Abstract
Chromosome 1 open reading frame 10 (C1orf10) is either down‐regulated or absent in esophageal squamous cell carcinoma (ESCC) tissues compared to its corresponding normal counterparts, and it is involved in heat shock and ethanol response and is expected to protect esophageal epithelium from damage. In the present study, we sequenced DNA samples from 32 individuals to search for genetic variants in the promoter region, coding region, and the untranslated region of C1orf10. Genotypes were analyzed in 991 patients and 984 controls. Odds ratios (ORs) and 95% confidence intervals (CIs) were estimated by logistic regression. Luciferase assays were carried out to find the functional SNPs. Six strongly linked single nucleotide polymorphisms (SNPs) spanning a region of 7 kb, –1747G/T, –1139G/C, –1079G/A, –900G/T, Gly480Ser, and 4666G/A were identified (D′= 1, r 2 = 1). Only one SNP –1139G/C was selected to analyze the association between C1orf10 genotypes and risk of ESCC. Subjects with the –1139CC genotype had a greater risk of developing ESCC compared with those with the –1139GG genotype (adjusted OR = 1.34; 95% CI, 1.02–1.76). There appears to be an interaction between the –1139G/C polymorphism and tobacco smoking that contributes to the risk for ESCC. However, we did not detect any obvious difference in reporter gene assay driven by each allele of C1orf10 promoter or 3′ UTR. These data showed that C1orf10 haplotypes containing –1747G/T, –1139G/C, –1079G/A, –900G/T, Gly480Ser, and 4666G/A are significantly associated with susceptibility to ESCC. (Cancer Sci 2009; 100: 1695–1700)
Environmental agents can influence tissue integrity, disease development, and related cancer development rate including those of the skin, breast, and gut.( 1 , 2 , 3 ) The cellular stress protein response system evolved and plays a key role in minimizing cell injury and maintaining tissue integrity in response to damaging levels of environmental agents.( 4 ) As such, the integrity of this system plays a role in modifying progression of diseases associated with aging, DNA or protein damage, and chronic injury. Cells of the normal human esophageal squamous epithelium are under relatively unique environmental stresses including bacterial infestation, viruses, thermal stresses, refluxed acid, oxidizing chemicals, and bile adducts, which contribute to tissue damage, promote aging, and initiate diseases.( 5 , 6 , 7 ) To defend these environmental stresses and maintain tissue integrity, a novel type of stress protein system has evolved in mammals.( 8 ) Chromosome 1 open reading frame 10 (C1orf10), a novel human gene identified by modified differential display PCR in 2000, is just one of the novel genes within the stress proteins.( 9 , 10 ) It maps to chromosome 1q21 which is commonly suppressed in ESCC by undefined general mechanism, and is suggested to be one member of the EDC.( 11 , 12 , 13 ) Besides, molecular characterization of C1orf10 revealed its role as a stress responsive protein, including its upregulated expression by stimulation of heat or acidic reagent,( 8 , 14 ) the activity as an anti‐apoptotic factor which attenuates the death induced by exposure of cells to normally lethal levels of DCA.( 15 ) As we know, esophageal cancer is caused by genetic and environmental factors, and stress proteins play important roles in the later process.( 7 ) As a member of the stress proteins in esophagus, the role of C1orf10 in stress response suggests that abnormalities of its expression may result in the abnormal response of esophageal epithelium to environmental stimulation which possibly leads to cancer. A previous study has shown that the expression of C1orf10 is either dramatically reduced or absent in esophageal cancer cell lines as well as primary esophageal cancer tissues compared with the corresponding normal esophageal mucosa.( 9 , 16 ) We then hypothesized whether the genetic variants of this gene are associated with ESCC.
Common variants which alter the amount of protein expression, for instance by affecting transcriptional regulation, or subtly alter the activity of protein itself, would be expected to have a quantitative influence on disease activity. In this study, we sought to identify new and functional polymorphisms in the promoter region, coding region, and the untranslated region of the C1orf10 gene. We further carried out a large scale case‐control study in the Han Chinese population to examine whether the haplotypes were associated with susceptibility to esophageal cancer.
Materials and Methods
Primer design for scanning the C1orf10 gene. Referring to the human C1orf10 sequence (GenBank accession no. AL135842), four overlapping PCR amplicons were designed to amplify the promoter sequence (2000 bp), whereas three PCR fragments were generated to scan the 5′ UTR, coding region, and 3′ UTR (Table 1).
Table 1.
Primers used to amplify regions of the human C1orf10 gene for DNA sequencing analysis
| Primer | Sequence (5′–3′) | Orientation | Ref. position | Amplification size | Region |
|---|---|---|---|---|---|
| P1 | CTTCAAACACCACCACAG | Forward | –2000 to –1983 | 782 | Promoter |
| P2 | TCTCCCTAGCAATCCTCT | Reverse | –1236 to –1219 | ||
| P3 | TTGGCTTCTGAGCTCAGAAAACTTC | Forward | –1485 to –1461 | 519 | Promoter |
| P4 | AGGCTTACCATGATCATGTGGTAAC | Reverse | –991 to –967 | ||
| P5 | TGCTGATTAGCCACCAGTTGAG | Forward | –1011 to –990 | 542 | Promoter |
| P6 | AGCAATTCCCTGCCTCAGACTC | Reverse | –490 to –469 | ||
| P7 | GGTCAGGAGTTCAAGATCAGC | Forward | –578 to –558 | 674 | Promoter 5′‐UTR Exon 1 |
| P8 | GCTCAGATCACCAAGACACTG | Reverse | +75 to +95 | ||
| P9 | GAAAGACCTAGCAGCAGACG | Forward | –97 (beginning of exon 2) | 361 | Exon 2 |
| P10 | AAATTCCCCAGGTCAAGGC | Reverse | +86 (beginning of intron 2) | ||
| P11 | GCTGCTGCTCTAGCCGTAT | Forward | –130 (beginning of exon 3) | 1100 | Exon 3 |
| P12 | CCTCCGTGGCTTACAGTTT | Reverse | +970 (beginning of exon 3) | ||
| P13 | AAGAACCAGCAAGCAGACAC | Forward | +633 (exon 3) | 778 | Exon 3 |
| P14 | TGCTCTTCAAAGGTAGGGAC | Reverse | +1800 (exon 3) |
Nucleotide positions are referenced relative to the major transcription start‐site of the human Chromosome 1 open reading frame 10 (C1orf10) sequence.
Scanning the C1orf10 gene. Genomic DNA was purified from a panel of 32 unrelated healthy subjects. PCR was carried out in the DNA Engine (Bio‐Rad, Hercules, CA, USA) with a 25‐µL volume containing about 50 ng genomic DNA, 6 pmol of each primer, 6 µmol of each dNTP, 1.5 mM Mg2+, and 1 U rTaq DNA polymerase with 1 × reaction buffer (Takara, Shiga, Japan). Thermal cycles were 95°C for 2 min, then 35 cycles of 95°C for 30 s, 61–70°C for 30 s, and 72°C for 45 s, followed by extension at 72°C for 7 min. The PCR products were analyzed by DNA sequencing.
Subjects for the case‐control study. A total of 991 patients with ESCC and 984 healthy controls were recruited in this study. All subjects were unrelated Han Chinese, living in Beijing and the surrounding regions. Patients with ESCC were consecutively recruited between 7 July 2000 and 20 December 2006, at the Cancer Hospital, Chinese Academy of Medical Sciences (Beijing, China). The pathological stage of ESCC at the time of diagnosis was classified by senior pathologists of the hospital on the basis of postoperative histopathological examination or biopsy according to the International Union Against Cancer (UICC) Tumor‐Node‐Metastasis classification. Tumor grade was classified into low (well differentiated), intermediate (moderately differentiated), and high (poorly differentiated) according to the World Health Organization grade classification. The control subjects were randomly selected from a pool of cancer‐free subjects recruited from a nutritional survey conducted in the same region during the same time period as the cases were collected. The selection criteria include no prior history of cancer and frequency matched to the cases by age (±5 years) and sex. At recruitment, informed consent was obtained from each subject, and each participant was then interviewed to collect detailed information on demographic characteristics and lifetime history of tobacco use. This study was approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Institute.
Genotype analysis. The C1orf10 genotypes at the –1139G/C site were determined by PCR‐based restriction fragment length polymorphism (PCR‐RFLP). The primers used were: –1139F, 5′‐GTC TCC GCT TCA CAG AGT GG‐3′; –1139R, 5′‐GCC ATC CCA GGG GTA AGT GC‐3′. PCR was performed with a 12.5‐µL mixture reaction containing 50 ng genomic DNA, 0.1 µM of each primer, 0.2 mM of dNTPs, 1.5 mM Mg2+, 4% DMSO, 4% 100 × BSA, and 0.5 U rTaq DNA polymerase with 1 × reaction buffer (Takara). The reaction was conducted under the following conditions: an initial melting step of 2 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 52°C, and 45 s at 72°C, and a final elongation of 7 min at 72°C. The 197 bp PCR products were digested with restriction enzyme Hha I (New England Biolabs, Ipswich, MA, USA) at 37°C overnight and separated on a 3% agarose gel (Fig. 1a) to determine the G/C polymorphism. Due to loss of the Hha I restriction site, the G/G allele produces only one fragment of 197 bp, whereas the C/C allele generates two fragments of 19 bp and 178 bp, and the G/C heterozygote has all of the three bands. Then, DNA sequencing was done to confirm the result revealed by PCR‐RFLP analysis (Fig. 1b). A 10% masked, random sample of cases and controls was tested twice by different persons and all results had 100% concordance.
Figure 1.
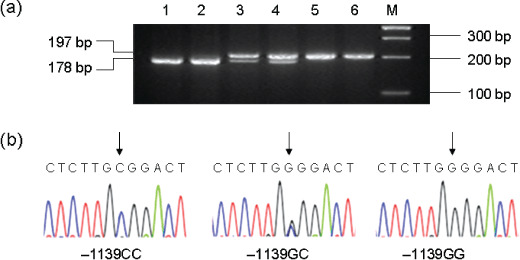
Analysis of the Chromosome 1 open reading frame 10 (C1orf10) –1139G/C polymorphism. (a) Representative gel picture showing PCR‐based restriction fragment length polymorphism (PCR‐RFLP) analysis of the C1orf10 genotypes in genomic DNAs of study subjects with the restriction enzyme Hha I. M, DNA size markers; subjects 1 and 2, –1139CC genotype; subjects 3 and 4, –1139GC genotype; subjects 5 and 6, –1139GG genotype. (b) Partial DNA sequence of three different allelic PCR products analyzed directly with an automatic sequencer showing a G to C transversion at the nucleotide location at which the arrow points.
Statistical analysis. χ2‐test or unpaired t‐test was used to examine differences in demographic variables, smoking, and distributions of genotypes between cases and controls. The associations between C1orf10 genotype and risk of the occurrence of ESCC were estimated by ORs and their 95% CIs, which were calculated by unconditional logistic regression models. The ORs were adjusted for age, sex, and pack‐years smoked. We tested the null hypotheses of multiplicative gene‐smoking interaction and evaluated departures from the multiplicative joint effect by including main effect variables and their product terms in the logistic regression model. A more‐than‐multiplicative interaction was suggested when OR 11 > OR 10 × OR 01. The homogeneity test was done to compare the difference between smoking‐related ORs among different genotypes or between the product of related ORs and joint effect OR. All analysis was carried out with Statistical Analysis System software (version 9.0; SAS Institute, Cary, NC, USA). Linkage disequilibrium coefficient and haplotype frequencies were estimated using Haploview and Haplo.stats software, respectively.
Construction of luciferase reporter plasmids. To distinguish the different transcriptional activity of two haplotypes in the C1orf10 promoter, we generated two reporter plasmids encompassing –2014 to + 32 bp of the C1orf10 gene. The reporter constructs were prepared by amplifying the promoter region from subjects homozygous for –1747G/T, –1139G/C, –1079G/A, –900G/T using –2014F, 5′‐GAA GCT AGC CCC ATC AGA AAA AGC TTC A‐3′ and + 32R, 5′‐GAA CTC GAG CCA GGT GGG ATG AAA CA‐3′. The PCR product was digested with Nhe I and Xho I and ligated into an appropriately digested pGL3‐Basic vector. Besides to test weather the SNP in the 3′ UTR influence the stability of mRNA, the 3′ UTR of the C1orf10 with 4666G/A were cloned into the Sac I/Xba I digested pIS0 vector (http://www.addgene.org/pgvec1?f=c&cmd=findpl&identifier=12178) respectively.( 17 ) The primers used were + 4658F, 5′‐GGC CGA GCT CCT TCC CCG ACT CCA ATG TC‐3′, 5′‐GGC CGA GCT CCT TCC CCA ACT CCA ATG TC–3′ and + 4990R, 5′‐GGC CTC TAG ATT GAT GCA TTA GGG TAG ATG‐3′. The PCR was in combination with Pyrobest DNA polymerase (Takara) to ensure high fidelity amplification, and complete DNA sequencing was then performed to confirm the integrity and authenticity of each construct.
Transient transfections and luciferase assays. Human ESCC cells EC9706 were cultured in RPMI‐1640 in a humidified, 5% CO2 incubator at 37°C. For transient transfection experiments, 5 × 103 cells were plated in 96‐multiwell plates and grown to 80–90% confluence. Then cells were co‐transfected with 200 ng reporter plasmid and 0.6 ng pRL‐CMV vector (Luciferase Assay System; Promega, Madison, WI, USA) by 0.5‐µL Lipofectamine 2000 per well. The pRL‐CMV vector was used as an internal control for transfection efficiency and the pGL3 Basic vector or pIS0 vector without an insert was used as a negative control. Six hours after transfection, the transfection reaction mixture was removed and cells were placed in complete medium with 10% FBS. Luciferase activity was determined using a Dual‐Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Fold increase was calculated by defining the activity of empty pGL3 Basic vector or pIS0 vector as 1. Differences were determined by t‐test, and P < 0.01 was considered significant.
Results
Identification of genetic variants in the C1orf10 gene in a Chinese population. Fourteen different primers were designed to screen the C1orf10 gene including promoter region, 5′ UTR, coding region, and 3′ UTR from a panel of 32 unrelated healthy Chinese individuals (providing at least a 95% confidence level to detect alleles with frequencies >5%). Four allelic variants, rs3753446 (–1747G/T), rs3753444 (–1139G/C), rs3753443 (–1079G/A), and rs4285700 (–900G/T) were detected in the promoter, one allelic variant rs3829868 (Gly480Ser) in exon 3, and one allelic variant rs10888486 (4666G/A) in the 3′ UTR (Fig. 2). All the six SNPs can be found in the National Center for Biotechnology Information (NCBI) SNP database and no novel variant was found in this study. We estimated the degree of linkage disequilibrium (LD) between SNPs as quantified by the disequilibrium coefficient D′ and r 2, which represent the proportion or representative of the maximum possible disequilibrium given observed allele frequencies. Complete linkage was observed among the six SNPs, with a D′ of 1 (r 2= 1). The frequencies of the G–1747‐G–1139‐ G–1079‐G–900‐Gly480‐G4666 and T–1747‐C–1139‐A–1079‐T–900‐Ser480‐A4666 haplotypes were 0.625 and 0.375, respectively.
Figure 2.
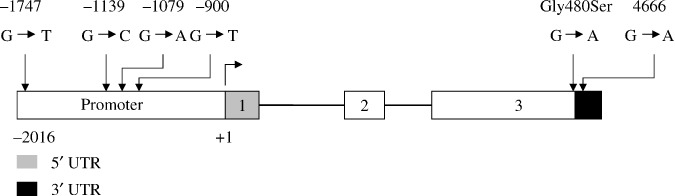
Genetic variants in the Chromosome 1 open reading frame 10 (C1orf10) gene. Summary of the location of single base substitutions identified in the promoter region, 5′ UTR, coding region, and 3′ UTR from a panel of 32 unrelated healthy Chinese Han individuals. Nucleotide positions are referenced relative to the major transcription start site, and amino acids are designated by the standard 1‐letter code.
Subject characteristics. The relevant characteristics of study subjects are shown in Table 2. The distributions of age and gender status between patients with ESCC and control subjects were not statistically different (P= 0.991 and 0.667, respectively), suggesting that the frequency matching was adequate. However, as expected, more smokers were presented among cases compared with controls (61.6%vs 37.6%; P < 0.001). In addition, cases had a higher value of pack‐years smoked than controls; 59.2% of smokers among cases smoked >22 pack‐years compared with 48.9% among controls (P = 0.002). Of the 991 patients, 98 (9.9%) had stage I, 209 (21.1%) had stage IIA, 126 (12.7%) had stage IIB, and 415 (41.9%) had stage III ESCC, whereas only 65 (6.6%) patients had stage IV disease. In total, 154 (15.5%) patients were classified into well‐differentiated ESCC (low grade), and 425 (42.9%) and 144 (14.5%) patients were classified into moderately differentiated (intermediate grade) or poorly differentiated (high grade) ESCC, respectively.
Table 2.
Distributions of select characteristics by case‐control status
| Variable | Cases (n = 991) No. (%) | Controls (n = 984) No. (%) | P‐values † |
|---|---|---|---|
| Sex | |||
| Male | 797 (80.4) | 784 (79.9) | 0.667 |
| Female | 194 (19.6) | 200 (20.3) | |
| Age (years) | 0.991 | ||
| ≤ 0 | 213 (21.5) | 217 (22.1) | |
| 51–60 | 355 (35.8) | 349 (35.5) | |
| 61–70 | 332 (33.5) | 330 (33.5) | |
| >70 | 91 (9.2) | 88 (8.9) | |
| Smoking status | <0.001 | ||
| Non‐smoker | 373 (37.6) | 546 (55.5) | |
| Smoker | 610 (61.6) | 370 (37.6) | |
| No data | 8 (0.8) | 68 (6.9) | |
| Smoking lever, pack‐years | 0.002 | ||
| ≤22 pack‐years | 249 (40.8) | 189 (51.1) | |
| >22 pack‐years | 361 (59.2) | 181 (48.9) | |
| Tumor stage ‡ | |||
| I | 98 (9.9) | ||
| IIA | 209 (21.1) | ||
| IIB | 126 (12.7) | ||
| III | 415 (41.9) | ||
| IV | 65 (6.6) | ||
| No data | 78 (7.8) | ||
| Tumor grade § | |||
| Low | 154 (15.5) | ||
| Intermediate | 425 (42.9) | ||
| High | 144 (14.5) | ||
| No data | 268 (27.1) | ||
χ2‐test for the difference between cases and controls.
According to the International Union Against Cancer (UICC) Tumor‐Node‐Metastasis classification.
According to the World Health Organization classification. Low, well‐differentiated carcinoma; intermediate, moderately differentiated carcinoma; high, poorly differentiated carcinoma.
The association between C1orf10 polymorphisms and risk of ESCC. Since close associations between the variations were observed throughout the entire C1orf10 gene, the region sequenced was analyzed as a single LD block for the haplotype inference. Only one SNP (rs3753444) was selected to analyze the association between C1orf10 genotypes and risk of ESCC. We considered that the genotype of this SNP could represent the other five SNPs. Table 3 shows allele frequencies and genotype distributions of C1orf10 –1139G/C in cases and controls. The allele frequencies for the G and C were found to be 0.590 and 0.410 among the control population, compared with 0.558 and 0.442 among cases, respectively. The genotype distribution among controls were 34.6% (GG), 48.9% (GC), and 16.5% (CC), which were not deviate from those expected from the Hardy–Weinberg equilibrium (P= 0.71). However, they are quite different from those (GG 31.5%, GC 48.6%, CC 19.9%) among patients. We noted that the –1139CC genotype was more prevalent in patients than in controls, and multivariate logistic regression analysis showed that subjects with the –1139CC genotype had increased risk of developing esophageal cancer compared with subjects with the –1139GG genotype (adjusted OR = 1.34; 95% CI, 1.02–1.76). However, the –1139GC genotype was not associated with increased risk (adjusted OR = 1.11; 95% CI, 0.90–1.37), suggesting a recessive effect of this SNP on esophageal cancer susceptibility.
Table 3.
Genotype distribution of C1orf10–1139G/C polymorphism between cases and controls and their association with ESCC
| Genotype | Cases (n = 991) No. (%) | Controls (n = 984) No. (%) | OR † (95% CI) |
|---|---|---|---|
| –1139 G/C | |||
| GG | 312 (31.5) | 340 (34.6) | 1 (ref) |
| GC | 482 (48.6) | 481 (48.9) | 1.11 (0.90–1.37) |
| CC | 197 (19.9) | 163 (16.5) | 1.34 (1.02–1.76) |
| C allele frequency | 0.442 | 0.410 | |
Data were calculated by unconditional logistic regression in comparison with the reference group carrying the GG genotype and adjusted for sex, age, and smoking status. C1orf10, Chromosome 1 open reading frame 10; CI, confidence interval; OR, odds ratio.
The risk of ESCC related to C1orf10 polymorphism was further examined separately by stratification by sex, age, and smoking status (Table 4). In the subgroup above 58 years, compared with subjects with the –1139GG genotype, subjects with the –1139CC genotype manifested significant association with substantially increased risk (OR = 1.70; 95% CI, 1.15–2.52). It was found that among non‐smokers, the adjusted OR of esophageal cancer for subjects carrying the –1139CC genotype was 1.28 (95% CI, 0.87–1.90). Among smokers, subjects carrying the –1139CC genotype had an OR of 1.48 (95% CI, 1.00–2.19). Furthermore, the risk of ESCC related to C1orf10 polymorphism was further examined separately by stratification by pack‐years (Table 5). Compared with non‐smokers carrying the –1139GG genotype, the risk for the presence of both smoking and the –1139CC genotype (OR = 4.63; 95% CI, 2.98–7.17) was greater than the produce of the OR for smoking (OR = 2.91; 95% CI 2.04–4.14) and the OR for the –1139CC genotype (OR = 1.28; 95% CI, 0.87–1.90), with statistical significance in the test for interaction (P= 0.049). These data suggested a potential multiplicative joint effect between smoking and the –1139CC genotype. Moreover, when the risk associated with the polymorphism was further valuated with smoking levels (≤22 and >22 pack‐years smoked), it increased consistently with cumulative smoking dose among smokers, in which a multiplicative joint effect between the susceptible genotype and categories of pack‐years smoked was also observed. As compared with the reference group, ORs (95% CI) of the –1139CC genotype for non‐smokers, smokers who smoked ≤22, and >22 pack‐years were 1.28 (0.87–1.90), 3.87 (2.20–6.83), and 5.29 (3.09–9.07), respectively (P < 0.001 for trend test). Thus, there appears to be an interaction between the –1139G/C polymorphism and tobacco smoking that contributes to the risk for ESCC in the population. However, no association was found between the C1orf10 polymorphism and the stage and grade of ESCC (data not shown).
Table 4.
Risk of esophageal cancer related to C1orf10–1139G/C genotypes by sex, age, and smoking status
| Characters | Genotype of C1orf10 –1139G/C (Number of cases/number of controls) | |||||||
|---|---|---|---|---|---|---|---|---|
| GG | OR (95% CI) | GC | OR † (95% CI) | P | CC | OR † (95% CI) | P | |
| Sex | ||||||||
| Male | 262/266 | 1 (ref) | 380/386 | 1.00 (0.80–1.28) | 0.971 | 155/132 | 1.24 (0.92–1.68) | 0.162 |
| Female | 50/74 | 1 (ref) | 102/95 | 1.75 (1.07–2.86) | 0.025 | 42/31 | 1.70 (0.92–3.17) | 0.093 |
| Age | ||||||||
| ≤58 years | 156/166 | 1 (ref) | 251/260 | 1.03 (0.77–1.38) | 0.840 | 84/89 | 1.02 (0.69–1.50) | 0.873 |
| >58 years | 156/174 | 1 (ref) | 231/221 | 1.19 (0.88–1.61) | 0.265 | 113/74 | 1.70 (1.15–2.52) ‡ | 0.007 |
| Smoking status | ||||||||
| Non‐smoker | 106/185 | 1 (ref) | 192/263 | 1.24 (0.91–1.69) | 0.167 | 75/98 | 1.28 (0.87–1.90) | 0.209 |
| Smoker | 203/133 | 1 (ref) | 287/182 | 1.05 (0.79–1.41) | 0.726 | 120/55 | 1.48 (1.00–2.19) § | 0.049 |
Data were calculated by unconditional logistic regression in comparison with the reference group carrying the GG genotype and adjusted for sex, age, and pack‐years within the strata.
P < 0.05, test for homogeneity between age‐related ORs among GG and CC genotype.
P < 0.05, test for homogeneity between smoking‐related ORs among GG and CC genotype.
C1orf10, Chromosome 1 open reading frame 10; CI, confidence interval; OR, odds ratio.
Table 5.
Risk of esophageal cancer related to C1orf10–1139G/C genotypes by smoking status and pack‐years smoked
| Characters | Genotype of C1orf10 –1139G/C (Number of cases/number of controls) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| GG | OR (95% CI) | P‐values | GC | OR † (95% CI) | P‐values | CC | OR † (95% CI) | P‐values | |
| Smoking status | |||||||||
| Non‐smoker | 106/185 | 1 (ref) | 192/263 | 1.24 (0.91–1.69) | 0.167 | 75/98 | 1.28 (0.87–1.90) | 0.209 | |
| Smoker | 203/133 | 2.91 (2.04–4.14) | <0.001 | 287/182 | 3.34 (2.36–4.72) | <0.001 | 120/55 | 4.63 (2.98–7.17) ‡ | <0.001 |
| =22 pack‐years | 93/72 | 2.54 (1.67–3.85) | <0.001 | 109/88 | 2.79 (1.84–4.24) | <0.001 | 47/29 | 3.87 (2.20–6.83) | <0.001 |
| >22 pack‐years | 110/61 | 3.48 (2.27–5.34) | <0.001 | 178/94 | 3.85 (2.61–5.67) | <0.001 | 73/26 | 5.29 (3.09–9.07) | <0.001 |
Data were calculated by unconditional logistic regression in comparison with the reference group of non‐smokers carrying the GG genotype, and adjusted for sex and age.
P < 0.05, test for homogeneity between smoking‐related ORs among the CC and GG genotype.
C1orf10, Chromosome 1 open reading frame 10; CI, confidence interval; OR, odds ratio.
Generation of reporter gene constructs to measure differences in allelic expression between C1orf10 promoter or 3′ UTR variants. To test whether the allelic variations in the promoter region influence the expression level of C1orf10, we generated reporter gene constructs in the context of the regulatory region containing the two haplotypes (Fig. 3a), and transiently transfected EC9706 cells using these allelic variations. No significant difference was observed in reporter gene expression driven by each haplotype of C1orf10 promoter. (Fig. 3b). We also examined the influence of 4666G/A on the stability of C1orf10 mRNA by construction of a reporter gene combining with C1orf10 3′ UTR (Fig. 4a). However, we did not detect any statistical difference (Fig. 4b). These results indicated that the four variations in the promoter region and 4666G/A in the 3′ UTR may not influence the C1orf10 expression level.
Figure 3.
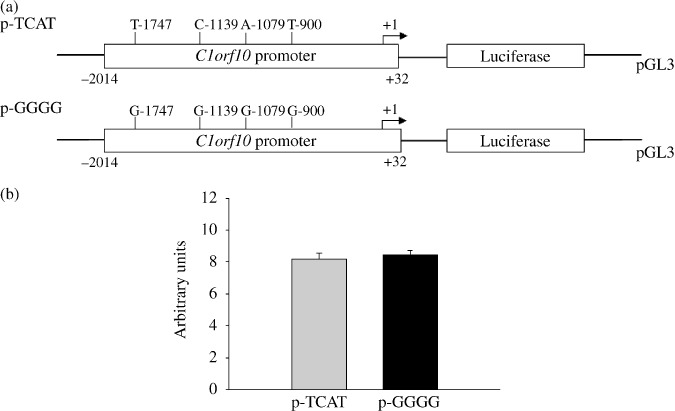
Transient reporter gene expression assays with constructs containing full‐length Chromosome 1 open reading frame 10 (C1orf10) C1orf10 promoter. (a), Schematic presentation of reporter gene constructs, used in transient transfections, containing a 2‐kb C1orf10 promoter with two haplotypes (T–1747‐C–1139‐A–1079‐T–900 and G–1747‐G–1139‐G–1079‐G–900). (b) EC9706 cells were transiently transfected as described. Fold increase was measured by defining the activity of the empty pGL‐3 Basic vector as 1. Data shown are mean fold increase ± SD from three independent transfection experiments, each done in triplicate.
Figure 4.
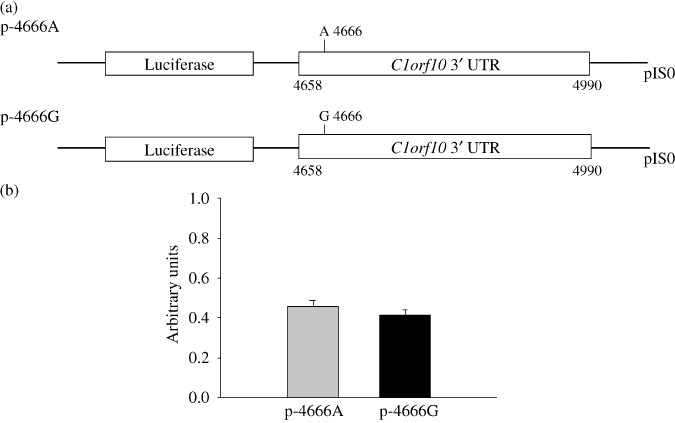
Transient reporter gene expression assays with constructs containing full‐length Chromosome 1 open reading frame 10 (C1orf10) 3′ UTR. (a) Schematic presentation of reporter gene constructs containing the C1orf10 3′ UTR with 4666 A/G. (b), EC9706 cells were transiently transfected as described. Luciferase reporter activity for EC9706 cells transfected with pIS0 C1orf10 3′ UTR expressed relative to pIS0. Data shown are mean ± SD from three independent transfection experiments, each done in triplicate.
Discussion
In the present study, we sought to identify functional SNPs in C1orf10 in a Chinese population and to investigate their association with the risk of developing ESCC. To summarize, by sequencing the full‐length of C1orf10 from a subset of 32 normal Han Chinese subjects, we found six SNPs, four of which (–1747G/T, –1139G/C, –1079G/A, and –900G/T) were in the promoter region, and two of which (Gly480Ser and 4666G/A) were in the exon 3 and 3′ UTR respectively. All the six SNPs have been deposited in the NCBI SNP database. Haplotype analysis showed that the six SNPs covering a region of 7 kb are in linkage disequilibrium. The results of case‐control analysis of 991 ESCC patients and 984 controls showed that the –1139CC genotype was associated with increased risk of ESCC. Moreover, we observed a potential multiplicative joint effect between the genotype and smoking, which was associated with even greater risk of ESCC. We observed a more than 4‐fold increased ESCC risk associated with the –1139CC genotype among smokers but not non‐smokers, suggesting a possible gene–environment interaction between the C1orf10 polymorphism and smoking in the etiology of ESCC in this Chinese population. In addition to tobacco smoking, other environmental factors such as alcohol consumption have been suggested to be associated with risk of ESCC, and this association might also be modulated by C1orf10 genotype. Unfortunately, the information on exposures other than smoking is not available in the present study, which prevents more comprehensive evaluation of the role of gene–environment interaction in ESCC development.
Based on these results and the previous observation of abnormalities in C1orf10 expression in ESCC, we speculated that the polymorphisms in the promoter region may affect the expression level of the gene; however, the two haplotypes did not show any significant difference by reporter assay. Moreover, the 4666G/A polymorphism in 3′ UTR had no difference on the transcription of reporter gene. Thus, the SNPs in promoter region and 3′ UTR may not contribute to susceptibility of ESCC by influencing expression level of C1orf10. Besides, no significant difference in C1orf10 mRNA expression levels was found when –1139G/C SNP (rs3753444) genotypes were compared by real‐time PCR analysis of C1orf10 mRNA in individual esophageal tissues (data not shown). We also noticed Gly480Ser, a SNP which may alter the activity of C1orf10. However, the function of C1orf10 is not clear so far, and not much information is available for discussing potential function of this SNP.
It is very clear that C1orf10 expression in esophageal cancer tissues and cell lines is much lower than that in the corresponding normal esophageal mucosa, whereas the precise function of C1orf10 in ESCC development remains unknown. C1orf10 is involved in heat shock response as one novel member of the tissue‐specific and atypical stress response system.( 8 ) Moreover, it might allow cells to tolerate normally lethal levels of DCA and protect from the toxic effect of bile acid as a survival factor.( 15 ) Subsequent studies have also identified C1orf10 as a candidate component of epithelial immunity based on its strong signature of adaptive evolution on DNA sequence of a type that is commonly associated with a coevolutionary arms race with a pathogen.( 10 ) Thus C1orf10 protein expression will presumably help maintain the barrier function in squamous epithelium in response to injury and function as a tumor suppressor. Our data of the case‐control analysis showed that the polymorphism of C1orf10 was associated with the risk of ESCC. It is possible that the genetic variants of C1orf10 change the expression level or activity of the gene, and then influence its function in ESCC development. It is also possible that the association between the C1orf10 polymorphism and ESCC might not really relate to the C1orf10 activity, but only to a secondary effect due to the linkage disequilibrium with a yet unidentified, but tightly linked, ESCC locus. In this way the polymorphism of C1orf10 might only serve as a haplotype tag. It is also possible that influence of several tightly linked polymorphisms within or close to the C1orf10 gene lead a combinatory contribution to the case‐control results. In fact, based on the HapMap CHB database (HapMap data, Rel 22/phase II Apr07, on NCBI B36 assembly, dbSNP b126; population: Han Chinese in Beijing, China; MAF are >0.05) in region chr1: 149,129,663.149,214,350, we find that all SNPs reported in this article are located in a region (84 kb) of high LD which contain two known genes: C1orf10 and IFPS. Thus, further studies would be warranted to address these possibilities.
In conclusion, we have found six SNPs that lie in the C1orf10 gene in a Han Chinese population. The six SNPs were strongly linked, spanning a region of 7 kb, and they demonstrated an increased risk of ESCC associated with the –1139CC genotype. These findings suggest a role for genetic determinants of stress protein in the development of esophageal cancer in a Chinese population. Since this is the first report demonstrating the contribution of C1orf10 polymorphism to ESCC, additional studies on ESCC would be warranted in different ethnic populations.
Abbreviations
| C1orf10 | Chromosome 1 open reading frame 10 |
| CI | Confidence interval |
| DCA | Deoxycholic acid |
| EDC | Epidermal differentiation complex |
| ESCC | Esophageal squamous cell carcinoma |
| OR | Odds ratio |
| SNP | Single nucleotide polymorphism |
Disclosure Statement
None.
Acknowledgments
This work was supported by National 863 High Tech Project of China (no. 2006AA02Z467), National Natural Science Foundation of China (no. 30772525), and Program for New Century Excellent Talents in University (no. NCET‐05‐0171). We would like to thank Dr David P. Bartel of the Whitehead Institute for Biomedical Research for the gift of pIS0 vector.
References
- 1. Dixon AJ, Dixon BF. Ultraviolet radiation from welding and possible risk of skin and ocular malignancy. Med J Aust 2004; 181: 155–7. [DOI] [PubMed] [Google Scholar]
- 2. Brody JG, Rudel RA, Michels KB et al . Environmental pollutants, diet, physical activity, body size, and breast cancer: where do we stand in research to identify opportunities for prevention? Cancer 2007; 109: 2627–34. [DOI] [PubMed] [Google Scholar]
- 3. Waterland RA. Epigenetic mechanisms and gastrointestinal development. J Pediatr 2006; 149: S137–42. [DOI] [PubMed] [Google Scholar]
- 4. Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene 2004; 23: 2907–18. [DOI] [PubMed] [Google Scholar]
- 5. Suzuki H, Kajiwara M, Miura S et al . Ethanol‐induced injury and GI bleeding. Nippon Rinsho 1998; 56: 2269–75. [PubMed] [Google Scholar]
- 6. Bulger EM, Helton WS. Nutrient antioxidants in gastrointestinal diseases. Gastroenterol Clin North Am 1998; 27: 403–19. [DOI] [PubMed] [Google Scholar]
- 7. Goldstein JL, Watkins JL, Greager JA et al . The esophageal mucosal resistance: structure and function of an unique gastrointestinal epithelial barrier. J Lab Clin Med 1994; 123: 653–9. [PubMed] [Google Scholar]
- 8. Yagui‐Beltran A, Craig AL, Lawrie L et al . The human oesophageal squamous epithelium exhibits a novel type of heat shock protein response. Eur J Biochem 2001; 268: 5343–55. [DOI] [PubMed] [Google Scholar]
- 9. Xu ZX, Wang MR, Xu X et al . Novel human esophagus‐specific gene C1orf10: cDNA cloning, gene structure, and frequent loss of expression in esophageal cancer. Genomics 2000; 69: 322–30. [DOI] [PubMed] [Google Scholar]
- 10. Little TJ, Nelson L, Hupp T. Adaptive evolution of a stress response protein. PLoS ONE 2007; 2: e1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Contzler R, Favre B, Huber M et al . A new member of the ‘fused gene’ family, is expressed during epidermal differentiation. J Invest Dermatol 2005; 124: 990–7. [DOI] [PubMed] [Google Scholar]
- 12. Mischke D, Korge BP, Marenholz I et al . Genes encoding structural proteins of epidermal cornification and S100 calcium‐binding proteins form a gene complex (‘epidermal differentiation complex’) on human chromosome 1q21. J Invest Dermatol 1996; 106: 989–92. [DOI] [PubMed] [Google Scholar]
- 13. Koon N, Zaika A, Moskaluk CA et al . Clustering of molecular alterations in gastroesophageal carcinomas. Neoplasia 2004; 6: 143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nelson L, Anderson S, Archibald AL et al . An animal model to evaluate the function and regulation of the adaptively evolving stress protein SEP53 in oesophageal bile damage responses. Cell Stress Chaperones 2008; 13: 375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darragh J, Hunter M, Pohler E et al . The calcium‐binding domain of the stress protein SEP53 is required for survival in response to deoxycholic acid‐mediated injury. FEBS 2006; 273: 1930–47. [DOI] [PubMed] [Google Scholar]
- 16. Luo A, Kong J, Hu G et al . Discovery of Ca2+‐relevant and differentiation‐associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene 2004; 23: 1291–9. [DOI] [PubMed] [Google Scholar]
- 17. Yekta S, Shih IH, Bartel DP. MicroRNA‐directed cleavage of HOXB8 mRNA. Science 2004; 304: 594–6. [DOI] [PubMed] [Google Scholar]


