P‐value < 0.05 was considered statistically significant.
Results
The demographic characteristics of the study subjects are summarized in Table 2. There was no significant difference in the distribution of age (P = 0.892), sex (P = 0.888), or alcohol use (P = 0.629). However, there were more regular smokers among the cases (42.3%) than among the controls (34.9%), and the difference was borderline significant (P = 0.056). Furthermore, the number of patients with cancer of the gastric cardia and non‐cardia were 171 (55.0%) and 140 (45.0%), respectively. The histological types were 170 (55.0%) for intestinal and 139 (45.0%) for diffuse gastric cancer; positive lymph nodes were identified in 148 (48.0%) cases. For depth of tumor infiltration, 73 (24.7%), 64 (21.6%), 111 (37.5%), and 48 (16.2%) were T1, T2, T3, and T4, respectively.
Table 2.
Frequency distribution of selected variables between gastric cancer cases and cancer‐free controls
| Characteristics | Cases (n= 312) | Controls (n= 329) | P‐value* | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Age (years) | |||||
| <65 | 179 | 57.4 | 187 | 56.8 | 0.892 |
| ≥65 | 133 | 42.6 | 142 | 43.2 | |
| Sex | |||||
| Male | 207 | 66.4 | 220 | 68.9 | 0.888 |
| Female | 105 | 33.6 | 109 | 31.1 | |
| Smoking status | |||||
| Never | 180 | 57.7 | 214 | 65.1 | 0.056 |
| Regular | 132 | 42.3 | 115 | 34.9 | |
| Alcohol use | |||||
| Never | 217 | 69.6 | 223 | 67.8 | 0.629 |
| Regular | 95 | 30.4 | 106 | 32.2 | |
| Tumor site † | |||||
| Cardia | 171 | 55.0 | |||
| Non‐cardia | 140 | 45.0 | |||
| Histological type † | |||||
| Intestinal | 170 | 55.0 | |||
| Diffuse | 139 | 45.0 | |||
| Depth of tumor infiltration † | |||||
| T1 | 73 | 24.7 | |||
| T2 | 64 | 21.6 | |||
| T3 | 111 | 37.5 | |||
| T4 | 48 | 16.2 | |||
| Lymph node metastasis † | |||||
| Negative | 160 | 52.0 | |||
| Positive | 148 | 48.0 | |||
Two‐sided χ2‐test for the frequency distribution of selected variables between gastric cancer cases and cancer‐free controls.
The numbers of subjects in cases (n= 311 for tumor site, n= 309 for histological types, n= 296 for depth of tumor infiltration, and n= 308 for lymph node metastasis) were less than the total case number (n= 312) because some information was not gained.
The primary information of ten tSNPs for CHB is shown in Table 1. The observed genotype frequencies of ten tSNPs among the control subjects were all in agreement with the Hardy–Weinberg equilibrium (all P 0.05). The allele frequencies of the genotyped tSNPs in controls were consistent with those of the International HapMap Project database for CHB. The single SNPs allele analysis indicated that the allele frequencies of two tSNPs, SNP3 rs11249206 and SNP7 rs760805, were significantly different between the cases and controls (P = 0.005 for SNP3 and P = 0.008 for SNP7).
The genotype frequencies of the ten tSNPs and their associations with gastric cancer risk are presented in Table 3. The single locus analyses revealed that the genotype frequencies of three tSNPs (SNP3 rs11249206, SNP7 rs760805, and SNP8 rs2236852) were significantly different between the cases and controls (P < 0.001 for SNP3, P = 0.022 for SNP7, and P < 0.001 for SNP8). Multivariate logistic regression analysis indicated that the variant TC and CC genotypes of SNP3 rs11249206 were associated with a significantly increased risk of gastric cancer compared with the wild‐type TT genotype (adjusted OR = 2.61, 95% CI = 1.79–3.80 for TC and adjusted OR = 1.75, 95% CI = 1.03–2.99 for CC). For the SNP7 rs760805, compared with the TT, the variant AA genotype was associated with a statistically significantly increased risk of gastric cancer (adjusted OR = 1.82, 95% CI = 1.14–2.92). Similarly, a significantly increased risk of gastric cancer was observed for the GA genotype (adjusted OR = 2.49, 95% CI = 1.61–3.84) and GG genotype (adjusted OR = 1.69, 95% CI = 1.05–2.72) in SNP8 rs2236852, compared with the AA genotype (Table 3).
Table 3.
Genotype distributions of the RUNX3 tSNPs in gastric cancer cases and controls and risk estimates
| SNP no. | SNP ID | Genotype | Cases | Controls | P‐value* | Adjusted OR (95% CI) † | ||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | |||||
| 1 | rs6672420 | TT | 149 | 47.7 | 161 | 49.4 | 0.290 | 1.00 (ref.) |
| TA | 130 | 41.7 | 142 | 43.6 | 1.00 (0.72–1.39) | |||
| AA | 33 | 10.6 | 23 | 7.0 | 1.64 (0.91–2.93) | |||
| 2 | rs11249208 | AA | 162 | 51.9 | 175 | 53.9 | 0.874 | 1.00 (ref.) |
| AG | 133 | 42.6 | 134 | 41.2 | 1.08 (0.78–1.50) | |||
| GG | 17 | 5.5 | 16 | 4.9 | 1.21 (0.59–2.49) | |||
| 3 | rs11249206 | TT | 60 | 19.3 | 119 | 36.5 | <0.001 | 1.00 (ref.) |
| TC | 212 | 68.2 | 162 | 49.7 | 2.61 (1.79–3.80) | |||
| CC | 39 | 12.5 | 45 | 13.8 | 1.75 (1.03–2.99) | |||
| 4 | rs7551188 | TT | 96 | 30.8 | 102 | 31.3 | 0.107 | 1.00 (ref.) |
| CT | 178 | 57.0 | 166 | 50.9 | 1.11 (0.78–1.58) | |||
| CC | 38 | 12.2 | 58 | 17.8 | 0.69 (0.42–1.13) | |||
| 5 | rs1395621 | CC | 104 | 33.3 | 115 | 35.0 | 0.491 | 1.00 (ref.) |
| CT | 154 | 49.4 | 148 | 45.0 | 1.13 (0.80–1.61) | |||
| TT | 54 | 17.3 | 66 | 20.0 | 0.92 (0.59–1.45) | |||
| 6 | rs906296 | GG | 175 | 56.1 | 171 | 52.6 | 0.520 | 1.00 (ref.) |
| GC | 118 | 37.8 | 137 | 42.2 | 0.85 (0.61–1.17) | |||
| CC | 19 | 6.1 | 17 | 5.2 | 1.06 (0.53–2.12) | |||
| 7 | rs760805 | TT | 47 | 15.1 | 67 | 20.4 | 0.022 | 1.00 (ref.) |
| AT | 151 | 48.4 | 173 | 52.6 | 1.21 (0.78–1.88) | |||
| AA | 114 | 36.5 | 89 | 27.0 | 1.82 (1.14–2.92) | |||
| 8 | rs2236852 | AA | 42 | 13.5 | 81 | 24.8 | <0.001 | 1.00 (ref.) |
| GA | 187 | 59.9 | 146 | 44.8 | 2.49 (1.61–3.84) | |||
| GG | 83 | 26.6 | 99 | 30.4 | 1.69 (1.05–2.72) | |||
| 9 | rs9438876 | AA | 158 | 50.6 | 164 | 49.8 | 0.974 | 1.00 (ref.) |
| AG | 130 | 41.7 | 140 | 42.6 | 0.96 (0.69–1.33) | |||
| GG | 24 | 7.7 | 25 | 7.6 | 0.97 (0.53–1.78) | |||
| 10 | rs2282718 | GG | 115 | 36.8 | 129 | 39.6 | 0.388 | 1.00 (ref.) |
| GA | 155 | 49.7 | 145 | 44.5 | 1.20 (0.85–1.69) | |||
| AA | 42 | 13.5 | 52 | 15.9 | 0.89 (0.55–1.44) | |||
Two‐sided χ2 test for the frequency distribution.
Adjusted for age, sex, smoking status, and alcohol use.
Considering potential interactions of the tSNPs on the risk of gastric cancer, we combined these three tSNPs based on the numbers of variant (risk) alleles (i.e. SNP3 C, SNP7 A, and SNP8 G alleles). As shown in Table 4, the combined genotypes with one to two variant (risk) alleles were more common (0.040 and 0.318, respectively) and those with three to five variant (risk) alleles were less common (0.417, 0.170, and 0.046, respectively) among the controls than among the cases (0.01, 0.148, 0.476, 0.289, and 0.077, respectively), and these differences were statistically significant (P < 0.001). When these combined genotypes were dichotomized into two groups (i.e. one to three vs four to six variant [risk] alleles), their distributions differed significantly between the cases and controls (P < 0.001). In the association analysis, we found that the individuals with four to six variant (risk) alleles had a significantly higher risk of gastric cancer (adjusted OR = 2.00, 95% CI = 1.41–2.85) than those with one to three risk alleles (Table 5). Further stratification analysis showed that this increased risk was more pronounced among subgroups of age ≥65 years (adjusted OR = 3.58, 95% CI = 2.00–6.38), never smokers (adjusted OR = 2.48, 95% CI = 1.58–3.89), and never drinkers (adjusted OR = 2.61, 95% CI = 1.70–3.99). However, no statistical evidence was observed for interactions between the combined genotypes and the variables (i.e. age, sex, tobacco smoking, and alcohol use) (data not shown).
Table 4.
Frequency distributions of the combined genotypes of RUNX3 SNP3, SNP7, and SNP8 between the cases and controls
| No. variant (risk) alleles of the combined genotypes † | Cases ‡ (n= 311) | Controls ‡ (n= 324) | P‐value* | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| 1 | 3 | 1.0 | 13 | 4.0 | <0.001 |
| 2 | 46 | 14.8 | 103 | 31.8 | |
| 3 | 148 | 47.6 | 135 | 41.7 | |
| 4 | 90 | 28.9 | 55 | 17.0 | |
| 5 | 24 | 7.7 | 15 | 4.6 | |
| 6 | 0 | 0.0 | 3 | 0.9 | |
| Dichotomized groups | |||||
| 1–3 | 197 | 63.3 | 251 | 77.5 | <0.001 |
| 4–6 | 114 | 36.7 | 73 | 22.5 | |
The number represents the numbers of variants within the combined genotypes, i.e. 1–6 = 1–6 variant (risk) alleles; the variant (risk) alleles used for the calculation were the SNP3 C, SNP7 A, and SNP8 G alleles.
The numbers of subjects in cases and controls were less than the total number (total case numbers = 312; total control numbers = 329) of subjects because some genotypes were not available.
Two‐sided χ2‐test for the frequency distributions between the cases and controls.
Table 5.
Stratification analyses between the combined genotypes of the RUNX3 tSNPs (SNP3, SNP7, and SNP8) and gastric cancer risk
| Variables | n (case/control) | Combined genotypes (case/control) | Crude OR (95% CI) | Adjusted OR (95% CI) † | |||
|---|---|---|---|---|---|---|---|
| 1–3 risk alleles | 4–6 risk alleles | ||||||
| n | % | n | % | ||||
| Total | 311/324 | 197/251 | 63.3/77.5 | 114/73 | 36.7/22.5 | 1.99 (1.41–2.82) | 2.00 (1.41–2.85) |
| Age (years) | |||||||
| <65 | 178/183 | 118/132 | 66.3/72.1 | 60/51 | 33.7/27.9 | 1.32 (0.84–2.06) | 1.34 (0.85–2.10) |
| ≥65 | 133/141 | 79/119 | 59.4/84.4 | 54/22 | 40.6/15.6 | 3.69 (2.09–6.54) | 3.58 (2.00–6.38) |
| Sex | |||||||
| Male | 207/217 | 137/172 | 66.2/79.3 | 70/45 | 33.8/20.7 | 1.95 (1.26–3.02) | 1.94 (1.25–3.02) |
| Female | 104/107 | 60/79 | 57.7/73.8 | 44/28 | 42.3/26.2 | 2.07 (1.16–3.70) | 2.21 (1.20–4.05) |
| Smoking status | |||||||
| Never | 179/210 | 106/164 | 59.2/78.1 | 73/64 | 40.8/21.9 | 2.45 (1.58–3.82) | 2.48 (1.58–3.89) |
| Regular | 132/114 | 91/87 | 68.9/76.3 | 41/27 | 31.1/23.7 | 1.45 (0.82–2.56) | 1.44 (0.81–2.54) |
| Alcohol use | |||||||
| Never | 216/220 | 129/175 | 59.7/79.5 | 87/45 | 40.3/20.5 | 2.62 (1.71–4.01) | 2.61 (1.70–3.99) |
| Regular | 95/104 | 68/76 | 71.6/73.1 | 27/28 | 28.4/26.9 | 1.08 (0.58–2.01) | 1.16 (0.60–2.25) |
Odds ratios were obtained from a logistic regression model with adjustment for age, sex, smoking status, and alcohol use.
In this study, we also analyzed the correlations between the RUNX3 variant genotypes and clinicopathological characteristics of gastric cancer, which included the tumor sites (i.e. cardia and non‐cardia), histological types (i.e. intestinal and diffuse), tumor infiltration (i.e. T1, T2, T3, and T4), and lymph node metastasis (i.e. negative and positive). However, no significant association was observed (data not shown).
Discussion
In this hospital‐based case‐control study, we found that the variant genotypes of three RUNX3 tSNP (i.e. SNP3 CC, SNP7 AA, and SNP8 GG genotypes) were associated with a statistically significantly increased risk of gastric cancer in a Chinese population. The result provided evidence that RUNX3 is a potential candidate gene for gastric cancer susceptibility, suggesting that the RUNX3 polymorphisms may play a role in the risk of developing gastric cancer in a Chinese population.
The human RUNX3 gene is located on chromosome 1p36, a region that is thought to carry many tumor suppressor genes implicated in various types of cancer.( 21 , 22 ) The three tSNPs (i.e. SNP3, SNP7, and SNP8) that were associated with an increased risk of gastric cancer are located in introns 1, 3, and 4, respectively. Recently, some published association studies have observed that the intron SNP were also associated with diverse diseases.( 23 , 24 , 25 , 26 6) For example, Huang et al. demonstrated that the epidermal growth factor receptor (EGFR) intron 1 polymorphism is associated with the occurrence of skin rash and speculated that the intron polymorphism might be more important than promoter polymorphism in regulation of EGFR expression.( 26 ) Our finding further provids evidence that polymorphisms within the introns of the RUNX3 gene may contribute to the etiology of gastric cancer.
To date, the mechanism responsible for the correlation of intron SNPs with disease development is not clearly understood. Notably, some SNPs in introns have been identified with function in gene transcription and protein expression.( 27 , 28 , 29 ) When we analyzed the putative transcription factors of RUNX3 using the Genomatix program (http://www.genomatix.de), we found that the SNP7 forecast result was identical to a previous observation in which myelin transcription factor 1 was predicted.( 16 ) Therefore, further functional studies are needed to validate the prediction results.
To date, few studies on RUNX3 polymorphisms and cancer susceptibility have been reported.( 16 , 30 ) In our previous study, we reported that the SNP7 AA genotype is associated with a significantly increased risk of bladder cancer in a Chinese population, which is consistent with our present findings.( 16 ) In contrast, Hu et al. failed to find a significant association between the RUNX3 364C>T (Arg122Cys) polymorphism within the conserved Runt domain and risk of gastric carcinoma in small sample sizes (86 cases and 169 controls).( 30 )
In the present study, we found that the combined genotypes of these three RUNX3 tSNPs were associated with a significantly increased risk of gastric cancer, suggesting that these RUNX3 polymorphisms may play a joint role in the development of gastric cancer. Our data further support the notion that a single polymorphism may only contribute a modest effect and the combined variants of a gene may provide a more comprehensive evaluation of genetic susceptibility in candidate genes with low penetration. However, considering that SNP3, SNP7, and SNP8 may simply reflect overfitting of the data, we conducted a repeated analysis (data not shown). Though the results showed the same trend with the former outcomes, a two‐stage study with larger sample size is needed to validate our findings.
In addition, we also found that the combined variants were associated with increased risk of gastric cancer among subgroups of older subjects (age ≥65 years), never smokers, and never drinkers. One possible explanation is that the older people may be exposed to accumulated environmental risk factors involved in the etiology of gastric cancer such as consumption of salt‐preserved food and pesticide residue vegetables. It is possible that never smokers and never drinkers may have been exposed to other carcinogens. Another reason might be that the genetic susceptibility may be masked by overwhelming exposure to tobacco smoking or alcohol consumption.( 31 ) However, our sample size is not large enough and the selected bias could not be excluded completely, so the finding might be due to chance and should be treated with caution.
Wei et al. found that loss of RUNX3 expression significantly affected the clinical outcome of gastric cancer patients.( 11 ) Moreover, its restoration causes drastic suppression of tumor growth and metastasis. Li et al. found that RUNX3 expression was reduced in 40% of early stage carcinomas, and the level increased to nearly 90% with the advancement of the cancer stage.( 10 ) Our analysis failed to find an association of RUNX3 polymorphism with tumor clinicopathological characteristics. The reasons for this result may be due to different ethnicities and the relatively small sample size. Larger studies with different ethnic populations are needed to determine whether RUNX3 genetic variants are associated with gastric cancer clinical characteristics.
Our study has several strengths: (1) the samples were recruited from Yangzhong and Yixing, two areas with high incidence and mortality of gastric cancer in China; (2) the cases and controls were adequately matched with age and sex; and (3) it is the first study to investigated the association between the RUNX3 tSNP and risk of gastric cancer. At the same time, there are some limitations in this study that need to be improved in future study. First, the sample size may not be large enough to identify significant genotype–clinical phenotype and gene–environment interactions, though we had 80% power at a 0.05 or smaller level to detect an OR of 1.6 or greater and 0.6 or smaller with an exposure frequency of 30% given our current study sample size. Second, our study was a hospital‐based case‐control study. Thus, the selection bias cannot be excluded. However, the bias might be minimized by matching the controls to the cases on sex and age and by further adjustment for the confounding factors in statistical analyses. Third, H. pylori is an important risk factor for gastric cancer. Because the H. pylori infection data was not complete, we did not analyze stratification of H. pylori infection on gastric cancer risk. Further studies with H. pylori infection information are needed. Finally, although three of the ten tSNPs were statistically associated with gastric cancer risk, it is possible that one or more of them will not be significant after multiple comparisons with Bonferroni correction (e.g. P = 0.220 for SNP7 after the Bonferroni correction). Therefore, replication studies with diverse ethnic groups and larger samples in gastric cancer are warranted.
In conclusion, the three intronic SNPs (i.e. SNP3 rs11249206, SNP7 rs760805, and SNP8 rs2236852) of the RUNX3 gene had an effect on risk of gastric cancer; and the combined genotypes of these three RUNX3 polymorphisms were associated with a significantly increased risk of gastric cancer. Larger studies with functional research and more detailed environmental exposure data are needed to confirm our findings.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (30271105, 30571583, 30800926, and 30872084), the PhD Programs Foundation of Ministry of Education of China (20060312002), the Natural Science Foundation of Jiangsu Province (BK2006231), the Key Project of Nanjing Medical University (2005NYDZD09), and the Medical Research Foundation from the Health Department of Jiangsu Province (H200767). We thank Dr Yang Zhao for part of the data management. We also thank Dr Jianwei Zhou for the help in part of the sample collection.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 2. Inoue K, Shiga T, Ito Y. Runx transcription factors in neuronal development. Neural Dev 2008; 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 2007; 204: 1945–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 2005; 5: 376–87. [DOI] [PubMed] [Google Scholar]
- 5. Ito Y. Oncogenic potential of the RUNX gene family: ‘overview’. Oncogene 2004; 23: 4198–208. [DOI] [PubMed] [Google Scholar]
- 6. Bae SC, Choi JK. Tumor suppressor activity of RUNX3. Oncogene 2004; 23: 4336–40. [DOI] [PubMed] [Google Scholar]
- 7. Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF‐beta superfamily signaling. Curr Opin Genet Dev 2003; 13: 43–7. [DOI] [PubMed] [Google Scholar]
- 8. Yanagawa N, Tamura G, Oizumi H et al . Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non‐small cell lung cancers. Lung Cancer 2007; 58: 131–8. [DOI] [PubMed] [Google Scholar]
- 9. Kim EJ, Kim YJ, Jeong P, Ha YS, Bae SC, Kim WJ. Methylation of the RUNX3 promoter as a potential prognostic marker for bladder tumor. J Urol 2008; 180: 1141–5. [DOI] [PubMed] [Google Scholar]
- 10. Li QL, Ito K, Sakakura C et al . Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002; 109: 113–24. [DOI] [PubMed] [Google Scholar]
- 11. Wei D, Gong W, Oh SC et al . Loss of RUNX3 expression significantly affects the clinical outcome of gastric cancer patients and its restoration causes drastic suppression of tumor growth and metastasis. Cancer Res 2005; 65: 4809–16. [DOI] [PubMed] [Google Scholar]
- 12. Tokumaru Y, Nomoto S, Jeronimo C et al . Biallelic inactivation of the RIZ1 gene in human gastric cancer. Oncogene 2003; 22: 6954–8. [DOI] [PubMed] [Google Scholar]
- 13. Goel A, Arnold CN, Tassone P et al . Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer 2004; 112: 754–9. [DOI] [PubMed] [Google Scholar]
- 14. Kim TY, Lee HJ, Hwang KS et al . Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest 2004; 84: 479–84. [DOI] [PubMed] [Google Scholar]
- 15. Kim WJ. RUNX3 inactivation by point mutations and aberrant DNA methylation in bladder tumors. Cancer Res 2005; 65: 9347–54. [DOI] [PubMed] [Google Scholar]
- 16. Zhang Z, Wang S, Wang M, Tong N, Fu G, Zhang Z. Genetic variants in RUNX3 and risk of bladder cancer: a haplotype‐based analysis. Carcinogenesis 2008; 29: 1973–8. [DOI] [PubMed] [Google Scholar]
- 17. Hohenberger P, Gretschel S. Gastric cancer. Lancet 2003; 362: 305–15. [DOI] [PubMed] [Google Scholar]
- 18. Green FL, Page DL, Fleming ID et al . Stomach of the digestive system. In: AJCC Cancer Staging Handbook, 6th edn. New York: Springer Press, 2002; 111–8. [Google Scholar]
- 19. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–5. [DOI] [PubMed] [Google Scholar]
- 20. Sinha R, Hiller M, Pudimat R, Gausmann U, Platzer M, Backofen R. Improved identification of conserved cassette exons using Bayesian networks. BMC Bioinformatics 2008; 9: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Munirajan AK, Ando K, Mukai A et al . KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem 2008; 283: 24 426–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res 2008; 68: 2551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pagani F, Buratti E, Stuani C, Bendix R, Dork T, Baralle FE. A new type of mutation causes a splicing defect in ATM. Nat Genet 2002; 30: 426–9. [DOI] [PubMed] [Google Scholar]
- 24. Tokuhiro S, Yamada R, Chang X et al . An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet 2003; 35: 341–8. [DOI] [PubMed] [Google Scholar]
- 25. Weersma RK, Zhou L, Nolte IM et al . Runt‐related transcription factor 3 is associated with ulcerative colitis and shows epistasis with solute carrier family 22, members 4 and 5. Inflamm Bowel Dis 2008; 14: 1615–22. [DOI] [PubMed] [Google Scholar]
- 26. Huang CL, Yang CH, Yeh KH et al . EGFR intron 1 dinucleotide repeat polymorphism is associated with the occurrence of skin rash with gefitinib treatment. Lung Cancer 2009; 64: 346–51. [DOI] [PubMed] [Google Scholar]
- 27. Kereszturi E, Kiraly O, Sahin‐Toth M. Minigene analysis of intronic variants in common SPINK1 haplotypes associated with chronic pancreatitis. Gut 2009; 58: 545–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi JW, Park CS, Hwang M et al . A common intronic variant of CXCR3 is functionally associated with gene expression levels and the polymorphic immune cell responses to stimuli. J Allergy Clin Immunol 2008; 122: 1119–26 e7. [DOI] [PubMed] [Google Scholar]
- 29. Lewandowska MA, Stuani C, Parvizpur A, Baralle FE, Pagani F. Functional studies on the ATM intronic splicing processing element. Nucleic Acids Res 2005; 33: 4007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu S, Ke YH, Hu PJ, Zeng ZR. Relationship between RUNX3 gene 364 locus C/T mutation and gastric cancer in Chinese. Chin J Pathophysiol 2005; 21: 1905–8. [Google Scholar]
- 31. Zhang Z, Xu Y, Zhou J et al . Polymorphisms of thymidylate synthase in the 5′‐ and 3′‐untranslated regions associated with risk of gastric cancer in South China: a case‐control analysis. Carcinogenesis 2005; 26: 1764–9. [DOI] [PubMed] [Google Scholar]


