Abstract
In the past several years, the importance of microRNA (miRNA) in cancer cells has been recognized. Proper control of miRNA expression is essential for maintaining a steady state of the cellular machinery. Recently, it was discovered that extracellular miRNAs circulate in the blood of both healthy and diseased patients, although ribonuclease is present in both plasma and serum. Most of the circulating miRNAs are included in lipid or lipoprotein complexes, such as apoptotic bodies, microvesicles, or exosomes, and are, therefore, highly stable. The existence of circulating miRNAs in the blood of cancer patients has raised the possibility that miRNAs may serve as a novel diagnostic marker. However, the secretory mechanism and biological function, as well as the meaning of the existence of extracellular miRNAs, remain largely unclear. In this review, we summarize the usefulness of circulating miRNA for cancer diagnosis, prognosis, and therapeutics. Furthermore, we propose a mechanism for the secretion and incorporation of miRNA into the cells. (Cancer Sci 2010)
In 1993, Ambros and colleagues discovered a gene, lin‐4, which affected development in Caenorhabditis elegans (C. elegans); they found that its product was a small non‐protein coding RNA, microRNA (miRNA).( 1 ) miRNAs are small regulatory RNA molecules that modulate the expression of their target genes and play important roles in a variety of physiological and pathological processes, such as development, differentiation, cell proliferation, apoptosis, and stress responses.( 2 ) miRNA biogenesis requires several post‐transcriptional processing steps to yield the functional mature miRNA.( 3 ) Currently, there are 940 mature human miRNA sequences listed in the miRNA registry (Sanger miRBase release 15; http://www.mirbase.org/). Over the past several years, many miRNAs have been investigated in various human cancers.( 4 ) The deregulation of the expression of miRNAs has been shown to contribute to cancer development through various kinds of mechanisms, including deletions, amplifications, or mutations involving miRNA loci, epigenetic silencing, the dysregulation of transcription factors that target specific miRNAs, or the inhibition of processing. miRNA expression profiling is of increasing importance as a useful diagnostic and prognostic tool, and many studies have indicated that miRNAs act as either an oncogene or a tumor suppressor. Recently, the discovery of miRNAs as novel biomarkers in serum or plasma represented a new approach for diagnostic screening in blood. Since current approaches to cancer screening are invasive and it is difficult to detect cancer in its early stages, it is important to understand the characteristics of secretory miRNAs and their usefulness in cancer detection. In this article, we review and assess the potential usefulness of circulating miRNAs in cancer therapeutics and diagnosis.
Discovery of circulating miRNA in cancer patients
In several studies, miRNA expression profiles have been shown to have signatures related to tumor classification, diagnosis, and disease progression. Since a single miRNA is said to be able to target several mRNAs, aberrant miRNA expression is capable of disrupting the expression of several mRNAs and proteins. For instance, Rosenfeld et al. ( 5 ) showed that miRNA expression profiles have been useful in detecting the tissue of origin for cancers of unknown primary origin. Furthermore, Lu et al. ( 6 ) demonstrated that the expression analysis of 217 miRNAs in various human cancers clearly reflects the developmental lineage and differentiation state of the tumors, and they also confirmed a general down‐regulation of miRNAs in tumors compared with normal tissues. These findings highlight the potential of miRNA profiling in cancer diagnosis. Most diagnostic expression profiling of miRNAs has been conducted using samples from tumor tissues; however, several studies have shown the diagnostic and prognostic usefulness of circulating miRNAs (see Table 1).( 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 )
Table 1.
Serum miRNAs as a biomarker
| Type of cancer | Biomarker candidate | Reference |
|---|---|---|
| Diffuse large B‐cell lymphoma (DLBCL) | Expression levels of miR‐155, miR‐210 and miR‐21 were higher in DLBCL patient than control sera High miR‐21 expression was associated with relapse‐free survival | 7 |
| Prostate cancer | Serum levels of miR‐141 can distinguish patients with prostate cancer from healthy controls | 8 |
| Ovarian cancer | The levels of the 8 specific miRNAs were similar between cellular and exosomal miRNAs. Exosomal miRNA from ovarian cancer patients exhibited similar profiles, which were significantly distinct from profiles observed in benign disease miR‐21, ‐92, ‐93, ‐126 and ‐29a were significantly overexpressed in the serum from cancer patients compared to controls | 9 10 |
| Non small cell lung cancer | Eleven serum miRNAs were found to be altered more than 5‐fold between longer‐survival and shorter‐survival groups, and levels of four miRNAs were significantly associated with overall survival | 11 |
| Acute myeloid leukemia (AML) Acute lymphoblastic leukemia (ALL) | miR‐92a decreased in the plasmas of acute leukemia patients | 12 |
| Breast cancer | Increased miR‐195 levels in patients were reflected in tumors, and circulating levels of miR‐195 and let‐7a decreased in cancer patients postoperatively, to levels comparable with control subjects miR‐155 was differentially expressed in the serum of women with hormone‐sensitive compared to women with hormone‐insensitive breast cancer | 13 14 |
| Gastric cancer | The plasma concentrations of miR‐17‐5p, miR‐21, miR‐106a, and miR‐106b were significantly higher in patients than controls, whereas let‐7a was lower in patients | 15 |
| Pancreatic cancer | Circulating miR‐210 levels are elevated in pancreatic cancer patients | 16 |
| Pancreatic ductal adenocarcinoma | The combined analyses of four miRNAs (miR‐21, miR‐210, miR‐155, and miR‐196a) in plasma can discriminate patients from normal healthy individuals | 17 |
| Squamous cell carcinoma (SCC) of tongue | Plasma miR‐184 levels were significantly higher in tongue SCC patients in comparison with normal individuals, and the levels were significantly reduced after surgical removal of the primary tumors | 18 |
| Colorectal cancer | Both miR‐17‐3p and miR‐92 were significantly elevated in the patients, and the plasma levels of these miRNAs were reduced after surgery | 19 |
| Hepatocellular carcinoma (HCC) | An increased amount of miR‐500 was found in the sera of the HCC patients, and its levels in sera returned to normal after the surgical treatment | 20 |
One of the first studies measuring miRNA levels in serum was reported by Lawrie et al. ( 7 ) who demonstrated that the serum levels of miR‐21 were associated with relapse‐free survival in patients with diffuse large B‐cell lymphoma; thus, miR‐21 may have potential as a diagnostic biomarker for this disease. Mitchell et al. ( 8 ) found that, by measuring the serum levels of miR‐141, they could distinguish patients with prostate cancer from healthy subjects. In that study, they also demonstrated the presence of circulating tumor‐derived miRNAs in blood by using a mouse prostate cancer xenograft model. Furthermore, these circulating miRNAs were also found in the serum of rats, mice, calves, bovine fetuses, and horses, indicating that circulating miRNAs were commonly discovered in mammalian species.( 21 )
Chen et al. ( 21 ) showed the miRNA expression profiles for lung cancer, colorectal cancer, and diabetes patients in comparison to those of healthy subjects and found that cancer patients had serum levels of miR‐25 and miR‐223 that were more elevated than those of healthy subjects. On the other hand, several serum miRNAs were significantly more overexpressed in patients than in healthy subjects in a study of ovarian cancer.( 10 ) In addition, Ng et al. ( 19 ) showed that miR‐92 is more significantly increased in colorectal cancer than in gastric cancer and inflammatory bowel disease as well as normal subjects and can be used as a potential biomarker to detect colorectal cancer in plasma samples. Recently, Hu et al. ( 11 ) performed a screening to detect serum miRNA to predict the prognosis of non‐small‐cell lung cancer (NSCLC) using Solexa sequencing followed by an extensively self‐validated study in a cohort of 303 patients with stage I to IIIa NSCLC. Eleven serum miRNAs were found to be altered more than 5‐fold between longer‐survival and shorter‐survival groups, and the levels of four miRNAs (miR‐486, miR‐30d, miR‐1, and miR‐499) were significantly associated with overall survival. The four‐miRNA signature was also an independent predictor of overall survival for both training and testing samples.
Previously, Yamamoto et al. ( 20 ) found that miR‐500 is an oncofetal miRNA in liver cancer using a global miRNA expression profile in mouse liver development. The expression of miR‐500 is high in fetal liver and down‐regulated in the developmental process and then up‐regulated in the process of liver cirrhosis. miR‐500 was abundantly expressed in several human liver cancer cell lines and 45% of human hepatocellular carcinoma (HCC) tissue. Most importantly, an increased amount of miR‐500 was found in the sera of HCC patients, which means that liver cancer‐specific miRNA, such as miR‐500, is circulating in the peripheral blood and can be a novel diagnostic marker. Furthermore, elevated serum levels of miR‐500 in HCC patients were significantly reduced after surgery and returned to normal levels. These results reveal that the abundance of miR‐500 in the serum of the HCC patients might reflect physiological and/or pathological conditions. Wong et al. ( 18 ) reported that miR‐184 showed significantly higher expression in tongue squamous cell carcinoma (SCC) cells than in the paired normal cells. In addition, the plasma level of miR‐184 was much higher in cancer patients with early and advanced tongue SCC than in normal individuals. Moreover, the mean plasma levels of miR‐184 were reduced in the patients after the surgical removal of the primary tumor. After the tumor was resected, it was important to use a serum biomarker for patients to monitor any recurrence of the tumor. Using serum miRNAs, such as miR‐500 and miR‐184, helps determine the next option for treatment in the early stage of cancer and metastasis.
Circulating miRNA carried in particles
Although serum contains ribonuclease, the existence of serum miRNAs suggests that these miRNAs are resistant to RNase digestion. Chen et al. ( 21 ) showed that serum miRNAs remained stable after being subjected to harsh conditions under which most RNA would be degraded. El‐Hefnawy et al. ( 22 ) showed that plasma RNA is protected from degradation by inclusion in lipid or lipoprotein complexes. Recent studies have revealed the novel genetic exchange between cells using miRNA either in microvesicles (up to 1 μm) or in small membrane vesicles of endocytic origin called exosomes (50–100 nm) (Table 2).( 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 ) One of the first reports showing the existence of miRNA in exosomes was studied by Valadi et al. ( 32 ) who reported that exosomes released from human and murine mast cell lines contain mRNAs and miRNAs. Hunter et al. ( 33 ) demonstrated that miRNAs contained in the microvesicles from blood were known to regulate the cellular differentiation of blood cells and metabolic pathways and to modulate immune function. On the other hand, evidence of the presence of tumor‐derived exosomes in the peripheral circulation was provided by Taylor et al. ( 9 ) Furthermore, they compared tumor‐derived miRNA profiles and peripheral blood‐derived exosomal miRNAs and showed that they were not significantly different. In addition, Rabinowits et al. ( 34 ) reported that the protein concentration of circulating exosomes was significantly higher in lung adenocarcinoma than in a control group. Significant differences were also found between the mean exosomal miRNA concentrations of the lung adenocarcinoma group and the control group. Interestingly, in four cases of lung adenocarcinoma in which paired tumor and plasma samples were examined, there was a close correlation between the circulating miRNAs of tumor‐derived exosomes and tumor miRNAs. Skog et al. ( 26 ) demonstrated that brain microvascular endothelial cells take up exosomes, which contain mRNA, miRNA, and angiogenic proteins released by glioblastoma cells. Moreover, miR‐21, known to be overexpressed in glioblastoma tumors, was more elevated in serum microvesicles from glioblastoma patients than in healthy controls.
Table 2.
Particles in body fluid and their roles in recipient cells
| Particles | Typical size | Origin of particle | Reported function of particles to recipient cells | Reference |
|---|---|---|---|---|
| Microvesicle | 0.1–1 μm | BCLL Melanoma cells | Stimulate bone marrow stromal cells to induce the production of VEGF Increased TGF‐β1 production by cultured macrophages Enhanced the metastatic potential of melanoma cell lines in vivo | 24 25 |
| Exosome | 10–100 nm | Glioblastoma cells Mammary adenocarcinoma cells | Stimulate tubule formation by endothelial cells Induce potent CD8+ T‐cell–dependent antitumor effects by dendritic cells | 26 27 |
| Prostasome | 50–500 nm | Prostatic ductal epithelial cells Prostate cencer cells | Boosting survivability, motility of spermatozoa and modulate acrosomal reactivity Protect prostatic malignant cells from complement attack | 28 29 |
| Apoptotic body | 0.5–2 μm | EBV‐carrying Burkitt’s lymphoma cell line Endothelial cells | Induce the expression of EBV‐specific markers in the recipient cells Induce the expression of CXCL12 in endothelial cells | 30 31 |
BCLL, B‐cell chronic lymphocytic leukemia; CXCL12, chemokine CXC motif ligand 12; EBV, Epstein–Barr virus; TGF‐β1, transforming growth factor β1; VEGF, vascular endothelial growth factor.
Considering that exosomes and microvesicles are evident in several types of body fluid from cancer patients, miRNA surely be able to be found not only in serum/plasma but also in other body fluid. Indeed, Michael et al. ( 35 ) showed the presence of miRNAs within exosomes isolated from human saliva. Furthermore, Park et al. ( 36 ) found that miR‐125a and miR‐200a were present in significantly lower levels in the saliva of oral SCC patients than in control subjects. Recently, we found miRNA presence in human breast milk and detected high expression levels of immune‐related miRNAs in the first 6 months of lactation.( 37 ) It is noteworthy that these miRNA molecules are stable even in very acidic conditions, indicating that breast milk allows the dietary intake of miRNAs by infants. Thus, these reports raise the possibility that circulating miRNAs could be used as non‐invasive diagnostic markers.
Mechanism of miRNA secretion and incorporation
We have summarized recent reports that show the existence of circulating miRNAs in the blood of cancer patients (Fig. 1). To use circulating miRNAs as a diagnostic marker, we need to gain a better understanding of the mechanisms by which miRNAs are released in the bloodstream. In some organisms, such as C. elegans and Drosophila melanogaster, small RNA pathways can spread from cell to cell. However, the secretory mechanism and incorporation of extracellular miRNAs in mammalian cells remain unclear.
Figure 1.
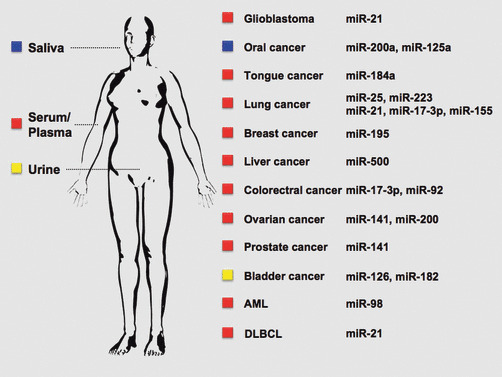
miRNAs in human body fluids are non‐invasive diagnostic markers for cancers. Many kinds of circulating miRNAs have been reported in various types of cancers. However, certain cancers cannot be diagnosed by known serum biomarkers. In such cases, circulating miRNAs in serum, saliva, and urine are good candidates for future use. AML, acute myeloid leukemia; DLBCL, diffuse large B‐cell lymphoma.
Rechavi et al. ( 38 ) showed that T cells receive small RNAs from B cells that can affect the expression of target genes in the recipient T cells upon cell contact. Furthermore, Pegtel et al. ( 39 ) demonstrated that miRNAs secreted by Epstein–Barr virus (EBV)‐infected cells are transferred to and act in uninfected recipient cells through exosomes. They also showed that these EBV‐miRNAs repressed confirmed EBV target genes. Importantly, although EBV DNA is restricted to the circulating B‐cell population, Pegtel et al. also showed that EBV BamHI‐A Rightward Transcript (BART) miRNAs are present in both B‐cell and non‐B‐cell fractions in peripheral blood mononuclear cells from patients with increased EBV. In addition, Yuan et al. ( 40 ) showed that embryonic stem (ES) cell microvesicles contain abundant miRNA and that they can transfer a subset of miRNAs to mouse embryonic fibroblasts in vitro, suggesting that stem cells can affect the expression of genes in neighboring cells by transferring miRNAs contained in microvesicles. As in these reports, several other studies have already shown the intercellular transfer of miRNAs from donor cells to recipient cells; however, there are only a few reports showing the mechanism of secretion of miRNAs. Recently, we showed that miRNAs are released through a ceramide‐dependent secretory machinery and the secretory miRNAs are transferable and functional in the recipient cells.( 41 ) We clarified that neutral sphingomyelinase 2 (nSMase2), which regulates ceramide biosynthesis, controls the secretion of miRNAs outside the cells. This enzyme is already known for the secretion of exosomes. However, the endosomal sorting complex required for the transport (ESCRT) system, which is another regulator of exosome secretion, is unnecessary for the release of miRNAs. Furthermore, a tumor‐suppressive miRNA secreted via a ceramide‐dependent pathway suppressed its target gene in the recipient cells, thereby leading to cell growth inhibition. In this report, we showed the importance of nSMase2 in the secretion of miRNA; however, there might be another mechanism for the secretion of miRNAs from cells. One candidate is an apoptotic body, namely, the membranous microvesicles shed from cells during apoptosis. Zernecke et al. ( 31 ) showed that endothelial cell‐derived apoptotic bodies are generated during atherosclerosis and carry paracrine alarm signals to recipient vascular cells, which trigger the production of chemokine CXC motif ligand 12 (CXCL12). CXCL12 production was mediated by miR‐126. miR‐126 was enriched in apoptotic bodies and repressed its target gene in recipient cells. This study showed an important observation, namely, that not only exosomes but also apoptotic bodies can transfer miRNAs between the cells. Interestingly, a recent report showed the relationship between an exosome and an apoptotic body. Phosphatidylserine, which is exposed on the surface of apoptotic cells and works as an “eat me” signal for phagocytes, is also expressed on the surface of exosomes. Miyanishi et al. ( 42 ) found an antibody that inhibited the phosphatidylserine‐dependent engulfment of apoptotic cells, and the antigen recognized by the antibody was identified by expression cloning as a type I transmembrane protein called Tim4 (T‐cell immunoglobulin‐ and mucin‐domain‐containing molecule). The expression of Tim4 in fibroblasts enhanced their ability to engulf apoptotic cells, and Tim4‐expressing cell lines were bound by exosomes via phosphatidylserine. These results indicate that Tim4 is a phosphatidylserine receptor for the engulfment of apoptotic cells and may also be involved in exosome‐mediated intercellular signaling.
As reported above, RNA interference (RNAi), which silences genes of a matching sequence by double‐strand RNA (dsRNA), spreads systemically in plants and nematodes to silence gene expression distant from the site of initiation. Passive dsRNA uptake has been uniquely observed in C. elegans due to the expression of systemic RNA interference defective‐1 (SID‐1). Tsang et al. ( 43 ) showed that ectopic expression of SID‐1 induced the cellular uptake of siRNA or dsRNA in mouse ES cells. The mammalian SID‐1 homologue localizes to the cell membrane of human cells and enhances their uptake of siRNA, resulting in an increased siRNA‐mediated gene silencing effect. Recently, Wolfrum et al. ( 44 ) reported that cholesterol‐conjugated siRNAs could silence gene expression in vivo. These reports clarify that not only are the low‐density lipoprotein (LDL)‐ and scavenger receptor class B type I (SR‐BI) receptors essential for siRNA delivery by LDL particles and high‐density lipoprotein (HDL)‐bound siRNAs but also SID‐1 is required for cellular uptake. There are still no answers with regard to the uptake of miRNA from donor cells; however, we might have a tightly regulated mechanism for the control of miRNA incorporation.
The phenomenon of secretion and incorporation of miRNAs seems to be a general biological event for organisms. Interestingly, recent studies have shown the importance of communication between cancer cells and their surroundings through exosomes (Fig. 2). For instance, epidermal growth factor receptor variant III (EGFR vIII) proteins were transferred into glioma cells lacking EGFRvIII via secretory membrane microvesicles.( 45 ) Because many tumors have a remarkable ability to mold their stromal environment to their own advantage, exosomes of cancer cells can contribute to the horizontal propagation of oncogenic miRNAs and their associated transforming phenotype among subsets of cancer cells. Although the mechanism of secretion and incorporation of miRNAs has not been clarified, secretory miRNAs may play a pivotal and general role as a signaling molecule in physiological and pathological events.
Figure 2.
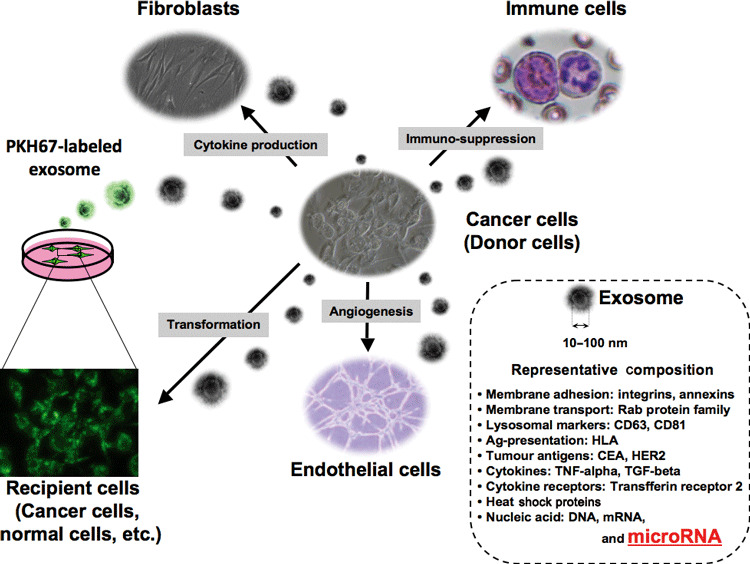
Secreted miRNAs contained in exosomes potentially influence microenvironmental cells, including immune cells, endothelial cells, and fibroblast cells. Soluble factors, such as cytokines and chemokines, have been shown to be intercellular communication tools between cancer and microenvironmental cells. In addition, recent observations have indicated that exosomes also function as an intercellular communication tool between them. For instance, exosomes, which are secreted by living tumor cells, contain and transfer tumor antigens to dendritic cells, and dendritic cells then induce potent CD8+ T‐cell‐dependent antitumor effects on syngenic and allogeneic established mouse tumors. In addition, this transforming activity can possibly be transferred via exosome. Furthermore, miRNA, which regulates multiple target genes, can also be transferred from tumor cells to tumor cells or tumor cells to normal cells. When purified PKH67‐labeled exosomes from donor cancer cells were incubated with recipient cancer cells, they became fluorescent, indicating that an oncogene can be propagated horizontally through exosomes. CEA, carcinoembryonic antigen; HLA, human leukocyte antigen; HER2, human epidermal growth factor receptor 2; TGF; transforming growth factor; TNF, tumor necrosis factor.
Discussion
Since cancer is fundamentally a dysregulation of gene expression, it is difficult to distinguish tumors which are morphologically similar but molecularly different by pathological assessment. For the earliest diagnosis, it is necessary to find non‐invasive cancer biomarkers to monitor molecular differences in tumors, which may assist in the selection of the best possible treatment for individual cancer patients. These cancer biomarkers include carcinoembryonic antigen (CEA), which is a commonly used marker of colon cancer, alpha‐fetoprotein (AFP), which is an associated marker for HCC, and the prostate‐specific antigen (PSA), a protein normally present at low levels in the blood of adult men that has a high association with prostate cancer. However, these cancer biomarkers are also elevated in benign conditions. For instance, prostate inflammation and benign prostatic hypertrophy result in increased PSA blood levels. Furthermore, blood levels of CEA and AFP are also elevated in benign diseases, such as cirrhosis, inflammatory bowel disorder, chronic lung disease and pancreatitis (CEA), and hepatitis and cirrhosis (AFP). Several current reports suggest that deregulation of miRNAs is tightly linked to cancer incidence and, in particular, that some miRNAs are closely associated with clinical prognosis. Therefore, it is anticipated that circulating miRNAs in plasma and/or serum may become novel methods for reducing both false‐positive and false‐negative results when undertaken by the conventional diagnostic method. Although the analysis of circulating miRNAs has just begun, the indications that such circulating miRNAs may have a biological role and may be involved in transforming cells suggest that these miRNAs may have potential as diagnostic, prognostic, and predictive biomarkers and may also be considered as therapeutic targets.
In this review, we have shown that circulating miRNAs are promising biomarkers for cancer diagnosis and prognosis; however, there have been conflicting findings about circulating miRNAs from the same tumor reported from different studies (Table 1). This might be due to the lack of an established endogenous miRNA control to normalize for circulating miRNA levels. In addition, a different extraction and quantification method among the studies also produced conflicting results. It is very difficult to determine which endogenous control is suitable for measuring circulating miRNAs because the expression profile of circulating RNA may change depending on the condition of the cancer patient, such as, for example, whether the patient is awaiting treatment, receiving chemotherapy, or post surgery. According to this view, it would be necessary to perform a well‐controlled analysis of circulating miRNAs in a large cohort of patients and healthy volunteers. These studies will provide further evidence in which miRNAs may be useful as serum biomarkers in clinical usage.
All the findings discussed in this report suggest that circulating miRNAs are promising as novel non‐invasive biomarkers useful for the elimination of false positives and false negatives of conventional various classifiers; however, the function of circulating miRNAs needs to be identified for the proper use of circulating miRNA biomarkers in evidence‐based medicine.
Acknowledgments
This work was supported by a Grant‐in‐Aid for the Third‐Term Comprehensive 10‐Year Strategy for Cancer Control, a Grant‐in‐Aid for Scientific Research on Priority Areas Cancer from the Ministry of Education, Culture, Sports, Science, and Technology, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio).
References
- 1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 1993; 75: 843–54. [DOI] [PubMed] [Google Scholar]
- 2. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 3. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10: 126–39. [DOI] [PubMed] [Google Scholar]
- 4. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10: 704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenfeld N, Aharonov R, Meiri E et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 2008; 26: 462–9. [DOI] [PubMed] [Google Scholar]
- 6. Lu J, Getz G, Miska EA et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–8. [DOI] [PubMed] [Google Scholar]
- 7. Lawrie CH, Gal S, Dunlop HM et al. Detection of elevated levels of tumour‐associated microRNAs in serum of patients with diffuse large B‐cell lymphoma. Br J Haematol 2008; 141: 672–5. [DOI] [PubMed] [Google Scholar]
- 8. Mitchell PS, Parkin RK, Kroh EM et al. Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci USA 2008; 105: 10513–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taylor DD, Gercel‐Taylor C. MicroRNA signatures of tumor‐derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008; 110: 13–21. [DOI] [PubMed] [Google Scholar]
- 10. Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real‐time PCR platform. Gynecol Oncol 2009; 112: 55–9. [DOI] [PubMed] [Google Scholar]
- 11. Hu Z, Chen X, Zhao Y et al. Serum microRNA signatures identified in a genome‐wide serum microRNA expression profiling predict survival of non‐small‐cell lung cancer. J Clin Oncol 2010; 28: 1721–6. [DOI] [PubMed] [Google Scholar]
- 12. Tanaka M, Oikawa K, Takanashi M et al. Down‐regulation of miR‐92 in human plasma is a novel marker for acute leukemia patients. PLoS ONE 2009; 4: e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg 2010; 251: 499–505. [DOI] [PubMed] [Google Scholar]
- 14. Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes 2009; 2: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsujiura M, Ichikawa D, Komatsu S et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 2010; 102: 1174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ho AS, Huang X, Cao H et al. Circulating miR‐210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol 2010; 3: 109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J, Chen J, Chang P et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood‐based biomarkers of disease. Cancer Prev Res (Phila Pa) 2009; 2: 807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR‐184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 2008; 14: 2588–92. [DOI] [PubMed] [Google Scholar]
- 19. Ng EK, Chong WW, Jin H et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009; 58: 1375–81. [DOI] [PubMed] [Google Scholar]
- 20. Yamamoto Y, Kosaka N, Tanaka M et al. MicroRNA‐500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009; 14: 529–38. [DOI] [PubMed] [Google Scholar]
- 21. Chen X, Ba Y, Ma L et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006. [DOI] [PubMed] [Google Scholar]
- 22. El‐Hefnawy T, Raja S, Kelly L et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem 2004; 50: 564–73. [DOI] [PubMed] [Google Scholar]
- 23. Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol 2009; 19: 43–51. [DOI] [PubMed] [Google Scholar]
- 24. Ghosh AK, Secreto CR, Knox TR, Ding W, Mukhopadhyay D, Kay NE. Circulating microvesicles in B‐cell chronic lymphocytic leukemia can stimulate marrow stromal cells: implications for disease progression. Blood 2010; 115: 1755–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lima LG, Chammas R, Monteiro RQ, Moreira ME, Barcinski MA. Tumor‐derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine‐dependent manner. Cancer Lett 2009; 283: 168–75. [DOI] [PubMed] [Google Scholar]
- 26. Skog J, Wurdinger T, Van Rijn S et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10: 1470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wolfers J, Lozier A, Raposo G et al. Tumor‐derived exosomes are a source of shared tumor rejection antigens for CTL cross‐priming. Nat Med 2001; 7: 297–303. [DOI] [PubMed] [Google Scholar]
- 28. Burden HP, Holmes CH, Persad R, Whittington K. Prostasomes‐‐their effects on human male reproduction and fertility. Hum Reprod Update 2006; 12: 283–92. [DOI] [PubMed] [Google Scholar]
- 29. Babiker AA, Nilsson B, Ronquist G, Carlsson L, Ekdahl KN. Transfer of functional prostasomal CD59 of metastatic prostatic cancer cell origin protects cells against complement attack. Prostate 2005; 62: 105–14. [DOI] [PubMed] [Google Scholar]
- 30. Holmgren L, Szeles A, Rajnavolgyi E et al. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood 1999; 93: 3956–63. [PubMed] [Google Scholar]
- 31. Zernecke A, Bidzhekov K, Noels H et al. Delivery of microRNA‐126 by apoptotic bodies induces CXCL12‐dependent vascular protection. Sci Signal 2009; 2: ra81. [DOI] [PubMed] [Google Scholar]
- 32. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654–9. [DOI] [PubMed] [Google Scholar]
- 33. Hunter MP, Ismail N, Zhang X et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008; 3: e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rabinowits G, Gercel‐Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009; 10: 42–6. [DOI] [PubMed] [Google Scholar]
- 35. Michael A, Bajracharya SD, Yuen PS et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis 2010; 16: 34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park NJ, Zhou H, Elashoff D et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 2009; 15: 5473–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune‐regulatory agent in breast milk. Silence 2010; 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rechavi O, Erlich Y, Amram H et al. Cell contact‐dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev 2009; 23: 1971–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pegtel DM, Cosmopoulos K, Thorley‐Lawson DA et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 2010; 107: 6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuan A, Farber EL, Rapoport AL et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS ONE 2009; 4: e4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010; 285: 17442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007; 450: 435–9. [DOI] [PubMed] [Google Scholar]
- 43. Tsang SY, Moore JC, Huizen RV, Chan CW, Li RA. Ectopic expression of systemic RNA interference defective protein in embryonic stem cells. Biochem Biophys Res Commun 2007; 357: 480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolfrum C, Shi S, Jayaprakash KN et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol 2007; 25: 1149–57. [DOI] [PubMed] [Google Scholar]
- 45. Al‐Nedawi K, Meehan B, Micallef J et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 2008; 10: 619–24. [DOI] [PubMed] [Google Scholar]


