Abstract
CD147 is a novel cancer‐associated biomarker that plays an important role in the invasion and metastasis of human lung cancer. In spite of its many known functions, little is known about CD147 transcriptional regulation. In this study, we explored the regulation of CD147 in human lung cancer tissues. Over 60% of the human lung cancer tissues expressed differential high levels of CD147. We then cloned the 5′‐flanking region of the human CD147 gene and identified a critical promoter region at −108 to –42 which contained one binding site for Sp1, which was essential in up‐regulating CD147 promoter activity. These results were proven by blocking Sp1 using RNAi or mithramycin A treatment and up‐regulating Sp1 using transfection with eukaryotic expression vector. Consistent with the CD147 transcription activation, a high level of Sp1 expression was detected in lung cancer cell lines overexpressing CD147. Chromatin immunoprecipitation assay showed that much more Sp1 could bind to the CD147 promoter in 95‐D with CD147 high expression than in SK‐MES‐1 with CD147 low expression. There was a significant positive correlation between CD147 expression and Sp1 expression level detected by immunohistochemistry (r = 0.831). Collectively, our results suggest that Sp1 is essential for regulating the CD147 gene expression in human lung cancer. (Cancer Sci 2010)
Lung cancer is the leading cause of cancer‐related death for both men and women in the USA, with an estimated 159 390 deaths in 2009.( 1 ) Depending on the stage and treatment, the 5‐year survival rate of lung cancer is only 14%,( 2 ) and this has improved little over the past two decades. One of the characteristics of lung cancer is its high potential for invasion and metastasis. Previous studies have reported that CD147 plays an important role in lung cancer invasion and metastasis.( 3 ) CD147 is a transmembrane glycoprotein present on the cancer cell surface belonging to the immunoglobulin superfamily, which was found to induce expression of matrix metalloproteinases (MMPs), particular MMP‐2 and MMP‐9 in fibroblasts or tumor cells.( 4 , 5 , 6 , 7 ) CD147 was originally isolated from the LX‐1 human pulmonary carcinoma cell line.( 8 , 9 ) Although it is essentially absent in normal adult lungs,( 10 ) pathological up‐regulation has been reported in lung cancers.( 11 , 12 ) CD147 was also an independent predictor prognosis for lung cancer patients.( 13 ) However, the mechanism of CD147 expression and its regulation in lung cancer are mostly unknown.
The regulation of gene expression by transcription factors is critical to many biological processes and carcinogenesis.( 14 ) One of the first transcription factors to be identified in mammalian cells was Sp1,( 15 ) a member of the zinc‐finger Sp family of proteins that includes the Kruppel‐like factor (KLF) family.( 16 ) Sp1 is expressed ubiquitously in various mammalian cells and is implicated in the transcription of many genes that contain GC boxes in their promoter,( 17 ) particularly housekeeping genes and those involved in cell growth and development.( 18 , 19 ) Initial analysis of the CD147 promoter region has revealed several potential transcription factor Sp1 binding sites, suggesting that Sp1 may be involved in CD147 transcription regulation.( 20 , 21 )
In this study, we investigated the expression level of CD147 and Sp1 in human lung cancer tissues and explored the importance of DNA sequence element, transcription factor Sp1 in regulating CD147 expression in human lung cancer cell lines. To the best of our knowledge, this is the first study to examine the role of Sp1 in CD147 transcription regulation in human lung cancer.
Materials and Methods
Lung cancer tissue collection. Forty‐seven paraffin‐embedded tissue specimens of lung cancer were obtained from the Department of Pathology, Xijing and Tangdu Hospital affiliated to Fourth Military Medical University (Xi’an, China) with signed informed consent. All histologically confirmed lung cancer patients had undergone surgical resections at Xijing Hospital. Study approval was obtained from the Xijing Hospital Institutional Review Board.
Immunohistochemical staining. Human lung cancer tissues were processed for immunostaining using anti‐CD147 antibody( 12 , 22 ) and anti‐Sp1( 23 ) antibody as described previously. Immunopositivity was independently evaluated by two pathologists, who were blinded to clinical data. Depending on the percentage of positive cells and staining intensity, CD147 and Sp1 staining were classified into four groups: negative, weak positive, moderate positive, and strong positive. Specifically, the percentage of positive cells was divided into five grades (percentage scores): <10% (0), 10–25% (1), 26–50% (2), 51–75% (3), and >75% (4). The intensity of the staining was divided into four grades (intensity scores): no staining (0), light brown (1), brown (2), and dark brown (3). CD147 and Sp1 staining positivity was determined using the following formula: overall score = percentage score × intensity score. An overall score of ≤1, >1 to ≤3, >3 to ≤6, and >6 was defined as negative, weak positive, moderate positive, and strong positive, respectively.( 24 )
Cell lines and culture conditions. The following human lung cancer cell lines were used in this study: A549 (lung adenocarcinoma cells), SK‐MES‐1 (lung squamous cell carcinoma cells), NCI‐H292 (lung mucoepidermoid pulmonary carcinoma cells), NCI‐H446 (small‐cell lung cancer cells), NCI‐H460 (large‐cell lung cancer cells), 95‐D (human highly metastatic lung adenocarcinoma cells), and SPC‐A‐1 (lung adenocarcinoma cells). HEK‐293 cells was were in transfection and the luciferase assay. All cell lines were purchased from the Shanghai Institute for Biological Sciences (Shanghai, China). All cell lines were routinely cultured in RPMI‐1640 medium (Hyclone Laboratories, Logan, UT, USA) supplemented with 10% fetal calf serum (Gibco, Rockville, MD, USA) at 37°C in a humidified atmosphere of 5% CO2.
Construction of reporter plasmids and mutagenesis. The 5′ region (−1761 to +37, relative to the transcription start site of the CD147 gene) of the CD147 gene was amplified by PCR using the Advantage‐GC Genomic PCR kit (Clontech, Palo Alto, CA, USA) and inserted into the pGL3‐Basic vector (Promega, Madison, WI, USA). Nested deletions of the CD147 promoter region were carried out using the plasmid P(−1761/+37) as the template for PCR amplifications, and the sequences of the products were sequenced by Shanghai Sangon (Shanghai, China). The sequences of all PCR primers are listed in Table 1.
Table 1.
Oligonucleotide sequence of PCR primers and siRNA fragments
| Primers for real‐time quantitative RT‐PCR | ||
|---|---|---|
| CD147 | 5′‐TCGCGCTGCTGGGCACC‐3′ | 5′‐TGGCGCTGTCATTCAAGGA‐3′ |
| Sp1 | 5′‐AATTTGCCTGCCCTGAGTGC‐3′ | 5′‐TTGGACCCATGCTACCTTGC‐3′ |
| GAPDH | 5′‐AGCAATGCCTCCTGCACCACCAAC‐3 | 5′‐CCGGAGGGGCCATCCACAGTCT‐3′ |
| Primers used for the generation of CD147 promoter/reporter and mutagenesis constructs | ||
| CD147P(−1761/+37) | 5′‐ATCGGCTAGCGGTCTAGAGGATCCCACCCTTCT‐3′ (Nhe I) | 5′‐ATCGAAGCTTGATTCCTATTCCTCGCCGGT‐3′ (Hind III) |
| CD147P(−1273/+37) | 5′‐ATCGGCTAGCTTCGGCTTAGTCTGCGGTCCTC‐3′ | |
| CD147P(−1018/+37) | 5′‐ATCGGCTAGCGTAACCGCCAGCCTCTCCTGA‐3′ | |
| CD147P(−644/+37) | 5′‐ATCGGCTAGCCCAACCAAATGGTGCACAGGAC‐3′ | |
| CD147P(−338/+37) | 5′‐ATCGGCTAGCCTGGGTAACACACTTTCAACGCTTC‐3′ | |
| CD147P(−217/+37) | 5′‐ATCGGCTAGCCCGTTTCCTAGCAACGCCG‐3′ | |
| CD147P(−178/+37) | 5′‐ATCGGCTAGCGGATTCCGTAGCGTGAGCC‐3′ | |
| CD147P(−108/+37) | 5′‐ATCGGCTAGCCGACCGGCGTCCCCGGCGCT‐3′ | |
| CD147P(−42/+37) | 5′‐ATCGGCTAGCGCCTCCGCCGCTTTTTATAG‐3′ | |
| CD147P(−108/+37Sp1mt) | 5′‐GGCGTCCCCGGCGCT A G A CCCGCCCCCGAGATG‐3′ | 5′‐CATCTCGGGGGCGGG T C T AGCGCCGGGGACGCC‐3′ |
| Primers used for Sp1 expression vector construct | ||
| Sp1 | 5′‐ATCGGGTACCATGAGCGACCAAGATCACTCCAT‐3′ (KpnI) | 5′‐ATCGCTCGAGTCAGAAGCCATTGCCACTGATAT‐3′ (Xho I) |
| siRNA designed to target Sp1 | ||
| Sp1‐si320 | 5′‐AAUGAGAACAGCAACAACUtt‐3′ | 5′‐AGUUGUUGCUGUUCUCAUU tt‐3′ |
| Sp1‐si590 | 5′‐CCUGGAGUGAUGCCUAAUAtt‐3′ | 5′‐UAUUAGGCAUCACUCCAGG tt‐3′ |
| Primers for ChIP assay promoter‐specific PCR | ||
| CD147 | 5′‐ACATATGAGCTCGAAGCGCCGGAAG‐3′ | 5′‐TAATAGCGGCCGCGAGGTGAGAAC‐3′ |
To generate site‐directed mutants of Sp1 binding element at −108 to +18, the QuickChange mutagenesis kit (Stratagene, La Jolla, CA, USA) was used according to the manufacturer’s instructions. The primers (mutations are shown in bold and italic throughout) for mutation of the Sp1 elements are listed in Table 1. The incorporation of mutation was verified by sequencing (Shanghai Sangon).
Transfection and luciferase assay. Wild‐type or mutant CD147 promoter plasmids containing firefly luciferase reporters were cotransfected in triplicate with an internal control pRL‐TK (Promega) using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The pRL‐TK contained a full length renilla luciferase gene under the control of a human β‐actin promoter. The amount of co‐transfected Sp1 was as indicated in the figures. Mithramycin A (Sigma, St. Louis, MO, USA) was added 10 h before harvesting the cells. Forty‐eight hours after transfection, cells were assayed for luciferase activity using a Dual‐Luciferase Reporter assay system (Promega).
Real‐time quantitative RT‐PCR. Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was carried out using a ReverTra Ace reagents kit (Toyobo, Osaka, Japan). Real‐time quantitative PCR was performed using a MiniOpticon real‐time PCR detection system (Bio‐Rad, Hercules, CA, USA) in SYBR Green mastermix (Takara, Otsu, Japan). The annealing temperature was 60°C for CD147 and GAPDH and 58°C for Sp1. All data were analyzed using Opticon Monitor software (version 3.1; Bio‐Rad) and the expression of CD147 was calculated as relative expression level to GAPDH using the delta‐Ct method as described previously.( 25 ) The sequences of all PCR primers are listed in Table 1.
Western blot analysis. Cell samples were lysed with RIPA buffer (Beyotime, NanTong, China). Equal amounts (10 μg) of total protein were loaded, and then subsequently immunoblotted with the primary antibodies, including CD147 monoclonal antibody prepared in our lab,( 22 ) anti‐Sp1 (sc‐59; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti‐tubulin monoclonal antibody (NeoMarkers, Freemont, CA, USA). The proteins were detected using the Amersham enhanced chemiluminescence system (Pierce, Rockford, IL, USA) according to the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP) assay. ChIP assay was carried out using the ChIP‐IT kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s protocol. Briefly, SK‐MES‐1 and 95‐D cells were fixed with formaldehyde and then were sonicated. Immunoprecipitation was carried out with 2 μg of anti‐Sp1 antibody (Santa Cruz Biotechnology) or rabbit IgG at 4°C overnight with rotation. The purified DNA was amplified by the promoter‐specific primers (Table 1). The PCR products were analyzed on 1% agarose gel. Three independent experiments were performed.
Sp1 transfection and knockdown. Small‐interfering RNAs designed to knock down Sp1( 26 , 27 ) and Sp1 eukaryotic expression vector were transfected into 95‐D and SK‐MES‐1 cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions, respectively. A final concentration of 0.2 μm Sp1 siRNA duplex was transfected into 40% confluent cells, which were harvested 36–44 h after transfection.
Statistical analysis. Each experiment was performed independently at least twice with similar results; one representative experiment was presented. All statistical analyses were performed using the SPSS 16.0 statistical software package (SPSS, Chicago, IL, USA). The significance of the data was determined using Student’s t‐test. Spearman’s rho was calculated to analyze the correlation of CD147 expression and Sp1 expression. All the statistical tests were 2‐sided and P‐values of <0.05 were considered to be significant.
Results
High expression level of CD147 in human lung cancer tissues. In the first set of experiments, CD147 expression was evaluated in the primary lung cancer tissue of 47 patients via immunohistochemistry. CD147 was strongly expressed in 10 (21.28%) cases; moderate, weak, and negative CD147 expressions were observed in 14 (29.79%), six (12.76%), and 17 (36.17%) cases, respectively. As shown in Figure 1, CD147 protein was predominantly localized in tumor epithelial cells, whereas little was detected in stroma. We also found that CD147 displayed positive membranous and cytoplasmic staining. The immunohistochemical results revealed that 63.83% lung cancer tissues showed CD147‐positive expression.
Figure 1.
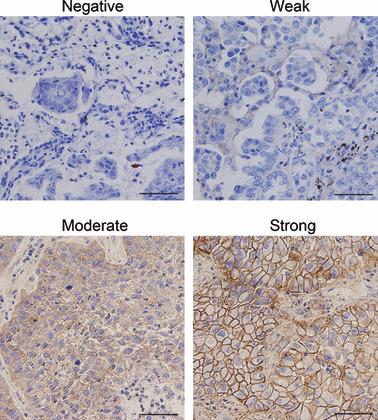
Differential expression of CD147 in lung cancer tissues. Expression of CD147 was detected by immunohistochemical staining analysis in lung cancer tissues. Pictures of representative areas are presented at different staining intensity (negative, weak, moderate, and strong). Scale bars, 50 μm.
Identification of the critical promoter region of CD147. To investigate the transcriptional regulation of the CD147 gene, we identified the promoter region of this gene. Based on the previous reports,( 28 ) we first constructed a luciferase reporter plasmid P(−1761/+37) that contained a 1798‐bp genomic DNA fragment spanning the 5′ upstream region of CD147. The cloned fragment exhibited obvious promoter activity, indicating that an active promoter sequence was isolated (Fig. 2a). Next, we developed a series of deletion constructs to identify the minimal promoter required for activation. Deletion of nucleotides from −338 to −217 on the 5′‐end led to moderated reduction of transcription activities; however, deletion in the CD147 region from −217 to +37 completely abolished the activity of the reporter gene. Thus, the most critical region for the basal transcriptional activity of the CD147 promoter is located within this 254‐bp region between positions −217 to +37 (Fig. 2a). The nucleotide sequence of the critical promoter region is shown in Figure 2(b). With the assistance of the TRANSFAC database, four Sp1 binding sites were found in the critical promoter of CD147.
Figure 2.
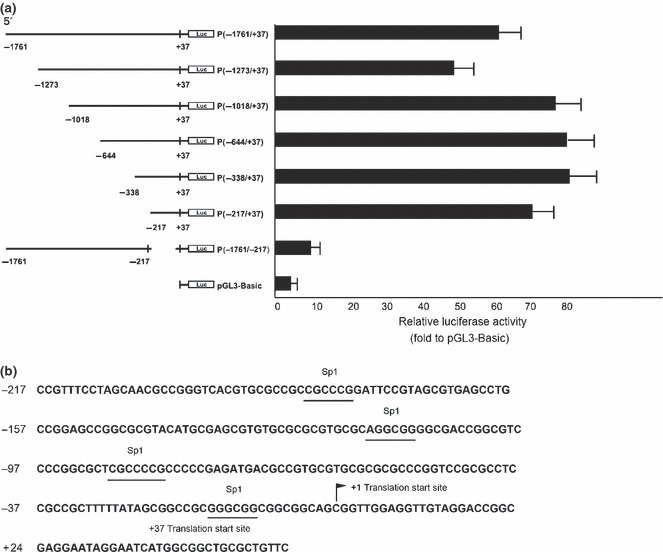
Identification of the critical promoter region of CD147. (a) Identification of the critical CD147 promoter region using luciferase assays. Numbers above the constructs indicate the positions relative to the translation start site of CD147. The activity of each promoter construct is shown as the luciferase activity relative to that of the pGL3‐Basic vector (a promoter‐less vector). Values were normalized for transfection efficiency by cotransfection with the Renilla expression plasmid and are shown as means ± SD for three independent experiments. (b) Nucleotide sequence of the CD147 critical promoter region. The consensus sequences for four potential Sp1 binding sites are underlined.
Analysis of the transcription factor Sp1 binding sites. Sp1 was known to play a role in the regulation of genes lacking a functional TATA box. The presence of numerous Sp1 binding sites in the critical promoter region of CD147 suggested that Sp1 might be involved in the regulation of CD147 activity. To further define the role of Sp1 in CD147 transcriptional regulation, small‐scale deletion mutants according to the four Sp1 binding sites were generated. Deletion of the region from −108 to −42 totally abrogated the promoter activity (Fig. 3a), which suggested that the region around −108 to −42 containing the third Sp1 binding site has the minimal essential elements to confer constitutive CD147 promoter activity.
Figure 3.

Analysis of the transcription factor Sp1 binding sites. (a) Identification of the minimal region in the CD147 promoter (−217/+37) necessary for basal CD147 promoter activity. Small‐scale deletion mutants according to the four Sp1 binding sites were generated. Luciferase activity is expressed as the percent activity of the pGL3‐Basic vector. Values were normalized for transfection efficiency by cotransfection with the Renilla expression plasmid and are shown as means ± SD for three independent experiments. (b) Sp1 binding motif was important for CD147 promoter activity. Values of luciferase activities due to the wild‐type construct P (−108/+37) and the mutated construct P (−108/+37Sp1mt) are shown as means ± SD for three independent experiments, each of which was performed using triplicate samples. Mutations are shown in bold above the histogram.
To determine whether the Sp1‐binding element is important in CD147 promoter activity, we generated a construct of CD147P(−108/+37Sp1mt) that contained mutations in the Sp1‐binding site by site directed mutagenesis. The promoter activity was completely abolished upon the mutations of the third Sp1 binding site (Fig. 3b). These results strongly indicate that Sp1 regulates CD147 promoter activity through one Sp1 binding site which is important to attain maximum activity.
Activation of the CD147 promoter activity by Sp1. Then, we cotransfected the CD147P(−108/+37) wild‐type construct and increasing amounts of Sp1‐expressing plasmid. We found a dose‐dependent increase in reporter activity (Fig. 4a). Moreover, we analyzed the effects of mithramycin A, a drug known to modify GC‐rich regions of the DNA and to inhibit Sp1 binding.( 29 ) As shown in Figure 4(b), treatment with mithramycin A down‐regulated the activity of CD147. Collectively, these results suggest a role for Sp1 in regulating CD147 promoter.
Figure 4.
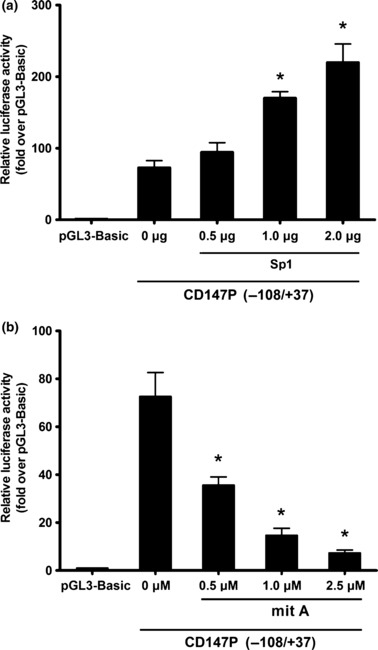
Sp1 up‐regulates the activity of CD147. (a) CD147 promoter (−108/+37) was cotransfected with indicated amount of Sp1. Empty vectors were added to ensure a constant input of DNA. (b) CD147 promoter (−108/+37) was transfected with mithramycin A at the indicated concentrations. Luciferase activity for both (a) and (b) is expressed as the percent activity of the pGL3‐Basic vector. Values were normalized for transfection efficiency by cotransfection with the Renilla expression plasmid and are shown as means ± SD for three independent experiments. *P < 0.05, assessed by Student’s t‐test.
CD147 and Sp1 expression in human lung cancer cell lines. A variety of established human lung cancer cell lines were cultured in vitro to determine the CD147 and Sp1 expression levels. We measured the steady‐state level of CD147 mRNA by real‐time quantitative RT‐PCR. As shown in Figure 5(a), all of the cell lines expressed a detectable level of CD147 mRNA. Specifically, NCI‐H460, 95‐D, NCI‐H446, and NCI‐H292 cells expressed a relatively high level of CD147 mRNA, whereas SK‐MES‐1, SPC‐A‐1, and A549 cells expressed a relatively low level of CD147 mRNA. Similar results were also observed in western blot analysis, indicating that the level of CD147 protein secretion (Fig. 5b) was correlated with the steady‐state level of CD147 mRNA (Fig. 5a).
Figure 5.
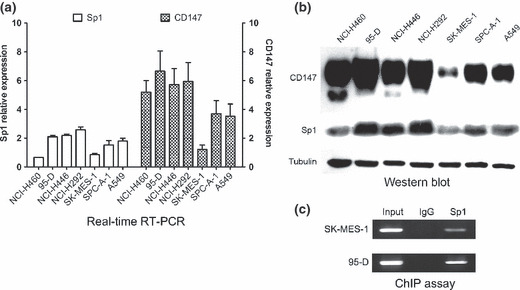
CD147 and Sp1 expression in human lung cancer cell lines. (a) mRNA expression of CD147 and Sp1 detected by real‐time quantitative RT‐PCR in seven lung cancer cell lines. The GAPDH gene was used as an internal control. (b) CD147 and Sp1 protein expression were determined by western blotting with tubulin as an internal control. (c) The binding of Sp1 to the CD147 promoter in vivo. ChIP assay using antibody against Sp1 was performed in SK‐MES‐1 and 95‐D cells. The normal rabbit IgG was used as a negative control and input indicates 5% input DNA, a positive amplification control.
Because of the role of Sp1 in regulating CD147 promoter activity, we sought to clarify whether the differential CD147 expression was due to the different level of Sp1 expression. Expressions of both mRNA and protein of Sp1were determined. As shown in Figure 5, a high level of Sp1 mRNA and protein were detected in NCI‐H460, 95‐D, NCI‐H446, and NCI‐H292 cells constitutively expressing a high level of CD147, whereas a low level of Sp1 mRNA and protein were detected in SK‐MES‐1, SPC‐A‐1, and A549 cells constitutively expressing a low of CD147. These results suggested that different CD147 expression levels may due to the Sp1 expression.
Determination of the specific binding of Sp1 to the CD147 promoter region. To directly demonstrate that Sp1 bind to the putative binding site within the critical CD147 promoter region, ChIP assays were performed in CD147‐low‐expressing SK‐MES‐1 cells and CD147‐high‐expressing 95‐D cells. We found that Sp1 bound to the CD147 promoter region but at significantly different levels. Very weak Sp1 binding was observed in SK‐MES‐1 cells, whereas strong Sp1 binding was shown in 95‐D cells (Fig. 5c). The ChIP results demonstrated that Sp1 bound specifically to the Sp1‐binding site in the CD147 critical promoter region and cells with higher CD147 expression bound more Sp1 protein.
Involvement of Sp1 in regulating CD147 expression. To illustrate the biological importance of Sp1 in CD147 gene regulation, we used RNA interference to knock down Sp1 expression in 95‐D with a high level of CD147 expression. In contrast, we used transient transfection to up‐regulate the Sp1 expression in SK‐MES‐1 with low level of CD147 expression. Then the CD147 mRNA and protein expressions were examined by RT‐PCR and western blot analysis. As shown in Figure 6(a), strongly reduced Sp1 mRNA and protein levels were observed, whereas control siRNA had no effect on Sp1 level. Meanwhile, the expression of endogenous CD147 RNA and protein (Fig. 6a) were effectively blocked by Sp1 siRNA transfection. In contrast, the expressions of Sp1 mRNA and protein were dramatically increased after transfection of the Sp1 eukaryotic expression plasmid in SK‐MES‐1 cells, and this up‐regulated expression of Sp1 resulted in a significant improvement in CD147 mRNA and protein expressions (Fig. 6b). These data demonstrate the involvement of Sp1 in regulating the human CD147 gene.
Figure 6.
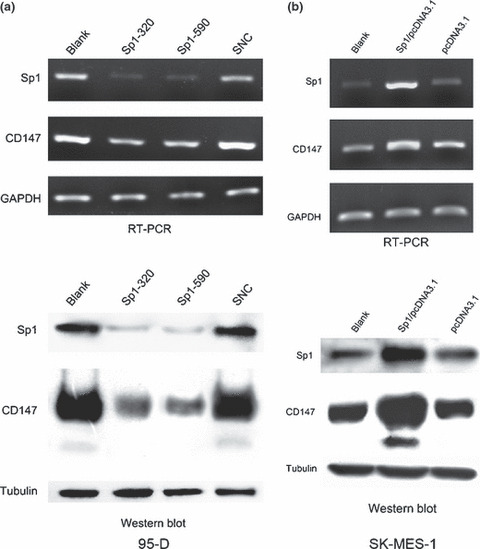
Involvement of Sp1 in regulating CD147 expression. (a) The 95‐D cell was transfected with siRNA targeting Sp1. (b) The SK‐MES‐1 cell was transfected with Sp1 expression vector. RT‐PCR and western blot analysis were used to detect the Sp1 and CD147 mRNA and protein expression levels. Control siRNA was used as negative control.
Correlation between Sp1 and CD147 expression in lung cancer tissues. To determine the biological relevance of Sp1‐mediated expression of CD147, Sp1 expression was also determined in the same 47 human lung cancer tissues. Sp1 protein was predominantly localized in cell nucleus. Sp1 was strongly expressed in 19 (40.43%) cases; moderate, weak, and negative Sp1 expressions were observed in 12 (25.53%), six (12.76%), and 10 (21.28%) cases, respectively. The immunohistochemical results revealed that 78.72% of lung cancer tissues showed Sp1‐positive expression. The representative staining of Sp1 is shown in Figure 7(a). Correlation analysis indicated that there was a significant positive correlation between Sp1 and CD147 expression levels with a correlation coefficient (r) = 0.831 and R 2 linear = 0.6905 (Fig. 7b). These data demonstrated clearly that constitutive expression of the transcription factor Sp1 contributes to CD147 overexpression, and hence to metastasis and progression of human lung cancer.
Figure 7.
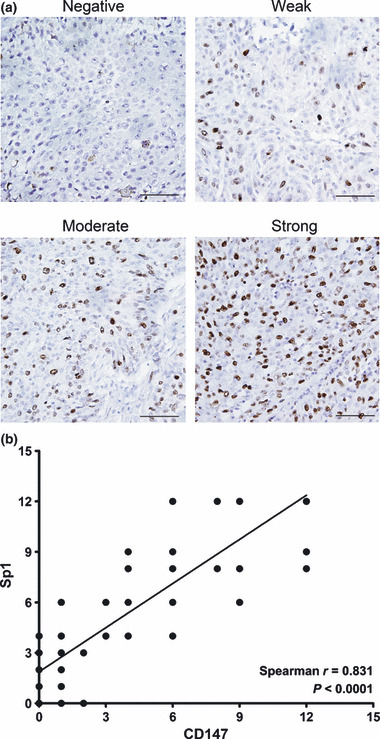
Correlation between Sp1 expression and CD147 expression in lung cancer tissues. (a) The differential expression of Sp1 in lung cancer tissues. Expression of Sp1 was detected by immunohistochemical staining analysis. Pictures of representative areas are presented at different staining intensity (negative, weak, moderate, and strong). Scale bars, 50 μm. (b) Analysis for correlation of Sp1 and CD147 expression in lung cancer tissues by calculating Sperman’s rho method.
Discussion
In our study, we demonstrated that the majority of the human lung cancer tissues and cell lines studied expressed CD147, but the expression level was differential. The CD147 expression appeared to result from its promoter activity, which correlated directly with the expression and activity of the transcription factor Sp1. There was a significant positive correlation between CD147 expression and Sp1 expression level. These findings suggest that elevated Sp1 activity is essential for CD147 expression in human lung cancer.
Our previous study systematically evaluated the CD147 expression profile in human normal and tumor tissues from 14 organs and found that epithelium‐derived carcinoma exhibited an overall CD147 positivity rate of 67.76%, while that of sarcomas was 27.34% and that of normal epithelial tissues was only 5.18%.( 12 ) In our study, CD147 expression was evaluated in 47 lung cancer tissues. CD147 was positively expressed in 63.83% lung cancer tissues, which was consistent with previous studies. But the expression level of CD147 was differential. Little is currently known about the transcriptional regulation leading to CD147 differential expression. Given the prominent role of CD147 in tumor progression,( 30 , 31 , 32 ) it is critical to understand the molecular basis of CD147 gene expression. The gene expression has been shown to be dynamically regulated by many factors at both the transcriptional and post‐transcriptional level. Among these mechanisms, the transcription factors seem to play the most crucial roles in gene regulation.( 23 ) In the present study, we aimed at defining the cis‐acting elements and transcription factors involved in CD147 expression in human lung cancer cells.
Functional analysis of the human CD147 promoter was initially performed. By using 5′ flanking region‐luciferase fusion plasmids, we identified the critical CD147 promoter as a 254‐bp region located between the base pairs −217 to +37 that contained four Sp1 binding sites. Sp1 is a well‐characterized, sequence‐specific, DNA‐binding protein that is important in the transcription of many cellular and viral genes that contain GC boxes in their promoter.( 33 ) Sp1 regulates many aspects of cancer biology, including cell growth, survival, invasion, angiogenesis, and metastasis.( 34 , 35 , 36 ) The Sp1 binding sites located in CD147 promoter activity have led us to hypothesize that transcription factor Sp1 may play an important role in CD147 gene regulation.
We therefore focused our current study of CD147 expression on this critical region of the CD147 promoter. Small‐scale deletion of the CD147P (−217/+37) reporter plasmid revealed that the proximal 66‐bp region (−108 to −42) is essential for CD147 promoter activity, which suggested that the region containing the third Sp1 binding site has the minimal essential elements to confer constitutive CD147 promoter activity. To define the contribution of the Sp1 binding element to CD147 promoter activity, site‐directed mutagenesis was performed. The activity of the CD147 promoter was greatly reduced; this finding demonstrated a crucial role of the Sp1 binding site in CD147 promoter activity. Then, we cotransfected the CD147P (−108/+37) wild‐type construct and increasing amounts of Sp1‐expressing plasmid. We found a dose‐dependent increase in reporter activity. Moreover, we found that treatment with mithramycin A down‐regulated the activity of CD147. Collectively, these results suggest a role for Sp1 in regulating the CD147 promoter.
Finally, we compared Sp1 and CD147 expression in different lung cancer cells. We found that the level of CD147 mRNA and protein expressions directly correlated with Sp1 mRNA and protein expressions. We found a high level of Sp1 protein in the cells expressing a high level of CD147, whereas a low level of Sp1 protein was seen in the cells expressing a low level of CD147. The ChIP results demonstrated that Sp1 bound specifically to the Sp1‐binding site in the CD147 critical promoter region and cells with higher CD147 expression bound more Sp1 protein. We further confirmed the role of Sp1 in the regulation of CD147 through down‐regulating Sp1 expression by RNA interference or up‐regulating Sp1 expression by cotransfected expression plasmid. Consistent with our reporter assays demonstrating a crucial role for Sp1 in CD147 promoter activity, changing Sp1 expression dramatically influenced CD147 expression. Data from lung cancer tissues were consistent with these biochemical findings. Most of the human lung cancer specimens overexpressed Sp1 protein; this was determined by immunostaining and correlated directly with CD147 overexpression. Because CD147 is a key cancer‐related antigen, it is expected that overexpression of Sp1 and subsequent overexpression of CD147 may play an important role in lung cancer metastasis and progression.
In summary, our present study was the first to demonstrate that differential Sp1 expression and activity directly regulate the levels of CD147 expression in human lung cancer, thus providing a novel mechanism for CD147 regulation. A further understanding of the molecular basis of Sp1 in CD147 regulation will have functional implications in suppressing lung cancer progression and will ultimately lead to the design of new therapeutic modalities.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (30530720, 30772479, and 30671099) and National Basic Research Program of China (2009CB521704).
References
- 1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin 2009; 59: 225–49. [DOI] [PubMed] [Google Scholar]
- 2. Minna JSJ. Harrison’s Principles of Internal Medicine. New York, USA: McGraw‐Hill, 2008; 551–62. [Google Scholar]
- 3. Zucker S, Hymowitz M, Rollo EE et al. Tumorigenic potential of extracellular matrix metalloproteinase inducer. Am J Pathol 2001; 158: 1921–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biswas C, Zhang Y, DeCastro R et al. The human tumor cell‐derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res 1995; 55: 434–9. [PubMed] [Google Scholar]
- 5. Chen ZNYZ, Mi L, Jiang JL, Guo XN. Structure and function analysis of hepatoma associated factor HAb18G. J Mol Cell Immunol 1999; 15: 34–9. [Google Scholar]
- 6. Kataoka H, DeCastro R, Zucker S, Biswas C. Tumor cell‐derived collagenase‐stimulatory factor increases expression of interstitial collagenase, stromelysin, and 72‐kDa gelatinase. Cancer Res 1993; 53: 3154–8. [PubMed] [Google Scholar]
- 7. Guo H, Li R, Zucker S, Toole BP. EMMPRIN (CD147), an inducer of matrix metalloproteinase synthesis, also binds interstitial collagenase to the tumor cell surface. Cancer Res 2000; 60: 888–91. [PubMed] [Google Scholar]
- 8. Ellis S, Nabeshima K, Biswas C. Monoclonal antibody preparation and purification of a tumor cell collagenase stimulatory factor. Cancer Res 1989; 49: 3385. [PubMed] [Google Scholar]
- 9. Nabeshima K, Lane W, Biswas C. Partial sequencing and characterization of the tumor cell‐derived collagenase stimulatory factor. Arch Biochem Biophys 1991; 285: 90. [DOI] [PubMed] [Google Scholar]
- 10. Greenlee KJ, Werb Z, Kheradmand F. Matrix metalloproteinases in lung: multiple, multifarious, and multifaceted. Physiol Rev 2007; 87: 69–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hakuma N, Betsuyaku T, Kinoshita I et al. High incidence of extracellular matrix metalloproteinase inducer expression in non‐small cell lung cancers. Association with clinicopathological parameters. Oncology 2007; 72: 197–204. [DOI] [PubMed] [Google Scholar]
- 12. Li Y, Xu J, Chen L et al. HAb18G (CD147), a cancer‐associated biomarker and its role in cancer detection. Histopathology 2009; 54: 677–87. [DOI] [PubMed] [Google Scholar]
- 13. Sienel W, Polzer B, Elshawi K et al. Cellular localization of EMMPRIN predicts prognosis of patients with operable lung adenocarcinoma independent from MMP‐2 and MMP‐9. Mod Pathol 2008; 21: 1130–8. [DOI] [PubMed] [Google Scholar]
- 14. Dynan WS, Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 1983; 32: 669–80. [DOI] [PubMed] [Google Scholar]
- 15. Dynan WS, Tjian R. The promoter‐specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 1983; 35: 79–87. [DOI] [PubMed] [Google Scholar]
- 16. Lomberk G, Urrutia R. The family feud: turning off Sp1 by Sp1‐like KLF proteins. Biochem J 2005; 392: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suske G. The Sp‐family of transcription factors. Gene 1999; 238: 291–300. [DOI] [PubMed] [Google Scholar]
- 18. Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 1997; 89: 619–28. [DOI] [PubMed] [Google Scholar]
- 19. Black AR, Black JD, Azizkhan‐Clifford J. Sp1 and kruppel‐like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol 2001; 188: 143–60. [DOI] [PubMed] [Google Scholar]
- 20. Liang L, Major T, Bocan T. Characterization of the promoter of human extracellular matrix metalloproteinase inducer (EMMPRIN). Gene 2002; 282: 75–86. [DOI] [PubMed] [Google Scholar]
- 21. Guo H, Majmudar G, Jensen TC, Biswas C, Toole BP, Gordon MK. Characterization of the gene for human EMMPRIN, a tumor cell surface inducer of matrix metalloproteinases. Gene 1998; 220: 99–108. [DOI] [PubMed] [Google Scholar]
- 22. Chen ZN. [Significance and application of anti‐malignant hepatoma MAb HAb18 in radioimmunal diagnosis of human hepatocellular carcinoma]. Zhonghua Zhong Liu Za Zhi 1992; 14: 9–12. [PubMed] [Google Scholar]
- 23. Shi Q, Le X, Abbruzzese JL et al. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res 2001; 61: 4143–54. [PubMed] [Google Scholar]
- 24. Yao J, Wang L, Wei D et al. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res 2004; 10: 4109. [DOI] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- 26. Hasegawa K, Wakino S, Tanaka T et al. Dimethylarginine dimethyl‐aminohydrolase 2 increases vascular endothelial growth factor expression through Sp1 transcription factor in endothelial cells. Arterioscler Thromb Vasc Biol 2006; 26: 1488–94. [DOI] [PubMed] [Google Scholar]
- 27. Li T, Chen YH, Liu TJ et al. Using DNA microarray to identify Sp1 as a transcriptional regulatory element of insulin‐like growth factor 1 in cardiac muscle cells. Circ Res 2003; 93: 1202–9. [DOI] [PubMed] [Google Scholar]
- 28. Faber PW, Van RooijHC, Schipper HJ, Brinkmann AO, Trapman J. Two different, overlapping pathways of transcription initiation are active on the TATA‐less human androgen receptor promoter. The role of Sp1. J Biol Chem 1993; 268: 9296–301. [PubMed] [Google Scholar]
- 29. Koga T, Suico M, Nakamura H et al. Sp1‐dependent regulation of Myeloid Elf‐1 like factor in human epithelial cells. FEBS Lett 2005; 579: 2811–6. [DOI] [PubMed] [Google Scholar]
- 30. Xu J, Xu HY, Zhang Q et al. HAb18G/CD147 functions in invasion and metastasis of hepatocellular carcinoma. Mol Cancer Res 2007; 5: 605–14. [DOI] [PubMed] [Google Scholar]
- 31. Li Y, Shang P, Qian AR, Wang L, Yang Y, Chen ZN. Inhibitory effects of antisense RNA of HAb18G/CD147 on invasion of hepatocellular carcinoma cells in vitro . World J Gastroenterol 2003; 9: 2174–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang JL, Zhou Q, Yu MK, Ho LS, Chen ZN, Chan HC. The involvement of HAb18G/CD147 in regulation of store‐operated calcium entry and metastasis of human hepatoma cells. J Biol Chem 2001; 276: 46870–7. [DOI] [PubMed] [Google Scholar]
- 33. Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii‐Kuriyama Y. A novel bHLH‐PAS factor with close sequence similarity to hypoxia‐inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U S A 1997; 94: 4273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer 2005; 41: 2438–48. [DOI] [PubMed] [Google Scholar]
- 35. Price SJ, Greaves DR, Watkins H. Identification of novel, functional genetic variants in the human matrix metalloproteinase‐2 gene: role of Sp1 in allele‐specific transcriptional regulation. J Biol Chem 2001; 276: 7549–58. [DOI] [PubMed] [Google Scholar]
- 36. Ibanez‐Tallon I, Ferrai C, Longobardi E, Facetti I, Blasi F, Crippa MP. Binding of Sp1 to the proximal promoter links constitutive expression of the human uPA gene and invasive potential of PC3 cells. Blood 2002; 100: 3325–32. [DOI] [PubMed] [Google Scholar]


