Abstract
The hedgehog (Hh) signaling pathway has been reported to be associated with the growth of pancreatic cancer, but its role in the invasive phenotype is poorly understood. Therefore, we investigated the role of the Hh pathway in pancreatic cancer cell invasiveness using a Matrigel invasion assay. Blockade of the Hh pathway by cyclopamine inhibited pancreatic cancer cell invasion in association with a decreased expression of matrix metalloproteinase (MMP)‐9. By contrast, activation of the Hh pathway by the addition of exogenous Sonic hedgehog increased cell invasion and MMP‐9 expression. Stable transfection of pancreatic cancer cells with Gli1 increased their invasiveness, which was associated with activation of MMP‐9. We also showed that inhibition of MMP‐9 by small interfering RNA blocked the increased invasiveness of Gli1‐transfected cells. Furthermore, inhibition of Gli1 by small interfering RNA suppressed the invasiveness and MMP‐9 expression of pancreatic cancer cells. Taken together, these findings suggest that members of the Hh pathway, especially Gli1, play an important role in the invasiveness of pancreatic cancer cells through the regulation of MMP‐9 expression. (Cancer Sci 2008; 99: 1377–1384)
Abbreviations:
- Dhh
Desert hedgehog
- GFP
green fluorescent protein
- Hh
hedgehog
- Ihh
Indian hedgehog
- Ig
immunoglobulin
- IL
interleukin
- MAPK
mitogen‐activated protein kinase
- MMP
matrix metalloproteinase
- Ptch
Patched
- rhShh
recombinant human sonic hedgehog
- RNAi
RNA interference
- RT‐PCR
reverse transcription–polymerase chain reaction
- SDS‐PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- Shh
sonic hedgehog
- siGli1
small interfering RNA targeting Gli1
- siRNA
small interfering RNA
- Smo
smoothened
Pancreatic cancer is one of the most lethal malignancies that has yet to be successfully controlled with therapy. Although various therapies such as surgery, chemotherapy, and radiation therapy have been carried out, few patients survive for 5 years.( 1 , 2 ) One reason for this lethality is its invasive and metastatic character. A better understanding of the mechanisms that underlie the development of pancreatic cancer would help to identify novel molecular targets for treatment.
The Hh signaling pathway is crucial to growth and patterning in a wide variety of tissues, including the pancreas, during embryonic development.( 3 , 4 , 5 , 6 ) Of three Hh ligands, Shh, Ihh, Dhh, the former is reported to play an essential role in the development of pancreatic cancer as well as pancreatic organogenesis. The response to Shh is mediated by two transmembrane proteins, Smo and Ptch, and by downstream transcription factors that are members of the Gli family. Three Gli genes have been identified. Gli2 and Gli3 have distinct context‐dependent repressor and activator functions.( 7 , 8 , 9 ) By contrast, Gli1 is an activator of target genes, and is itself a transcriptional target of the Hh pathway.( 10 , 11 , 12 ) Recent studies have reported an association between Hh pathway activation and initiation of human tumors.( 6 ) Recent studies have also shown cell autonomous ligand‐dependent activation of the Hh pathway in small cell lung cancer and carcinomas of the esophagus, stomach, biliary tract, and pancreas.( 13 , 14 , 15 ) It has been also shown that cyclopamine, a Smo antagonist, suppresses the growth of pancreatic cancer.( 14 , 15 ) These findings suggest that the Hh pathway could be a viable therapeutic target for the treatment of pancreatic cancer.
In the present study, we analyzed the relationship between Hh pathway activation, especially Gli1, and the invasive ability of pancreatic cancer cells.
Materials and Methods
Cell culture. Three human pancreatic ductal adenocarcinoma cell lines (AsPC‐1, SUIT‐2, and CFPAC‐1) were maintained in RPMI‐1640 (Life Technologies, Grand Island, NY) supplemented with 10% fetal calf serum (Life Technologies), at 37°C in 5% CO2.
Reagents and antibodies. Cyclopamine, purchased from Toronto Research Chemicals (Toronto, Canada), was diluted in 100% methanol. Mouse anti‐Shh blocking antibody (5E1), developed by Thomas M. Jessell, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health & Human Development and maintained by The University of Iowa, Department of Biological Sciences (Iowa City, IA). Control mouse IgG was purchased from Sigma‐Aldrich (St Louis, MO). rhShh NH2‐terminal peptide was purchased from R&D Systems (Minneapolis, MN). Recombinant human IL‐1β was purchased from Sigma‐Aldrich. Anti‐Gli1 (sc‐6153) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti‐MMP‐9 (#3852) was purchased from Cell Signaling Technology (Danvers, MA).
Matrigel invasion assay and migration assay. The invasiveness of pancreatic cancer cells was assessed based on the invasion of cells through Matrigel‐coated transwell inserts.( 16 ) In brief, the upper surface of a filter (pore size, 8.0 µm; BD Biosciences, Heidelberg, Germany) was coated with basement membrane Matrigel (BD Biosciences). Cells were suspended in RPMI‐1640 with 10% fetal calf serum containing the desired dose of reagents. Then 0.8 × 105 cells were added to the upper chamber and incubated for 16 h. After incubation, the filter was fixed and stained with Diff‐Quik reagent (International Reagents, Kobe, Japan). Cells that had migrated from the upper to the lower side of the filter were counted under a light microscope (BX50; Olympus, Tokyo, Japan) at a magnification of ×100. Tumor cell invasiveness was defined as the total number of invaded cells. Each experiment was carried out in triplicate wells.
The migration ability of pancreatic cancer cells was assessed in the same way as described above, but non‐Matrigel‐coated transwell inserts were used.
Proliferation assay. To examine the proliferation of pancreatic cancer cells, we carried out a 3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide assay.( 17 ) Cells were incubated under the same conditions as the Matrigel invasion assay, and their proliferation was assessed.
Real‐time RT‐PCR. For real‐time RT‐PCR, RNA (1 µg) was treated with DNase and reverse transcribed to cDNA using the QuantiTect Reverse Transcription kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Reactions were run with an iQ SYBR Green Supermix (Bio‐Rad Laboratories, Hercules, CA) on a DNA Engine Opticon 2 System (MJ Research, Waltham, MA). The amount of each target gene in a given sample was normalized to the level of β‐actin in that sample.
Gelatin zymography. Gelatinase secretion of cell culture supernatants was determined by SDS‐PAGE zymography using gelatin as a substrate for MMP‐2 and MMP‐9, as described by Sehgal et al.( 18 ) In brief, tumor cells were seeded at 5 × 105 cells/6‐well plate. After the cells reached 80% confluence, the medium was aspirated and 4 mL of fresh serum‐free RPMI‐1640 containing the desired dose of reagents was added to each well. Supernatants were collected after a 24‐h incubation and concentrated to 50 µL using a membrane dialysis concentrator (Millipore, Bedford, MA). The same volume of concentrated supernatants was mixed with SDS sample buffer without prior denaturation and run on 10% SDS‐PAGE gels containing 1 mg/mL gelatin. Gelatin Zymo MMP Marker (active‐MMP‐2, pro‐MMP‐2,‐9; Life Laboratory Company, Yamagata, Japan) was used as a positive control. After electrophoresis, the gels were washed and stained in a solution of 0.25% Coomassie Brilliant Blue G‐250 and destained with acetic acid and methanol. The gels were scanned using a GT‐X800 scanner (Epson, Tokyo, Japan) and semiquantitative analysis was carried out with ImageJ version 1.33 software.
Transient transfection. pIRES2‐hSHH‐EGFP (referred to as pSHH‐GFP) was kindly provided by Dr Aubie Shaw (Division of Urology, Department of Surgery, University of Wisconsin, MA( 19 )). Cells seeded in 6‐well plates were transfected with 2 µg of pGFP or pSHH‐GFP using TransFast reagent (Promega, Madison, WI) according to the manufacturer's instructions. Transfected cells were used for experiments 24 h after transfection.
Transfection and establishment of stable transfectants. CFPAC‐1 cells were resuspended in Nucleofector Solution (Amaxa Biosystems, Cologne, Germany) followed by addition of 2.5 µg pcDNA3‐Gli1 or pcDNA3 (a kind gift of Dr Y Sasaki, Riken, Kobe, Japan( 7 )). Cells were transfected by electroporation with a Nucleofector System (Amaxa Biosystems). G418 (Invitrogen, Carlsbad, CA) selection was started 24 h after transfection and maintained for 10–21 days; individual clones derived from G418‐resistant single cells were pooled and grown for Gli1 mRNA expression analyses. To confirm the levels of Gli1 mRNA in the G418‐resistant clones (pcDNA3‐Gli1‐ and pcDNA3‐transfected clones), real‐time RT‐PCR was carried out as described previously,( 20 ) pcDNA3‐Gli1 clones expressed 17.4‐fold higher levels of Gli1 mRNA than did AsPC‐1 cells, which constitutively expressed Gli1 mRNA. No clones obtained due to transfection with pcDNA3 expressed detectable levels of Gli1 mRNA (data not shown).
RNA interference. siRNA for MMP‐9 (ON‐TARGETplus SMART pool, L‐005970), siRNA for Gli1 (ON‐TARGETplus SMART pool, L‐003896) and negative control siRNA (ON‐TARGETplus siCONTROL non‐targeting pool, D‐001810) were purchased from Dharmacon RNA Technologies (Chicago, IL). Cells seeded in 6‐well plates were transfected with 100 nM siRNA using Lipofectamine RNAiMAX Reagent (Invitrogen) according to the manufacturer's instructions. Cells were used for experiments at the indicated hours after transfection.
Immunocytochemistry. Pancreatic cancer cells (5 × 105/well) were seeded onto pre‐underlaid poly L‐lysine‐coated cover glass (Asahi Techno Glass, Chiba, Japan) in 24‐well plates, and were incubated overnight in 10% fetal bovine serum–RPMI. Immunostaining was carried out as previously described with some modification.( 21 ) In brief, cells were fixed in 4% paraformaldehyde followed by permeabilization with 0.2% Triton X‐100, then incubated with primary followed by secondary antibodies. Cells were counterstained with 4′,6′‐diamidino‐2‐phenylindole dihydrochloride (Sigma‐Aldrich). After mounting in Vectorshield Mounting Medium (Vector Laboratories, Burlingame, CA), samples were examined by fluorescence microscopy (Axio Imager.A1; Carl Zeiss, Oberkochen, Germany). Antibodies and dilutions used were as follows: rabbit anti‐MMP‐9 (1:100); goat anti‐Gli1 (1:100); Alexa Fluor 488 chicken antirabbit IgG (1:400; Invitrogen); and Alexa Fluor 594 donkey antigoat IgG (1:400; Invitrogen).
Statistical analysis. Mann–Whitney's U‐test was used for statistical analysis. All results with a P < 0.05 were considered statistically significant.
Results
Inhibition of the Hh pathway suppressed the invasive ability of pancreatic cancer cells. We used two human pancreatic cancer cell lines, AsPC‐1 and SUIT‐2, in which constitutive activation of the Hh pathway has been confirmed.( 22 ) Cyclopamine (10 µM) significantly suppressed Gli1 mRNA expression, indicating inhibition of the Hh pathway, because Gli1 is a transactivator of target genes and is itself a transcriptional target of the Hh pathway( 10 , 11 , 12 ) (Fig. 1a). When these cells were cultured with cyclopamine of various concentrations (5–10 µM) for 16 h, cyclopamine below 10 µM did not affect the proliferation of these cells (Fig. 1b). Cyclopamine (10 µM) did not affect the migration ability of either cell line (data not shown). However, the number of cells that invaded from the upper to the lower chamber through the Matrigel‐coated filter was significantly decreased in a dose‐dependent manner (Fig. 1c). We also confirmed the ligand dependency of Hh‐related invasive ability using an anti‐Shh blocking antibody (5E1). When incubated with 5 µg/mL 5E1, the number of invaded cells was significantly decreased (Fig. 1d), whereas the addition of 5E1 did not affect the proliferation of these cells during the Matrigel invasion assay (data not shown). These results suggest that Hh pathway inhibition suppresses the invasive ability of these cells in a ligand‐dependent manner, independently of any inhibition of cell proliferation.
Figure 1.

Inhibition of the hedgehog pathway by cyclopamine (Cyc) in AsPC‐1 and SUIT‐2 human pancreatic ductal adenocarcinoma cells. (a) AsPC‐1 and SUIT‐2 cells were treated with 10 µM Cyc for 12 h and relative Gli1 mRNA expression was examined by real‐time reverse transcription–polymerase chain reaction. Gli1 mRNA expression was normalized to β‐actin mRNA expression. (b) After incubation with the indicated dose of Cyc for 16 h, cell viability was measured using a 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide assay detected at an absorbance at 570 nm. (c, d) AsPC‐1 and SUIT‐2 cells were seeded into a Matrigel‐coated invasion chamber with 5 or 10 µM Cyc (c) or with 5 µg/mL anti‐sonic hedgehog antibody (5E1) or mouse immunoglobulin G (as a negative control) (d), and incubated for 16 h. The number of cells was counted under a light microscope. Magnification, × 100; scale bars, 100 µm; columns, means of three independent experiments; bars, SD. +, addition of cyclopamine (a,c) or 5E1(d); –, no addition of cyclopamine (a,c) or 5E1(d). *P < 0.05; **P < 0.01. n.s., not significant.
Activation of the Hh pathway enhanced the invasive ability of pancreatic cancer cells. First, both cell lines were transfected with pSHH‐GFP to overexpress Shh or pGFP as a control, and the Gli1 mRNA level was monitored by real‐time RT‐PCR. The transfection efficiency was over 70% in both cell lines (data not shown). Real‐time RT‐PCR revealed that cells transfected with pSHH‐GFP expressed higher levels of Gli1 mRNA than cells transfected with pGFP (Fig. 2a, left). pSHH‐GFP transfection did not affect the proliferation of these cells during the 16‐h cultivation period (data not shown). The Matrigel invasion assay showed that the number of pSHH‐GFP transfected cells invading through the Matrigel‐coated filter was much larger than the number of invading pGFP transfected cells (Fig. 2a, right).
Figure 2.
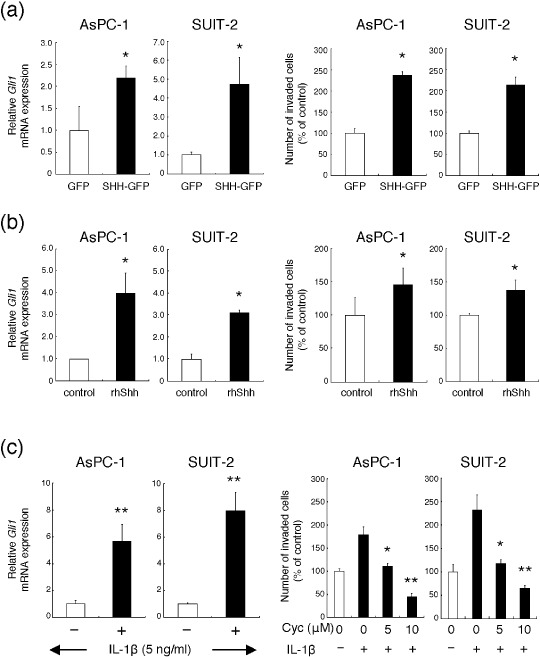
Activation of the hedgehog pathway enhances the invasive ability of AsPC‐1 and SUIT‐2 human pancreatic ductal adenocarcinoma cells. (a) AsPC‐1 and SUIT‐2 cells were transfected with pIRES2‐hSHH‐EGFP (pSHH‐GFP) or pGFP, then expression of Gli1 mRNA relative to that of β‐actin mRNA was assessed by real‐time reverse transcription–polymerase chain reaction (left). After transfection, the cells were seeded into a Matrigel‐coated invasion chamber and incubated for 16 h. Quantitation of the assay is shown in the graph (right). (b) AsPC‐1 and SUIT‐2 cells were treated with 0.4 µg/mL recombinant human sonic hedgehog NH2‐terminal peptide (rhShh) for 4 h. Expression of Gli1 mRNA was assessed (left). Cells were seeded into a Matrigel‐coated invasion chamber with 0.4 µg/mL rhShh and incubated for 16 h (right). (c) AsPC‐1 and SUIT‐2 cells were treated with 5 ng/mL interleukin‐1β (IL‐1β) for 6 h, then expression of Gli1 was assessed (left). Cells were seeded into a Matrigel‐coated invasion chamber with 5 ng/mL IL‐1β and 5 or 10 µM cyclopamine (Cyc) for 16 h (right). Columns, means of three independent experiments; bars, SD. +, addition of IL‐1β; –, no addition of IL‐1β. *P < 0.05; **P < 0.01.
Second, both cell lines were co‐cultured with rhShh to activate the Hh pathway during the Matrigel invasion assay. The expression of Gli1 mRNA was significantly increased by rhShh (Fig. 2b, left). rhShh did not affect proliferation of these cells during the Matrigel invasion assay (data not shown), but cells in which the Hh pathway was activated by rhShh invaded through the Matrigel‐coated filter more than control cells (Fig. 2b, right).
Finally, we used our IL‐1β–Hh pathway activation system reported previously.( 22 ) In brief, when both types of cancer cell were treated with 5 ng/mL IL‐1β, the Hh pathway was activated through increased Shh induction. Activation of the Hh pathway by IL‐1β was verified by the detection of increased expression of Gli1 mRNA using real‐time RT‐PCR (Fig. 2c, left). Although 5 ng/mL IL‐1β did not significantly affect the proliferation of these cells during a 16‐h incubation period (data not shown), IL‐1β increased the number of cells that invaded to the lower chamber, and cyclopamine decreased the increased number of invaded cells in a dose‐dependent manner in both cell lines (Fig. 2c, right).
These results confirmed that activation of the Hh pathway increases the invasive ability of pancreatic cancer cells in a ligand‐dependent manner, independently of an enhancement of proliferation.
Hh pathway regulation affected MMP‐9 expression of pancreatic cancer cells. These results strongly suggest that the Hh pathway contributes to the degradation of Matrigel, which includes type IV collagen. Therefore, we focused on the relationship between Hh pathway activity and MMP‐2 and MMP‐9 expression.
We carried out gelatin zymography to evaluate the enzymatic activity of MMPs. Inhibition of the Hh pathway by cyclopamine suppressed the enzymatic activity of both pro‐ and active‐MMP‐9 in both cell lines (Fig. 3a). However, we could find only pro‐MMP‐2, not active‐MMP‐2. Moreover, the expression of pro‐MMP‐2 was not suppressed significantly by cyclopamine. Then we carried out real‐time RT‐PCR to evaluate MMP‐9 mRNA expression. Inhibition of the Hh pathway by cyclopamine also suppressed MMP‐9 mRNA expression in both AsPC‐1 and SUIT‐2 cells (Fig. 3b).
Figure 3.

Regulation of hedgehog pathway activation affects matrix metalloproteinase (MMP)‐9 expression in AsPC‐1 and SUIT‐2 human pancreatic ductal adenocarcinoma cells. (a) AsPC‐1 and SUIT‐2 cells were incubated with 10 µM cyclopamine for 24 h in serum‐free medium, and MMP‐2 and MMP‐9 enzymatic activity was examined by gelatin zymography. (b) Cells were incubated with 10 µM cyclopamine for 12 h, then expression of MMP‐9 mRNA relative to that of β‐actin mRNA was assessed by real‐time reverse transcription–polymerase chain reaction. (c) Cells were incubated with 0.4 µg/mL rhShh for 24 h in serum‐free medium, and MMP‐2 and MMP‐9 enzymatic activity was examined. (d) Cells were incubated with 0.4 µg/mL recombinant human sonic hedgehog NH2‐terminal peptide (rhShh) for 12 h, then expression of MMP‐9 mRNA was examined. Band intensities of MMPs were quantified with ImageJ version 1.33 software. The control band intensity is indicated as 1.0. Marker indicates pro‐MMP‐9 and active‐MMP‐2. Columns, means of three independent experiments; bars, SD. n.s., not significant; *P < 0.05.
By contrast, activation of the Hh pathway by rhShh increased the enzymatic activity of both pro‐MMP‐9 and active‐MMP‐9 (Fig. 3c) in both cell lines. Here again, we could not find the active form of MMP‐2, and the expression of pro‐MMP‐2 was not increased by rhShh. Real time RT‐PCR showed that MMP‐9 mRNA expression was increased by rhShh (Fig. 3d). These results suggest a contribution of MMP‐9 to the Hh pathway‐related invasive ability of these cells.
Stable Gli1‐expressing clones showed increased invasive ability mediated by MMP‐9 activation. To better clarify the relationship between the Hh pathway, invasion and MMP‐9 expression, we established stable Gli1‐expressing clones using a human pancreatic cancer cell line (CFPAC‐1) that has not been reported to express Shh mRNA,( 20 ) as described in the Materials and Methods section. We used a clone (referred to as pcDNA‐Gli1 cells) expressing 17.4‐fold higher levels of Gli1 mRNA than AsPC‐1 cells. No clones obtained by transfection with pcDNA3 (referred to as pcDNA cells) expressed detectable levels of Gli1 mRNA. No significant difference in cell proliferation was observed between pcDNA‐Gli1 cells and pcDNA cells during the 16‐h cultivation period (data not shown). In the Matrigel invasion assay, the number of invaded pcDNA‐Gli1 cells was 4.2‐fold higher than that of control pcDNA cells (Fig. 4a). On that occasion, zymography showed that both pro‐MMP‐9 and active‐MMP‐9 enzymatic activity of pcDNA‐Gli1 cells were higher than that of pcDNA cells (Fig. 4b). Active‐MMP‐2 was not detected in CFPAC1 cells, and the expression of pro‐MMP‐2 was not significantly increased in pcDNA‐Gli1 cells. Real time RT‐PCR showed that MMP‐9 mRNA expression in pcDNA‐Gli1 cells was significantly higher than that in pcDNA cells (Fig. 4c).
Figure 4.

CFPAC‐1 human pancreatic ductal adenocarcinoma cells transfected with Gli1 showed increased invasive ability. (a) CFPAC‐1 cells were transfected with pcDNA‐Gli1 or pcDNA, and stable transfectants were established. These clones were seeded into a Matrigel‐coated chamber for 16 h. The number of cells was counted under a light microscope (top). Magnification: ×100; scale bars, 100 µm. Quantitation of the assay is shown in the graph (bottom). (b) Both groups of transfected cells were incubated in serum‐free medium, and the enzymatic activity of matrix metalloproteinase (MMP)‐2 and MMP‐9 was examined by gelatin zymography. Band intensities of MMP were quantified with ImageJ version 1.33 software. The control band intensity is indicated as 1.0. Marker indicates pro‐MMP‐9 and active‐MMP‐2. (c) Expression of MMP‐9 mRNA relative to that of β‐actin mRNA was assessed by real‐time reverse transcription–polymerase chain reaction. (d) pcDNA‐Gli1‐ or pcDNA‐transfected cells were transfected with small interfering RNA (siRNA) for MMP‐9 (siMMP‐9) or control siRNA. After transfection, the expression of MMP‐9 mRNA was assessed. (e) After transfection, the expression of MMP‐9 protein was assessed by immunocytochemistry. MMP‐9, green; 4′,6′‐diamidino‐2‐phenylindole dihydrochloride (DAPI), blue. Magnification, ×400; scale bars, 20 µm. (f) These cells were seeded into a Matrigel‐coated invasion chamber for 16 h. Columns, means of three independent experiments; bars, SD. *P < 0.05; **P < 0.01.
RNAi for MMP‐9 suppressed the increased invasive ability of pcDNA‐Gli1 cells. To confirm the role of MMP‐9 in the Hh pathway‐related invasive ability of pancreatic cancer cells, we knocked down MMP‐9 expression in pcDNA‐Gli1 cells and pcDNA cells by RNAi. Real‐time RT‐PCR showed that transfection of cells with siRNA targeting MMP‐9 (siMMP‐9) resulted in a 90% or greater knockdown of MMP‐9 mRNA expression (Fig. 4d). Immunocytochemistry showed that siMMP‐9 treatment decreased the MMP‐9 protein expression (Fig. 4e). We next carried out a Matrigel invasion assay using these siMMP‐9‐transfected cells. The number of siMMP‐9 transfected cells that migrated through the Matrigel‐coated filter was reduced by 70% in pcDNA‐Gli1 cells and by 25% in pcDNA cells (Fig. 4f). These results showed that inhibition of MMP‐9 blocked the increased invasiveness of pcDNA‐Gli1‐transfected cells in the Matrigel invasion assay.
RNAi for Gli1 suppressed the invasive ability of pancreatic cancer cells. We next focused on the role of Gli1, an activator of target genes and itself a transcriptional target of the Hh pathway, in the invasive ability of pancreatic cancer cells. To evaluate the role of Gli1, we knocked down Gli1 expression of pancreatic cancer cells. Real‐time RT‐PCR showed that transfection of AsPC‐1, SUIT‐2 and pcDNA‐Gli1 cells with siGli1 resulted in approximately 90% (in AsPC‐1 and SUIT‐2) and 60% (in pcDNA‐Gli1 cells) knockdown of Gli1 mRNA expression (Fig. 5a). Immunocytochemistry showed that siGli1 treatment decreased the Gli1 protein expression (Fig. 5b) and MMP‐9 mRNA expression was significantly decreased in all cells (Fig. 5c). Furthermore, in the Matrigel invasion assay, the number of invaded siGli1‐transfected cells was decreased compared with the control siRNA‐transfected cells (Fig. 5d).
Figure 5.
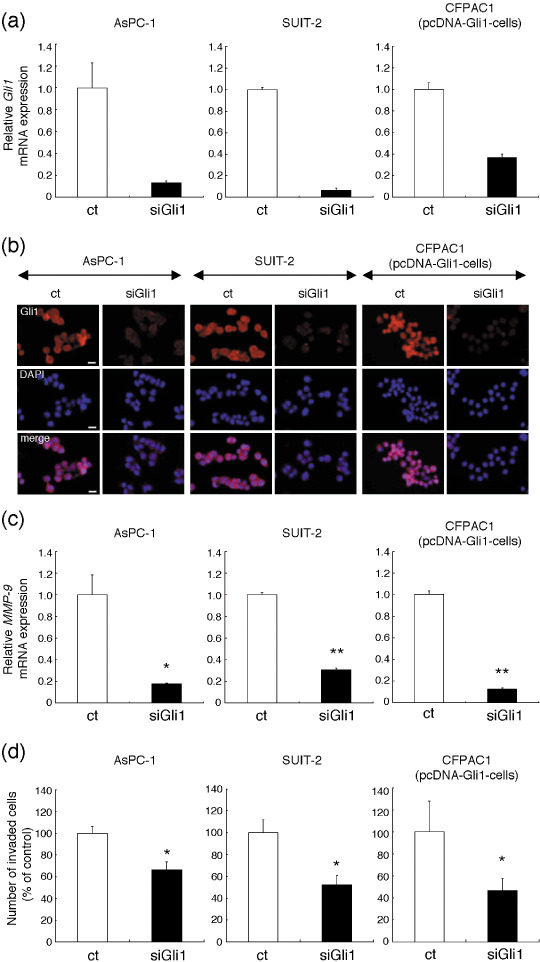
RNA interference for Gli1 inhibited the invasive ability of AsPC‐1‐, SUIT‐2‐, and pcDNA‐Gli1‐transfected CFPAC‐1 human pancreatic ductal adenocarcinoma cells. (a and b) AsPC‐1, SUIT‐2, and pcDNA‐Gli1‐transfected CFPAC‐1 cells (pcDNA‐Gli1‐cells) were transfected with small interfering RNA (siRNA) for Gli1 (siGli1) or control siRNA (ct) for 48 h, and the expression of Gli1 mRNA was assessed by real‐time reverse transcription–polymerase chain reaction (RT‐PCR). (a) The expression of Gli1 protein was assessed by immunocytochemistry. Gli1, red; 4′,6′‐diamidino‐2‐phenylindole dihydrochloride (DAPI), blue. Magnification, ×400; scale bars, 20 µm (b). (c) Matrix metalloproteinase (MMP)‐9 mRNA relative to that of β‐actin mRNA, was assessed by real‐time RT‐PCR. (d) After 48 h of transfection, these cells were seeded in a Matrigel‐coated invasion chamber for 16 h. Quantitation of the assay is shown in the graph. Columns, means of three independent experiments; bars, SD. *P < 0.05; **P < 0.01.
Discussion
Recent increasing data indicate a contribution of the Hh pathway not only to cell proliferation, but also to the repair of several tissues.( 23 ) For example, it has been shown that, during repair of bronchial epithelium, transient activation of Hh pathway occurs within the normally quiescent bronchial epithelium, and such a process might well occur during adult gut epithelial turnover.( 13 ) As we know, many kinds of molecules, including MMPs, take part in degradation of extracellular matrix are participating in tissue repair. This suggests that the Hh pathway might also contribute to cell invasion by mediating MMPs.
Based on these observations, we hypothesized that the Hh pathway plays an important role in the invasive ability of pancreatic cancer. In the present study, we selected two human cancer cells, AsPC‐1 and SUIT‐2. Blockade of the Hh pathway suppressed the invasive ability of both cell types (Fig. 1), but activation of the Hh pathway increased their invasive ability (Fig. 2). These findings indicate a close relationship between the Hh pathway and the invasive ability of pancreatic cancer cells.
How does the Hh pathway affect the invasive ability of pancreatic cancer cells? It is noteworthy that Hh pathway inhibition with cyclopamine affects neither the proliferation nor the migration ability of these cells, under the present experimental conditions (Fig. 1 and data not shown). These results indicate that the Hh pathway could play a relatively specific role in Matrigel degradation. Most MMPs probably participate in the process of matrix degradation. The degradation of basement membrane type IV collagen is a critical early event in tumor invasion, suggesting that MMP‐2 and MMP‐9 might play a particularly important role.( 24 , 25 ) Therefore, we focused on MMP‐2 and MMP‐9. Real‐time RT‐PCR and gelatin zymography showed that cyclopamine and rhShh decreased and increased MMP‐9 expression, respectively (Fig. 3). Furthermore, a Gli1‐expressing pancreatic cancer clones showed increased MMP‐9 mRNA expression and MMP‐9 enzymatic activity (Fig. 4b,c), and the increased invasive ability of these cells was abolished by RNAi against MMP‐9 (Fig. 4f). Thus, the Hh pathway contributes to the invasive ability of pancreatic cancer cells through MMP‐9 induction.
It has been reported that aberrant activation of MMP‐2 is more frequent in human pancreatic cancer than that of MMP‐9.( 26 ) Although we evaluated MMP‐2 expression by gelatin zymography, we could detect only pro‐MMP‐2, not active‐MMP‐2. Moreover, the expression of pro‐MMP‐2 was not significantly correlated with Hh pathway activity. Considering these results we had come to a conclusion that the Hh pathway did not affect MMP‐2 activity, at least not in the pancreatic cancer cells we used in this study.
It is important that stably Gli1‐expressing pancreatic cancer cell clones (pcDNA‐Gli1 cells) showed a marked increase in their invasive ability (Fig. 4a) and increased active‐MMP‐9 expression (Fig. 4b), and that knockdown of Gli1 resulted in suppression of the invasive abilities (Fig. 5d) and MMP‐9 mRNA expression (Fig. 5c). These data indicate that Gli1 might play a crucial role both in the invasive ability and in the MMP‐9 expression of these cells.
In our study, it is still unclear how the Hh pathway activates MMP‐9 expression. Recently, it has been reported that the Hh pathway interacts with the Ras/MAPK pathway.( 27 ) Because the MAPK pathway has been reported to regulate MMPs,( 28 ) we suggest that one of the possible mechanism of activating MMP‐9 is the Ras/MAPK cascade.
Interestingly, Feldmann et al.( 29 ) showed that the Hh pathway might play an important role in pancreatic cancer invasion and metastasis using a xenograft model. They focused on snail and E‐cadherin as the key molecule of the Hh‐related invasion, and suggested that epithelial‐to‐mesenchymal transition might participate in the Hh‐related invasion in pancreatic cancer. In the present study, we also showed an important role for the Hh pathway in the invasive ability of pancreatic cancer cells. We focused on MMP‐9 as the key molecule and suggested that degaradation of the extracellular matrix also participates in the Hh‐related invasion. We showed for the first time that MMP‐9 plays an important role in the invasive ability mediated by the Hh pathway in pancreatic cancer.
We also showed that the Hh pathway, especially Gli1, contributes to pancreatic cancer invasion through MMP‐9 induction. The results of the present study suggest that Gli1 might become a new therapeutic target for inhibiting the invasion of pancreatic cancer cells.
Acknowledgments
The authors thank Kaori Nomiyama and Miyuki Manabe for skillful technical assistance. This work was supported by General Scientific Research Grants 18390350 and 18659372 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1. Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2006; 20: 1218–49. [DOI] [PubMed] [Google Scholar]
- 2. Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet 2004; 363: 1049–57. [DOI] [PubMed] [Google Scholar]
- 3. Hebrok M. Hedgehog signaling in pancreas development. Mech Dev 2003; 120: 45–57. [DOI] [PubMed] [Google Scholar]
- 4. Cohen MM Jr. The hedgehog signaling network. Am J Med Genet A 2003; 123: 5–28. [DOI] [PubMed] [Google Scholar]
- 5. Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–87. [DOI] [PubMed] [Google Scholar]
- 6. Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer 2003; 3: 903–11. [DOI] [PubMed] [Google Scholar]
- 7. Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino‐terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 1999; 126: 3915–24. [DOI] [PubMed] [Google Scholar]
- 8. Ruiz i Altaba A. Gli proteins encode context‐dependent positive and negative functions. Implications for development and disease. Development 1999; 126: 3205–16. [DOI] [PubMed] [Google Scholar]
- 9. Aza‐Blanc P, Lin HY, Ruiz i Altaba A, Kornberg TB. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development 2000; 127: 4293–301. [DOI] [PubMed] [Google Scholar]
- 10. Lee J, Platt KA, Censullo P, Ruizi Altaba A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 1997; 124: 2537–52. [DOI] [PubMed] [Google Scholar]
- 11. Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem 2003; 278: 5116–22. [DOI] [PubMed] [Google Scholar]
- 12. Karlstrom RO, Tyurina OV, Kawakami A et al . Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development 2003; 130: 1549–64. [DOI] [PubMed] [Google Scholar]
- 13. Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small‐cell lung cancer. Nature 2003; 422: 313–7. [DOI] [PubMed] [Google Scholar]
- 14. Thayer SP, Di Magliano MP, Heiser PW et al . Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 2003; 425: 851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berman DM, Karhadkar SS, Maitra A et al . Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature 2003; 425: 846–51. [DOI] [PubMed] [Google Scholar]
- 16. Zhang H, Morisaki T, Matsunaga H et al . Protein‐bound polysaccharide PSK inhibits tumor invasiveness by down‐regulation of TGF‐β1 and MMPs. Clin Exp Metastasis 2000; 18: 343–52. [DOI] [PubMed] [Google Scholar]
- 17. Vistica DT, Skehan P, Scudiero D, Monks A, Pittman A, Boyd MR. Tetrazolium‐based assays for cellular viability: a critical examination of selected parameters affecting formazan production. Cancer Res 1991; 51: 2515–20. [PubMed] [Google Scholar]
- 18. Sehgal I, Baley PA, Thompson TC. Transforming growth factor β1 stimulates contrasting responses in metastatic versus primary mouse prostate cancer‐derived cell lines in vitro . Cancer Res 1996; 56: 3359–65. [PubMed] [Google Scholar]
- 19. Fan L, Pepicelli CV, Dibble CC et al . Hedgehog signaling promotes prostate xenograft tumor growth. Endocrinology 2004; 145: 3961–70. [DOI] [PubMed] [Google Scholar]
- 20. Ohuchida K, Mizumoto K, Fujita H et al . Sonic hedgehog is an early developmental marker of intraductal papillary mucinous neoplasms: clinical implications of mRNA levels in pancreatic juice. J Pathol 2006; 210: 42–8. [DOI] [PubMed] [Google Scholar]
- 21. Yamanaka N, Morisaki T, Nakashima H et al . Interleukin 1β enhances invasive ability of gastric carcinoma through nuclear factor‐κB activation. Clin Cancer Res 2004; 10: 1853–9. [DOI] [PubMed] [Google Scholar]
- 22. Nakashima H, Nakamura M, Yamaguchi H et al . Nuclear factor‐κB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res 2006; 66: 7041–9. [DOI] [PubMed] [Google Scholar]
- 23. Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature 2004; 432: 324–31. [DOI] [PubMed] [Google Scholar]
- 24. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2002; 2: 161–74. [DOI] [PubMed] [Google Scholar]
- 25. Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 2002; 295: 2387–92. [DOI] [PubMed] [Google Scholar]
- 26. Bloomston M, Zervos EE, Rosemurgy AS 2nd. Matrix metalloproteinases and their role in pancreatic cancer: a review of preclinical studies and clinical trials. Ann Surg Oncol 2002; 9: 668–74. [DOI] [PubMed] [Google Scholar]
- 27. Morton JP, Mongeau ME, Klimstra DS et al . Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc Natl Acad Sci U S A 2007; 104: 5103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mandal M, Mandal A, Das S, Chakraborti T, Sajal C. Clinical implications of matrix metalloproteinases. Mol Cell Biochem 2003; 252: 305–29. [DOI] [PubMed] [Google Scholar]
- 29. Feldmann G, Dhara S, Fendrich V et al . Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res 2007; 67: 2187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]


