Abstract
The role of insulin‐like growth factor binding protein 5 (IGFBP5) in tumorigenesis and development of cancer is not well‐defined. IGFBP5 has been shown to either stimulate or inhibit cell proliferation via an IGF‐dependent mechanism and to promote cell proliferation and migration in an IGF‐independent manner. In the authors’ previous study, IGFBP5 was found to be significantly up‐regulated in lymph node metastases compared with their paired primary breast cancers. To further determine the role of IGFBP5 in breast cancer development and to evaluate its clinical significance in breast cancer, the mRNA expression level was detected in 30 normal breast tissues, 108 primary tumors, and 30 lymph node metastases using real time reverse transcription–polymerase chain reaction. The expression levels were correlated with several clinical parameters, including clinical stage, pathologic tumor size, axillary lymph node status, nuclear grade, estrogen receptor status, Her2 status, and local relapse or distant metastasis of the patients. As a result, the expression of IGFBP5 mRNA correlated positively with the invasion of axillary lymph nodes and the status of hormonal receptor. Furthermore, overexpression of IGFBP5 was associated with poor outcome of breast cancer patients with positive lymph nodes and negative ER. Thus, the expression level of IGFBP5 may contribute to the development of breast cancer and is a prognostic factor for breast cancer. (Cancer Sci 2007; 98: 1592–1596)
The insulin‐like growth factor binding protein 5 (IGFBP5) belongs to a protein family of at least six members that binds to insulin‐like growth factor (IGF). In humans, the IGFBP5 gene spans 33 kb and is located on 2q33–q36. The mature IGFBP5 protein consists of 252 amino acids and has a molecular mass of approximately 29 kDa. IGFBP5 was initially recognized as a secreted protein that binds to IGF with high affinity by its N‐terminal motif and modulates their mitogenic and anti‐apoptotic effects by either inhibiting or augmenting the interaction of IGF with their cell‐surface receptor (IGF‐IR). IGFBP5 has been shown to inhibit the proliferative responses of skeletal muscle cells( 1 ) and breast cancer cells( 2 ) to IGF‐I. In contrast, in bone,( 3 ) fibroblast,( 4 ) osteoblasts,( 5 ) and vascular smooth muscle cells,( 6 ) IGFBP5 has been found to potentiate the effect of IGF on cell growth. The complexity of the cellular function of IGFBP and its regulation is further revealed by recent studies that have shown that IGFBP5 also stimulates cell growth,( 7 , 8 ) migration,( 6 , 9 , 10 ) and cell attachment to extracellular matrix (ECM)( 11 , 12 ) through IGF‐independent mechanisms. Thus, the role of IGFBP5 in cancer is far from being clear.
In the authors’ previous study of breast cancer tissues,( 13 ) it was found that IGFBP5 was significantly up‐regulated in lymph node metastases relative to their paired primary cancers, suggesting that IGFBP5 may play an important role in the metastasis of breast cancer. Additional studies have supported the notion that IGFBP5 overexpression contributes to metastasis and poor prognosis of cancer. Wang et al. have demonstrated a strong correlation between overexpression of IGFBP5 and the histologic grade of ovarian cancer and glioblastoma.( 14 , 15 ) And up‐regulated IGFBP5 was included in the ‘poor prognosis signature’ in a study by van't Veer et al. ( 16 ) To explore the role of IGFBP5 in the development of breast cancer, and to evaluate the clinical significance of IGFBP5 in the diagnosis and prognosis of breast cancer, the IGFBP5 mRNA expression levels were measured in 30 normal breast tissues, 108 primary tumors, and 30 lymph node metastases using real‐time reverse transcription–polymerase chain reaction (RT‐PCR). The association between IGFBP5 expression and several clinicopathologic parameters, including clinical stage, pathologic tumor size, axillary lymph node status, nuclear grade, estrogen receptor (ER) status, Her2 status, and local relapse or distant metastasis, was analyzed.
Materials and Methods
Clinical samples. All breast tissues, including 30 normal tissues, 108 primary tumors and 30 lymph node metastases samples were collected from 108 breast cancer patients undergoing complete dissection of breast and axillary lymph nodes (average 22 nodes resected) followed by adjuvant systematic therapy consisting of chemotherapy and hormonal therapy (adjuvant cyclophosphamide, methotrexate, 5‐fluorouracil [CMF]) or combined tamoxifen when with positive ER at Tianjin Cancer Hospital, China, between January 2002 and November 2003. Of these 108 cases, primary cancers and paired normal breast tissues were collected in 20 cases, primary cancers and paired lymph node metastases were obtained in another 20 cases, matched normal breast, primary cancers and lymph node metastases were collected in 10 cases, and only primary cancer samples were obtained in the other 58 cases. Tissue samples were snap‐frozen in liquid nitrogen and stored at –80°C. All samples were examined using HE staining, and only samples with 75% or more tumor cells in primary tumors and metastases samples were selected for real‐time RT‐PCR. The detailed clinical characteristics of these patients, including clinical stage, pathologic tumor size, lymph node status, nuclear grade, ER status, and existence of relapse or distant metastases are shown in Table 1. ER and progesterone receptor (PR) expression were determined using immunohistochemical staining (positive when more than 15% of the nuclei showed staining). Her2 was defined as positive when more than 10% of the membrane showed staining using immunohistochemical assay. The use of these tissues was approved by the Institutional Reviewing Board and the Research Committee.
Table 1.
Correlation between the mRNA levels of insulin‐like growth factor binding protein 5 (IGFBP5) and clinicopathologic factors
| Clinicopathologic factors | Cases | x̄ ± s | t/F | P‐value |
|---|---|---|---|---|
| Breast sample | ||||
| Normal breast | 30 | 1.45E‐04 ± 1.83E‐04 | –5.092 | 0.000 |
| Breast cancer | 108 | 1.08E‐02 ± 2.15E‐02 | ||
| Tumor size (cm) | ||||
| ≤2 | 50 | 7.95E‐03 ± 1.59E‐02 | 0.970 | 0.382 |
| 2–5 | 50 | 1.38E‐02 ± 2.69E‐02 | ||
| >5 | 8 | 8.37E‐03 ± 1.01E‐02 | ||
| Lymph node status | ||||
| Negative | 52 | 4.65E‐03 ± 9.11E‐03 | 2.995 | 0.004 |
| Positive | 56 | 1.63E‐02 ± 2.75E‐02 | ||
| Lymph node positive number | ||||
| 0 | 52 | 4.65E‐03 ± 9.11E‐03 | 3.811 | 0.012 |
| 1–3 | 22 | 2.00E‐02 ± 2.89E‐02 | ||
| 4–9 | 15 | 1.92E‐02 ± 3.29E‐02 | ||
| ≥10 | 19 | 9.71E‐03 ± 2.08E‐02 | ||
| Clinical stage | ||||
| I | 15 | 1.24E‐02 ± 1.81E‐02 | 0.475 | 0.700 |
| II | 67 | 8.90E‐03 ± 1.97E‐02 | ||
| III | 22 | 1.50E‐02 ± 2.93E‐02 | ||
| IV | 4 | 1.04E‐02 ± 1.48E‐02 | ||
| Nuclear grade † | ||||
| I | 12 | 8.91E‐03 ± 1.61E‐02 | 0.774 | 0.464 |
| II | 73 | 9.41E‐03 ± 1.92E‐02 | ||
| III | 23 | 1.56E‐02 ± 2.98E‐02 | ||
| ER ‡ | ||||
| Positive | 67 | 1.31E‐02 ± 2.63E‐02 | 2.209 | 0.030 |
| Negative | 37 | 5.61E‐03 ± 6.68E‐03 | ||
| Missing | 4 | |||
| PR ‡ | ||||
| Positive | 46 | 1.50E‐02 ± 2.84E‐02 | 1.835 | 0.071 |
| Negative | 58 | 6.71E‐03 ± 1.32E‐02 | ||
| Missing | 4 | |||
| Her2 § | ||||
| Positive | 28 | 1.05E‐02 ± 2.15E‐02 | 0.345 | 0.731 |
| Negative | 55 | 9.03E‐03 ± 1.81E‐02 | ||
| Missing | 25 | |||
| Relapse or distant metastasis | ||||
| Negative | 54 | 7.52E‐03 ± 1.22E‐02 | 0.938 | 0.359 |
| Positive | 19 | 1.33E‐02 ± 2.59E‐02 | ||
| Less than 3 years follow up | 35 | |||
Grade I nuclei indicates well‐differentiated breast cells that generally appear normal; Grade III shows poorly differentiated breast cells that do not appear normal; and Grade II has characteristics between Grade I and Grade III tumors with moderately differentiated breast cells.
Positive when more than 15% of the membrane showed staining by immunohistochemical staining.
Positive when more than 10% of the nuclei showed staining by immunohistochemical staining.
RNA extraction and cDNA preparation. RNA was extracted with TRIZOL reagent (Invitrogen, Gaithersburg, MD, USA) according to the manufacture's instructions. The RNA quality was assessed using formaldehyde agarose gel electrophoresis and was quantified spectrophotometrically. 5 µg of total RNA was used to perform RT for first‐strand cDNA synthesis. In brief, RNA was denatured for 5 min at 65°C and snap cooled on ice in the presence of 0.5 µg Oligo(dT) (Invitrogen) and 10 mmol dNTP mix (Invitrogen), followed by incubation at 4°C for 50 min with First‐Strand Buffer, 0.2 µmol DTT (Invitrogen), 40 U RNaseOUT ribonuclease inhibitor (Invitrogen) and 200 U SuperScript II (Invitrogen) in total volume 20 µL reaction system. Reactions were stopped by incubation at 70°C for 15 min.
Real‐time PCR. Real‐time PCR analysis was performed using the Platinum Quantitative PCR SuperMix‐UDG System (Invitrogen) according to the manufacturer's instructions. Transcripts of the housekeeping gene, glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), were quantified as control, as described.( 17 ) Primers and Taqman probes of IGFBP5 were 5′‐CGTGCTGTGTACCTGCCCAAT‐3′, 5′‐CACTGAAAGTCCCCG TCAAC‐3′, 5′(FAM)‐AGAGAAAGCAGTGCAAACCTTCCC‐(TAMRA)‐3′. Assays were performed with the ABI 7500 TaqMan system (Applied Biosystems, Foster City, CA, USA). PCR was carried out after incubation at 50°C for 2 min and pre‐denaturing at 95°C for 3 min, followed by 40 cycles at 95°C for 30 s and 65°C for 1 min. Quantification of expression of the target gene in samples was accomplished by measuring the fractional cycle number at which the amount of expression reaches a fixed threshold (CT). The relative quantification was given by the CT values, determined by triplicate reactions for test and reference samples for each target and for GAPDH. Triplicate CT values were averaged and the GAPDH CT subtracted to obtain ΔCT. The relative expression level of target gene was determined as 2−ΔCT.
Statistical analysis. Paired t‐test was used to analyze the mRNA expression differences between primary breast cancers and paired normal breast tissue, and differences between primary breast cancers and their matched lymph node metastases. Student's t‐test and one‐way analysis of variance (anova) were used to compare mRNA expression differences between/among different clinicopathologic groups. Cases with more than 3 years follow‐up were used to perform survival analysis. Disease‐free survival time was defined as the time interval from surgery to local relapse/first distant organ metastases or to the last follow‐up visit. Survival analysis was carried out according to Kaplan–Meier analysis. Multivariate survival analysis was performed by a forward stepwise Cox proportional hazards regression model. All calculations were performed with SPSS for Windows statistical software package (SPSS Inc, Chicago, IL, USA).
Results
The expression level of IGFBP5 mRNA in breast tissues. The mRNA level of IGFBP5 measured using real‐time PCR analyses ranged from 6.206E‐06 to 2.541E‐01 in all tissues including normal breast, primary breast cancers, and lymph node metastases. All samples were grouped into four groups according to the mRNA levels of IGFBP5: ‘negative IGFBP5/–’ (less than 1.0E‐03), ‘weakly positive IGFBP5/+’ (from 1.0E‐03 to 3.0E‐03), ‘moderately positive IGFBP5/++’ (from 3.0E‐03 to 1.0E‐02), and ‘strongly positive IGFBP5/+++’ (more than 1.0E‐02).
The mRNA expression differences of IGFBP5 between primary breast cancers and normal breast tissues. The mRNA of IGFBP5 was significantly up‐regulated (from 1.26 to 3146.79‐fold) in cancer tissue compared with paired normal breast in 29 of 30 cases (including 15 node‐negative cases and 15 node‐positive cases; t = 3.28, P = 0.003). IGFBP5 were negative (‘–’) in all of 30 normal breast tissues, but positive (‘+’ to ‘+++’) in most of the paired breast cancer tissues (24/30, 80%, Fig. 1a). In all of the 108 primary cancers, IGFBP5 were positive in 63% (33/52) cases with negative lymph nodes, and in 82% (46/56) cases with positive nodes (Fig. 1b). And IGFBP5 was significantly higher in all of the 108 primary cancers than in 30 normal breast tissues (t = 5.092, P = 0.000, Table 1).
Figure 1.
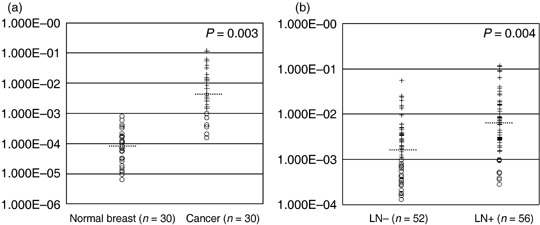
The expression of insulin‐like growth factor binding protein 5 (IGFBP5) mRNA in normal breast tissues and breast cancers. The circles represented cases with negative IGFBP5 (‘–’), and the crosses represented cases with positive IGFBP5 (‘+’ to ‘+++’). The dash line represents the median in each group. IGFBP5 was negative in all of 30 normal breast tissues, but positive in 67% (10/15) of primary cancers with negative lymph nodes and in 93% (14/15) of primary cancers with positive nodes. (a) The expression difference of IGFBP5 mRNA in 30 breast cancers and paired normal breast tissues. (b) The mRNA level of IGFBP5 was higher in cases with positive lymph nodes (LN+) than cases with negative nodes (LN–).
Correlation between the mRNA levels of IGFBP5 and lymph node metastases. In the present set of samples, IGFBP5 levels were up‐regulated in lymph node metastases in eight of 30 cases, but down‐regulated in 12 of 30 cases. Equal levels were seen in the rest of the 10 cases. However, the average mRNA level of IGFBP5 was 3.5‐fold higher in the primary cancers with positive lymph nodes than in the cases with negative lymph nodes (t = 2.995, P = 0.004), although the mRNA level of IGFBP5 could not separate the patients with different lymph node status (Fig. 1b). When these cases were classified into two groups: ‘group without palpable nodes’ (clinical N0) and ‘group with palpable nodes’ (clinical N1‐2), it was found that the false‐negative rate was increased with the increase of mRNA level of IGFBP5 in the ‘group without palpable nodes’ (Table 2), and the false‐positive rate was increased with the decrease of the mRNA level of IGFBP5 in the ‘group with palpable nodes’ (Table 2).
Table 2.
Correlation of insulin‐like growth factor binding protein 5 (IGFBP5) and false palpable lymph nodes
| IGFBP5 | Clinical palpable negative nodes | Clinical palpable positive nodes | Total | ||
|---|---|---|---|---|---|
| Pathologic negative (%) | Pathologic positive (false negative) (%) | Pathologic positive (%) | Pathologic negative (false positive) (%) | ||
| – | 15 (78.95) | 4 (21.05) | 6 (60.00) | 4 (40.00) | 29 |
| + | 9 (81.82) | 2 (18.18) | 8 (53.33) | 7 (46.67) | 26 |
| ++ | 6 (46.15) | 7 (53.85) | 10 (66.67) | 5 (33.33) | 28 |
| +++ | 4 (40.00) | 6 (60.00) | 13 (88.67) | 2 (13.33) | 25 |
| total | 34 | 19 | 37 | 18 | 108 |
Correlation between the mRNA level of IGFBP5 and clinicopathologic factors. No significant differences of IGFBP5 were found between the mRNA level of IGFBP5 and tumor size, clinical stage, nuclear grade and Her2 status (P > 0.05, Table 1). The mRNA of IGFBP5 was significantly lower in the ER‐negative group than in the ER‐positive group (P = 0.030, Table 1), and lower in the PR‐ negative group than in the PR‐positive group (P = 0.071, Table 1).
Prediction of disease‐free survival based on the mRNA level of IGFBP5. Seventy‐three cases were followed up for more than 3 years (from 36 to 58 months; median 47 months), including 19 cases that developed to local relapse (nine cases) or distant metastasis (three to lung, two to liver, two to brain, one to bone, one to the other breast, one to solar plexus). Disease‐free survival time of cases with negative/weakly positive IGFBP5 (–/+) was longer than in cases with moderately/strongly positive IGFBP5 (++/+++) (P = 0.008; Fig. 2a). Tumor size, clinical stage, nuclear grade, lymph node status, ER and PR, Her2 and IGFBP5 were analyzed using Cox's multivariate analysis. As a result, tumor size greater than 5 cm (odds ratio [OR] = 2.086 [95% CI = 1.074–4.053], P = 0.030), higher clinical stage (OR = 2.241 [95% CI = 0.881–5.702], P = 0.090), negative PR (OR = 4.080 [95% CI = 1.486–11.236], P = 0.006) and moderately/strongly positive IGFBP5 (OR = 2.700 [95% CI = 1.049–6.948], P = 0.039) were independent factors in predicting the outcome of breast cancer. Then disease‐free survival analyses of different ER status or axillary lymph node status were calculated. As a result, the mRNA level of IGFBP5 could be a prognostic predictor to breast cancer patients with negative ER (OR = 4.440 [95% CI 1.285–15.333], P = 0.008; Fig. 2c) and patients with positive lymph nodes (OR = 4.136 [95% CI 0.976–17.517], P = 0.033; Fig. 2d). But it was not a good prognostic predictor for patients with positive ER (P = 0.140; Fig. 2b) or without lymph nodes metastases (P = 0.900; Fig. 2e).
Figure 2.
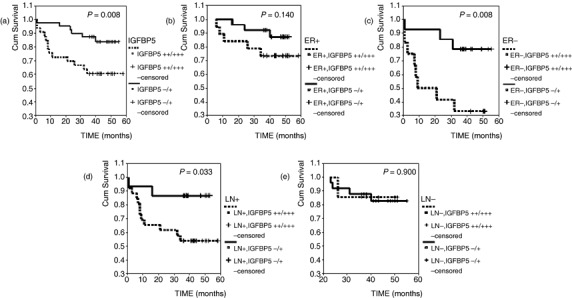
Results of survival analyses based on the mRNA level of insulin‐like growth factor binding protein 5 (IGFBP5). ER, estrogen receptor; LN, lymph node.
Discussion
In the present study, the mRNA level of IGFBP5 was found to be up‐regulated in breast cancers relative to normal breast tissues, overexpressed in breast cancers with positive axillary lymph nodes compared to cancers with negative lymph nodes, and positively correlated with the status of hormonal receptors. No significant difference of IGFBP5 was found between the mRNA levels of IGFBP5 and tumor size, clinical stage and nuclear grade. Furthermore, overexpression of IGFBP5 was associated with poor outcome of breast cancer patients with positive lymph nodes and negative ER.
IGFBP5 has been shown to affect cell proliferation and apoptosis differently under different conditions and contexts. IGFBP5 has been shown to inhibit proliferation of breast cancer via IGF‐dependent mechanism in vitro,( 2 ) whereas other studies have shown that IGFBP5 also stimulates cell proliferation( 8 ) of breast cancer in an IGF‐independent manner in vitro. IGFBP5 has been shown to induce apoptosis by inhibition of IGF‐signaling,( 2 ) enhance tumor necrosis factor (TNF)‐induced growth inhibition,( 18 ) increase expression of the pro‐apoptotic regulator BAX,( 2 ) and decrease the expression of the anti‐apoptotic BCL2.( 2 ) On the other hand, IGFBP5 has been shown to promote cell proliferation by inhibiting ceramide‐induced apoptosis,( 8 , 19 ) and to inhibit cell death induced by arginine–glycine–aspartate peptide (RGD).( 19 ) Moreover, IGFBP5 may facilitate metastasis by increasing cell attachment to the extracellular matrix (ECM) and base membrane,( 12 , 19 ) and by enhancing cell migration.( 11 ) These contrary molecular mechanisms suggest a potential balancing function of IGFBP5 in cell proliferation and apoptosis, and that the balance and functions cross‐talk with other cellular pathways in the cells. The present results suggest that overexpression of IGFBP5 mRNA may play a positive role in breast tumorigenesis and development, indicating that the proliferation and metastasis stimulation effects of IGFBP5 may be predominant over the anti‐proliferation effect in vivo.
Yee et al. showed a positive correlation between the expression of IGFBP5 protein and ER and PR expression status in axillary lymph node‐negative breast cancers, but showed no relationship between IGFBP5 expression and the disease‐free survival of lymph node‐negative breast cancer patients.( 20 ) Consistent with their report, the present results showed that the mRNA of IGFBP5 was down‐regulated in cases with negative ER/PR compared to cases with positive ER/PR, and that overexpression of IGFBP5 could not predict the disease‐free survival of node‐negative breast cancer patients. However, it was found that IGFBP5 could be a powerful prognostic factor for patients with positive lymph nodes and negative ER. Although Wang et al. demonstrated a strong correlation between overexpression of IGFBP5 and high histologic grade of ovarian cancer and glioblastoma,( 14 , 15 ) this correlation was not found in breast cancers for the first time in the present study.
IGFBP5 was up‐regulated in breast cancer and could distinguish most of the normal breast and cancer tissues, indicating that it might be a potential adjuvant molecular marker for diagnosis of breast cancer. Although the mRNA levels of IGFBP5 were not able to accurately classify axillary lymph node status, high levels of IGFBP5 were associated with higher false‐negative rates of clinical palpation in ‘group without palpable nodes,’ and low levels of IGFBP5 with higher false positive rate of clinical palpable nodes in the ‘group with palpable nodes.’ So if IGFBP5 mRNA in needle biopsy specimens could be examined preoperatively, it could serve as an adjuvant factor of clinical palpation to predict axillary lymph node status and to guide surgery and systemic therapy.
Breast cancer is a hormone‐dependent disease. ER/PR status reflects the level of estrogen and progesterone in vivo, and is a predictor of clinical outcome of breast cancer patients. Hewitt et al. found that the expression of IGFBP5 could be up‐regulated by estrogen, indicating that the signal regulation of estrogen and progesterone might be involved in the complex functions of IGFBP5 in the development of breast cancer.( 21 ) Furthermore, overexpression of IGFBP5 was a powerful prognostic predictor for breast cancer patients with positive lymph nodes and negative ER. Thus, IGFBP5 may play an important role in the breast cancer biologic process and might serve as a molecular marker to guide patient‐tailored therapy.
Acknowledgments
Grant support: The National High‐tech Research Development Plans of China (2002AA2Z2011); Applied Basic Research Programs of Science and Technology Commission Foundation of Tianjin (06YFJMJC1290); The Tissue Bank is jointly supported by the National Foundation for Cancer Research (USA).
References
- 1. Coolican SA, Samuel DS, Ewton DZ et al . The mitogenic and myogenic actions of insulin‐like growth factors utilize distinct signaling pathways. J Biol Chem 1997; 272: 6653–62. [DOI] [PubMed] [Google Scholar]
- 2. Butt AJ, Dickson KA, McDougall F et al . Insulin‐like growth factor‐binding protein‐5 inhibits the growth of human breast cancer cells in vitro and in vivo . J Biol Chem 2003; 278: 29 676–85. [DOI] [PubMed] [Google Scholar]
- 3. Mohan S, Nakao Y, Honda Y et al . Studies on the mechanisms by which insulin‐like growth factor (IGF) binding protein‐4 (IGFBP‐4) and IGFBP‐5 modulate IGF actions in bone cells. J Biol Chem 1995; 270: 20 424–31. [DOI] [PubMed] [Google Scholar]
- 4. Jones JI, Gockerman A, Busby WH Jr. et al . Extracellular matrix contains insulin‐like growth factor binding protein‐5: potentiation of the effects of IGF‐I. J Cell Biol 1993; 121: 679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andress DL, Loop SM, Zapf J et al . Carboxy‐truncated insulin‐like growth factor binding protein‐5 stimulates mitogenesis in osteoblast‐like cells. Biochem Biophys Res Commun 1993; 195: 25–30. [DOI] [PubMed] [Google Scholar]
- 6. Hsieh T, Gordon RE, Clemmons DR et al . Regulation of vascular smooth muscle cell responses to insulin‐like growth factor (IGF)‐I by local IGF‐binding proteins. J Biol Chem 2003; 278: 42 886–92. [DOI] [PubMed] [Google Scholar]
- 7. Salih DA, Tripathi G, Holding C et al . Insulin‐like growth factor‐binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proc Natl Acad Sci 2004; 101: 4314–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perks CM, McCaig C, Clarke JB et al . Effects of a non‐IGF binding mutant of IGFBP‐5 on cell death in human breast cancer cells. Biochem Biophys Res Commun 2002; 294: 995–1000. [DOI] [PubMed] [Google Scholar]
- 9. Berfield AK, Andress DL, Abrass CK. IGFBP‐5 (201–218) stimulates Cdc42GAP aggregation and filopodia formationin migrating mesangial cells. Kidney Int 2000; 57: 1991–2003. [DOI] [PubMed] [Google Scholar]
- 10. Abrass CK, Berfield AK, Andress DL. Heparin binding domain of insulin‐like growth factor binding protein‐5 stimulates mesangial cell migration. Am J Physiol 1997; 273: F899–906. [DOI] [PubMed] [Google Scholar]
- 11. Xu Q, Yan B, Li S et al . Fibronectin binds insulin‐like growth factor‐binding protein 5 and abolishes its ligand‐dependent action on cell migration. J Biol Chem 2004; 279: 4269–77. [DOI] [PubMed] [Google Scholar]
- 12. McCaig C, Perks CM, Holly JM. Intrinsic actions of IGFBP‐3 and IGFBP‐5 on Hs578T breast cancer epithelial cells: inhibition or accentuation of attachment and survival is dependent upon the presence of fibronectin. J Cell Sci 2002; 115: 4293–303. [DOI] [PubMed] [Google Scholar]
- 13. Hao X, Sun B, Hu L et al . Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer 2004; 100: 1110–22. [DOI] [PubMed] [Google Scholar]
- 14. Wang H, Wang H, Zhang W et al . Overexpression of IGFBP5, but not IGFBP3, correlates with the histologic grade of human diffuse glioma: a tissue microarray and immunohistochemical study. Technol Cancer Res Treat 2006; 5: 195–9. [DOI] [PubMed] [Google Scholar]
- 15. Wang H, Rosen DG, Wang H et al . Insulin‐like growth factor‐binding protein 2 and 5 are differentially regulated in ovarian cancer of different histologic types. Mod Pathol 2006; 19: 1149–56. [DOI] [PubMed] [Google Scholar]
- 16. Van‘t Veer LJ, Dai H, Van De Vijver MJ et al . Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415: 530–6. [DOI] [PubMed] [Google Scholar]
- 17. Hu Z, Bonifas JM, Aragon G et al . Evidence for lack of enhanced hedgehog target gene expression in common extracutaneous tumors. Cancer Res 2003; 63: 923–8. [PubMed] [Google Scholar]
- 18. Butt AJ, Dickson KA, Jambazov S et al . Enhancement of tumor necrosis factor‐alpha‐induced growth inhibition by insulin‐like growth factor‐binding protein‐5 (IGFBP‐5), but not IGFBP‐3 in human breast cancer cells. Endocrinology 2005; 146: 3113–22. [DOI] [PubMed] [Google Scholar]
- 19. McCaig C, Perks CM, Holly JM. Signalling pathways involved in the direct effects of IGFBP‐5 on breast epithelial cell attachment and survival. J Cell Biochem 2002; 84: 784–94. [DOI] [PubMed] [Google Scholar]
- 20. Yee D, Sharma J, Hilsenbeck SG. Prognostic significance of insulin‐like growth factor‐binding protein expression in axillary lymph node‐negative breast cancer. J Natl Cancer Inst 1994; 86: 1785–9. [DOI] [PubMed] [Google Scholar]
- 21. Hewitt SC, Deroo BJ, Hansen K et al . Estrogen receptor‐dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol Endocrinol 2003; 17: 2070–83. [DOI] [PubMed] [Google Scholar]


