Abstract
To elucidate the mechanism underlying suppression by curcumin of esophageal carcinogenesis induced by NMBA, we evaluated the CYP level and mutagenic activation of environmental carcinogens, by immunoblot analyses and Ames preincubation test, respectively, and bilirubin, 4‐nitrophenol and testosterone UDPGT activities in F344 rats treated with curcumin and/or NMBA. No significant alterations in the hepatic levels of constitutive CYP proteins, mutagenic activation by liver S9 or hepatic UDPGT activities were produced by subcutaneous treatment with 0.5 mg/kg NMBA for 5 weeks and/or feeding of 0.05% and 0.2% curcumin for 6 weeks. In contrast, gavage of 0.2% curcumin decreased esophageal CYP2B1 and 2E1 by up to 60%, compared with vehicle control. Similarly, intragastric treatment with 270 mg/kg curcumin decreased esophageal and gastric CYP2B1 and CYP2E1, but not in lung, kidney or intestine. Conversely, large intestinal CYP2B1 was 2.8‐fold higher in the treated rats than in control rats. Mutagenic activities of NOC, including NMBA, in the presence of esophagus and stomach S9 were markedly decreased in the treated rats, whereas those in the presence of large intestine S9 were 2.2–3.0‐fold above control. These results show that modifying effects of curcumin on esophageal carcinogenesis can be attributed to a decrease in metabolic activation of NMBA by esophageal CYP2B1 during the initiation phase, without the contribution of metabolic activation and inactivation by liver. Further, the present findings suggest the potential of curcumin for modification of gastric and intestinal carcinogenesis initiated with NOC. (Cancer Sci 2006; 97: 896–904)
Abbreviations:
- 4‐NP
4‐nitrophenol
- AFB1
aflatoxin B1
- BHP
N‐nitrosobis (2‐hydroxypropyl)amine
- BP
benzo[a]pyrene
- CYP
cytochrome P‐450
- DEN
N‐nitrosodiethylamine
- DMN
N‐nitrosodimethylamine
- DMSO
dimethyl sulfoxide
- EROD
ethoxyresorufin O‐deethylase
- G6P
glucose 6‐phosphate
- G6PDH
G6P dehydrogenase
- Glu‐P‐1
2‐amino‐6‐methyldipyrido[1,2‐a:3′,2′‐ d]imidazole
- GST
glutathione S‐transferase
- HCA
heterocyclic amine
- IQ
2‐amino‐3‐methylimidazo[4,5‐f]quinoline
- MC
3‐methylcholanthrene
- MeAαC
2‐amino‐3‐methyl‐9H‐pyrido[2,3‐b]indole
- MROD
methoxyresorufin O‐demethylase
- NDMM
N‐nitroso‐2,6‐dimethylmorpholine
- NMBA
N‐nitrosomethylbenzylamine
- NOC
N‐nitroso compound
- NPYR
N‐nitrosopyrrolidine
- PB
phenobarbital
- PROD
pentoxyresorufin O‐dealkylase
- SD
standard deviation
- Trp‐P‐2
3‐amino‐1‐methyl‐5H‐pyrido[4,3‐b]indole
- UDP
uridine diphosphate
- UDPGT
UDP‐glucuronyltransferase.
Human esophageal cancer has been closely associated with exposure to nitrate and nitrite, which are precursors of NOC, and exposure to NMBA is also associated with an increased risk of human cancer in the esophagus in China.( 1 , 2 , 3 , 4 ) In fact, NMBA is known to be the most potent carcinogen for rat esophagus, irrespective of its mode of administration.( 5 , 6 ) It has been shown that esophageal mucosa microsomes from male Sprague–Dawley rats can form benzaldehyde and formaldehyde from NMBA at rates 1/5 and 1/60 of hepatic levels of both metabolites, respectively, and the metabolisms are inhibited more than 95% by CO and 70% by SKF525A, typical CYP inhibitors.( 7 ) O 6‐methylated guanine level in rats given NMBA is six times higher in esophagus DNA than in liver DNA,( 8 ) and isothiocyanates markedly decrease the incidence and multiplicity of NMBA‐induced esophageal tumors, with inhibition of esophageal DNA methylation in F344 rats.( 9 ) Further, it has been shown that ethanol has an enhancing effect on DEN‐induced esophageal tumorigenesis in F344 rats,( 10 ) with enhancement of metabolic activation of DEN by hepatic CYP2E1.( 11 ) These findings indicate the importance of metabolic activation of NMBA by the target organ and/or liver during the initiation phase, and some NOC are known to be metabolized in the esophagus at a relatively high rate, often leading to high levels of esophageal DNA alkylation.( 8 , 12 , 13 ) The total CYP content of the esophagus microsomes is only 7% of that of liver microsomes,( 14 ) but little is known about the expression of CYP species in rat esophagus; CYP1A1 and 2A3 proteins are constitutively expressed in rat esophagus microsomes, but CYP2B1/2, 2E1 and 3A2 proteins are not.( 14 , 15 , 16 ) We have recently shown that NMBA is mutagenetically activated by hepatic CYP2B1 and 2B2, but not CYP2E1, in rats.( 11 )
Curcumin is the major yellow pigment in turmeric (the ground rhizome of Curcuma longa Linn), widely used as a spice and coloring agent in several foods, such as curry, mustard and potato chips as well as cosmetics and drugs. This natural dye has been reported to inhibit tumors in various organs, such as the tongue, skin, mammary gland, forestomach, liver, duodenum and colon from laboratory animals, during the initiation and/or promotion phases.( 17 ) In addition, Ushida et al.( 18 ) reported that esophageal tumorigenesis induced by NMBA is markedly suppressed by treatment with curcumin during the initiation and promotion phases in F344 rats. Curcumin is not toxic to humans up to 8 g/day when taken by mouth for 3 months,( 19 ) being suggested as an effective chemopreventive agent. Turmeric and curcumin are known to reduce BP‐ or 7,12‐dimethylbenz[a]anthracene‐derived DNA adducts in rat liver( 20 ) and in hamster buccal pouch.( 21 ) Turmeric and curcumin have been reported to be a competitive inhibitor for EROD and PROD,( 22 ) and to decrease total CYP content( 23 ) and EROD, MROD and PROD activities in liver, lung and stomach microsomes from rats.( 24 ) Curcumin also induces 4‐NP UDPGT( 25 , 26 ) and 1‐chloro‐2,4‐dinitrobenzene GST( 23 , 25 , 27 , 28 , 29 ) in rat liver. However, to our knowledge, no data have shown the effect of curcumin on hepatic levels of CYP enzymes, metabolic activation by CYP and UDPGT activities towards bilirubin and testosterone in rats, or on extrahepatic levels of CYP isoforms in any animal species.
In order to elucidate the mechanism(s) underlying suppression of NMBA‐induced esophageal tumorigenesis by curcumin, hepatic and extrahepatic levels of microsomal CYP enzymes known to activate typical environmental carcinogens, mutagenic activation of these carcinogens by tissue S9 and three kinds of hepatic UDPGT activities were assayed in male F344 rats treated with curcumin and/or NMBA.
Materials and Methods
Chemicals
BP, DMN, DEN, 4‐NP, testosterone, bilirubin, UDP‐glucuronic acid, Trp‐P‐2 and MeAαC acetates, Glu‐P‐1 hydrochloride and IQ were purchased from Wako Pure Chemicals (Osaka, Japan) and NMBA was from Sakai Laboratory (Fukui, Japan). Curcumin (>98% pure) was obtained from Nacalai Tesque (Kyoto, Japan), NPYR was from Aldrich Chemical (Milwaukee, WI, USA) and AFB1 was from Makor Chemicals (Jerusalem, Israel). UDP‐[14C(U)]glucuronic acid was purchased from American Radiolabeled Chemicals (St Louis, MO, USA) and G6P, G6PDH, NADP+, NADPH, NADH and ATP were from Oriental Yeast (Tokyo, Japan). All other commercial products were of the purest grade available. BHP and NDMM were synthesized in our laboratory as described previously.( 30 )
Animal treatment and tissue preparation
All animal experiments were undertaken following guidelines for the care and use of experimental animals set by Gifu Pharmaceutical University and Gifu University. Male 4‐ or 5‐week‐old F344 rats purchased from Japan SLC (Hamamatsu, Japan) were housed in wire cages (two or three rats/cage) and maintained under standard laboratory conditions. Thirty male rats, 5 weeks old, were divided into six groups consisting of five animals. Rats in Groups 3 and 5 were given 0.05% curcumin in the basal diet CE‐2 (CLEA Japan, Tokyo, Japan) and those in Groups 4 and 6 were given 0.2% curcumin throughout the experiment. One week after initiation of the respective diets, rats in Groups 1, 3 and 4 were subcutaneously treated with 20% DMSO three times per week for 5 weeks, and rats in Groups 2, 5 and 6 received 0.5 mg/kg NMBA dissolved in 20% DMSO.( 18 ) All the animals were decapitated 24 h after the last dose of the vehicle or NMBA. Alternatively, 40 male rats, 6 weeks old, were divided into four groups. Ten rats were given 20% DMSO, serving as a control, 20 rats were treated with 70 or 270 mg/kg curcumin through an intragastric tube, and 10 rats were subcutaneously treated with 0.5 mg/kg NMBA as a single dose. All animals were killed 6 h (NMBA‐treated rats) or 24 h after treatment. In addition, 32 rats, 6 weeks old, were divided into two groups and were given DMSO or 270 mg/kg curcumin as a single dose, then killed 24 h after injection. Livers, kidneys and lungs were perfused in situ with ice‐cold sterile 1.15% KCl and 25% homogenates in 1.15% KCl were prepared. Esophagus, stomach, small intestine and large intestine were rinsed in ice‐cold 1.15% KCl after harvesting, and these mucosae were then stripped off the submucosae and any muscular tissues. S9 and microsomal fractions from these tissues were prepared using established procedures.( 30 )
Assay of total CYP content
Total CYP content in liver and esophagus microsomes was spectroscopically determined by the method of Omura and Sato.( 31 )
Western blot analysis
Gel electrophoresis and blot analyses were carried out as described in detail previously,( 32 ) according to the established methods of Laemmli( 33 ) and Towbin et al.( 34 ), respectively. Separated proteins were transferred by semi‐dry electroblotting from the sodium dodecylsulfate–polyacrylamide gel to polyvinylidene difluoride membrane (Immobilon‐P; Millipore, Bedford, MA, USA) in 25 mM Tris buffer (pH = 8.3) containing 0.19 M glycine and 20% (v/v) methanol. The membranes were blocked by incubation with phosphate‐buffered saline containing 5% skim milk for 1 h. The blocked membranes were incubated with goat antirat polyclonal antibodies for CYP1A1/2, CYP2B1/2, CYP2E1 and CYP3A2 (Daiichi Pure Chemicals, Tokyo, Japan) then stained using biotinylated antigoat immunoglobulin G (Vector Laboratories, Burlingame, CA, USA) and a Wako ABC‐POD kit (Wako Pure Chemicals).
Mutation assay
All tests were carried out using the Ames preincubation assay.( 35 ) Six NOC were dissolved in 100 µL of water and all the other carcinogens in 50 µL of DMSO. The mutagenicities of IQ (dose, 0.03 µg/plate), Trp‐P‐2 (0.03 or 0.3 µg/plate), Glu‐P‐1 (0.3 µg/plate), MeAαC (10 µg/plate), BP (5 µg/plate), AFB1 (1 µg/plate), NPYR (0.25 mg/plate), NMBA (1 mg/plate), DMN and DEN (1 or 10 mg/plate), and BHP and NDMM (10 mg/plate) were checked in the presence of liver, esophagus, stomach or intestine S9 mix, using established procedures.( 11 , 36 , 37 , 38 , 39 ) The amount of tissue S9 fraction was 10 µL/plate for the HCAs, 50 µL for BP and 150 µL for the NOC and AFB1. Salmonella typhimurium tester strains TA100 and TA98 were used for the six NOC and the other carcinogens, respectively. The S9 mix contained 4 mM NADPH, 4 mM NADH, 0.5 U G6PDH, 5 mM G6P and 5 mM ATP, except for the NOC, for which 4 mM NADP+ and 5 mM G6P were used.( 32 )
Assay of UDPGT activity
UDPGT activities towards bilirubin and 4‐NP in liver microsomes were assayed according to the methods of Heirwegh et al.( 40 ) and Isselbacher et al.,( 41 ) respectively, and that towards testosterone was determined using UDP‐[14C(U)]glucuronic acid as described by Matern et al.( 42 )
Statistical analysis
Statistical analyses by Student's t‐test were carried out to determine the significance of the differences between groups. All statements of significance are either P < 0.05 or P < 0.01.
Results
Determination of total CYP content and CYP isoform in several tissues
Figure 1 shows immunoblots and levels of microsomal CYP proteins in male F344 rats treated with NMBA and curcumin for up to 6 weeks. Esophageal CYP2B1 and CYP2E1 were constitutively detected with an antibody against hepatic CYP2B1/2 and CYP2E1 in the vehicle group (Group 1). In Group 2 rats subcutaneously treated with 0.5 mg/kg NMBA for 5 weeks the CYP2B1 level was not changed, but that in Group 3 and 4 rats fed 0.05% and 0.2% curcumin, respectively, for 6 weeks was decreased by 30%−40% relative to Group 1 rats. Similarly, the constitutive CYP2B1 level was 40% and 60% lower in Group 5 and 6 rats, respectively, than in Group 1 rats. Treatment with NMBA and/or 0.05% curcumin caused no decrease in the constitutive CYP2E1 level, whereas that in Group 4 and 6 rats was decreased by 30% relative to Group 1 rats. However, there were no significant differences in hepatic levels of constitutively detected CYP2B2, 2E1, 1A2 or 3A2 among the six groups, and neither CYP2B1 nor 1A1 were expressed in any group of rats. In addition, no significant alterations in hepatic and esophageal levels of total CYP content spectroscopically determined were observed in the six groups (liver, 1.35–1.60 nmol/mg; esophagus, 0.30–0.39 nmol/mg).
Figure 1.
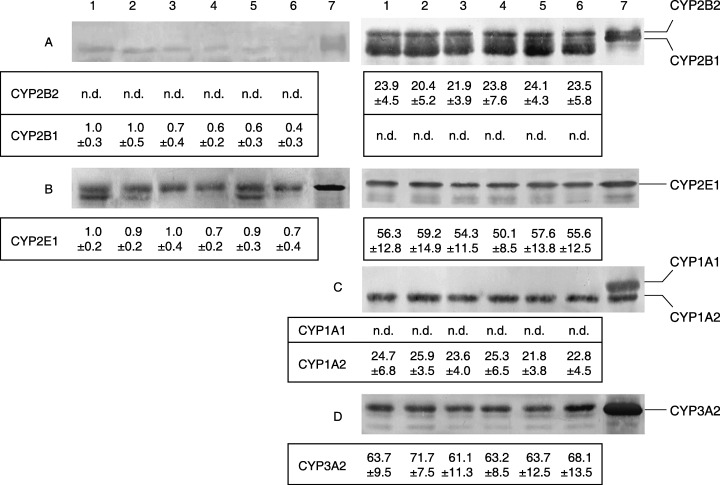
Immunoblots and densitometric determination of expression of CYP protein in esophagus (left) and liver (right) microsomes from rats treated with NMBA, curcumin or both. Both microsomes were pooled from five rats treated with vehicle (lane 1) or 0.5 mg/kg NMBA (lane 2) three times per week for 5 weeks, 0.05% (lane 3) or 0.2% (lane 4) curcumin for 6 weeks, and NMBA + 0.05% (lane 5) or + 0.2% (lane 6) curcumin. Lane 7 contains CYP standards from male Sprague–Dawley rats treated with PB (A and D), acetone (B) or MC (C). Liver microsomes contain 0.4 µg (A–C) and 1.0 µg (D) microsomal protein, and the values represent means ± SD of pmol/mg microsomal protein obtained from four experiments. Esophagus microsomes (A, B) contain 30 µg microsomal protein, and the values represent means ± SD of the ratio to arbitrary units obtained with the vehicle group. n.d., not detected.
In order to confirm the new evidence for CYP expression and suppression, the potency of curcumin was further checked in hepatic and extrahepatic microsomes from rats orally treated with 70 and 270 mg/kg curcumin, corresponding to the daily intake in the diet containing 0.05% and 0.2% curcumin, respectively, as a single dose. As shown in Figure 2, esophageal CYP1A1, 1A2 and 3A2, in addition to CYP2B1 and 2E1, were detected with antibodies against hepatic CYP in the vehicle group. The constitutive levels of CYP2B1 and 2E1 in rats given 270 mg/kg curcumin were significantly decreased by 40% (P < 0.01), relative to control rats, whereas levels of the other CYP isoforms were not changed, and NMBA produced no significant changes in the five CYP isoforms. There were also no significant differences in hepatic levels of constitutive CYP2B2, 2E1, 1A2 and 3A2 among the four groups, and neither CYP2B1 nor 1A1 were detected in any group of rats. Figure 3 shows immunoblots of CYP2B1 and 2E1 in six extrahepatic tissues, including the esophagus, from rats treated with 270 mg/kg curcumin as a single dose. CYP2B1 and 2E1 proteins, but not CYP2B2, were constitutively detected in all the extrahepatic tissues, except for CYP2E1 in the small intestine. The treatment with curcumin significantly decreased gastric and esophageal CYP2B1 by 30% and 40% (P < 0.05), respectively, relative to the vehicle group. It also decreased esophageal CYP2E1 by 60% (P < 0.01) and gastric CYP2E1 to an undetectable level. In contrast, the CYP2B1 level in the large intestine was 2.8‐fold higher (P < 0.01) in the curcumin‐treated rats than in control rats; there were no significant alterations in the levels of CYP2B1 or 2E1 observed in the small intestine, lung or kidney.
Figure 2.
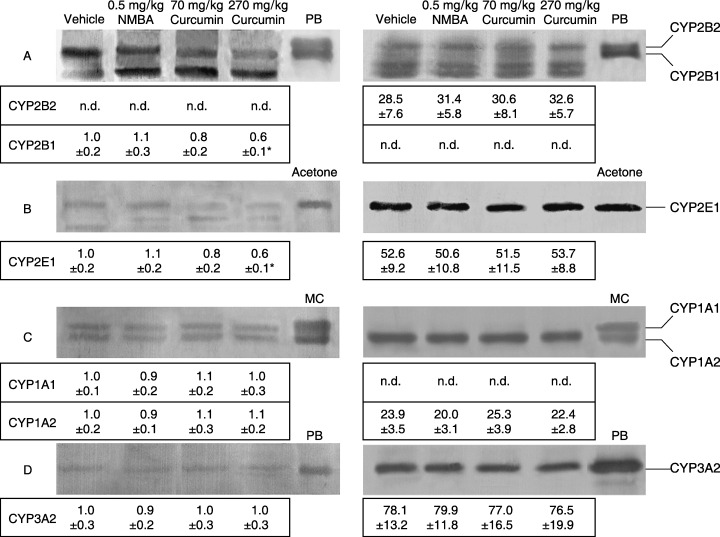
Immunoblots and densitometric determination of expression of CYP protein in esophagus (left) and liver (right) microsomes from rats treated orally with curcumin or subcutaneously with NMBA as a single dose. Both microsomes were pooled from 10 rats 24 h after each treatment. Lanes PB, acetone and MC contain CYP standards. The amounts of each microsomal protein, including (C) and (D) in the esophagus, and the means ± SD are identical to those described in Figure 1 legend. *P < 0.01, compared with the vehicle group (Student's t‐test). n.d., not detected.
Figure 3.
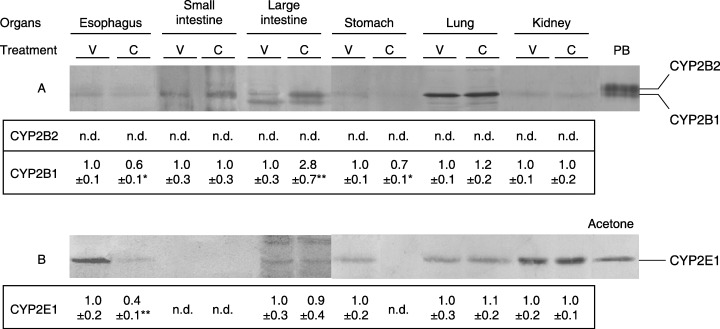
Immunoblots and densitometric determination of CYP2B (A) and 2E (B) expressions in extrahepatic tissue microsomes from rats orally treated with 270 mg/kg curcumin as a single dose. These microsomes were pooled from 16 rats 24 h after treatment with vehicle (V) or curcumin (C). Lanes PB and acetone contain CYP standards and the values represent means ± SD of the ratio to arbitrary units obtained with the vehicle group from between four and eight experiments. The assays of electrophoresis were carried out with 30 µg microsomal protein, except for lung CYP2B1/2 and kidney CYP2E1, for which 5 µg microsomal protein was loaded. *P < 0.05 and **P < 0.01, compared with the vehicle group (Student's t‐test). n.d., not detected.
Mutagenic activation of environmental carcinogens by tissue S9
To clarify the potential of liver S9 to mediate mutagenic activation of carcinogens, the mutagenicities of NMBA and 11 other carcinogens known to be metabolically activated by CYP2B1/2, 2E1, 1A1/2 and 3A2, were tested in Salmonella strains TA98 and TA100. Figure 4 shows the mutagenic activities of six NOC including NMBA, four HCA, BP and AFB1 in the presence of liver S9 mix from rats treated with NMBA and/or curcumin for up to 6 weeks (five groups excluding Group 5). There were no significant differences in the mutagenic activities of any NOC in the strain TA100, nor in HCAs, BP or AFB1 in the strain TA98, among the five groups. Similarly, the single treatment with 0.5 mg/kg NMBA or 270 mg/kg curcumin produced no significant alteration in mutagenicity of five NOC excluding BHP, Glu‐P‐1 and Trp‐P‐2 (data not shown).
Figure 4.
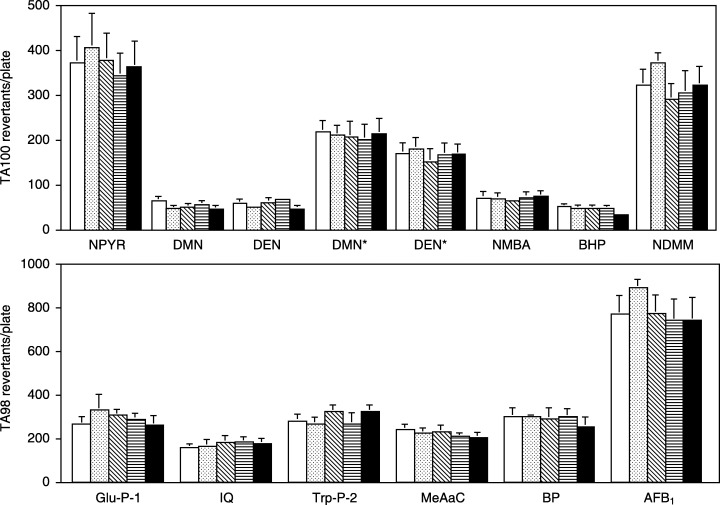
Mutagenic activities of various carcinogens in the Salmonella typhimurium tester strains TA100 (upper, six NOC) and TA98 (lower, four HCA, BP and AFB1) in the presence of liver S9 from rats treated with NMBA, curcumin or both. Each test was carried out in duplicate (four to eight plates) with liver S9 pooled from five rats each treated with vehicle (Group 1; clear), NMBA (Group 2; light stippled), 0.05% curcumin (Group 3; hatched), 0.2% curcumin (Group 4; horizontal) or NMBA + 0.2% curcumin (Group 6; solid) for up to 6 weeks. Each bar represents the means ± SD after subtraction of spontaneous revertants (TA100, 108–136; TA98, 9–14). DMN and DEN were assayed in both doses of 1 and *10 mg/plate, and Trp‐P‐2 was in 0.3 µg/plate.
Figure 5 shows the mutagenic activities of NMBA, DMN and DEN in the presence of esophagus, stomach or large intestine S9 from rats orally treated with the vehicle or 270 mg/kg curcumin as a single dose. In the curucumin‐treated rats the mutagenic activities of NMBA and DMN in the presence of esophagus S9 were decreased to almost half or less (P < 0.01) of those in control rats. In the presence of stomach S9 from the treated rats, the mutagenic activities of DMN and DEN were decreased by 82%−85% (P < 0.01) relative to control rats and that of NMBA to the spontaneous revertants. However, the mutagenic activities of NMBA and DMN in the presence of large intestine S9 were increased by curcumin treatment to 2.2‐ (P < 0.05) and 3.0‐fold (P < 0.01), respectively, relative to those in control rats.
Figure 5.
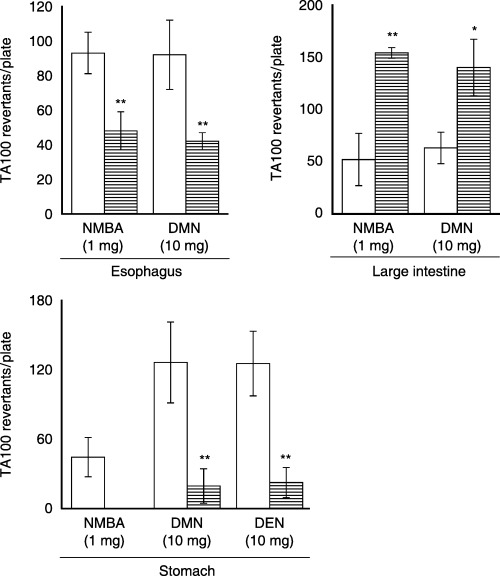
Effect of a single treatment with 270 mg/kg curcumin on the mutagenic activities of NMBA, DEN and DMN in the Salmonella typhimurium strain TA100 in the presence of esophagus, stomach or large intestine S9. Each test was carried out in duplicate (four to six plates) with tissue S9 pooled from 16 rats each treated with vehicle (clear) or 270 mg/kg curcumin (horizontal). Each bar represents the means ± SD after subtraction of spontaneous revertants (111–124). *P < 0.05 and **P < 0.01, compared with the vehicle group (Student's t‐test).
Three kinds of hepatic UDPGT activities
Table 1 summarizes the effects of NMBA and curcumin treatment for up to 6 weeks on hepatic UDPGT activities towards bilirubin, 4‐NP and testosterone in liver microsomes (Groups 1–6). There were no significant differences in three kinds of UDPGT activities among the six groups. Similarly, no significant alteration in these activities was observed in the single treatments with 0.5 mg/kg NMBA or 270 mg/kg curcumin (data not shown).
Table 1.
UDPGT activities in liver microsomes from male F344 rats repeatedly treated with NMBA, curcumin or both for up to 6 weeks
| Group | Treatment | UDPGT activity (nmol/min/mg protein) | ||
|---|---|---|---|---|
| Bilirubin | 4‐Nitrophenol | Testosterone | ||
| 1 | Vehicle | 1.10 ± 0.07 | 26.8 ± 7.8 | 0.10 ± 0.03 |
| 2 | NMBA | 1.05 ± 0.14 | 32.3 ± 4.5 | 0.12 ± 0.01 |
| 3 | 0.05% curcumin | 1.07 ± 0.05 | 27.7 ± 5.8 | 0.12 ± 0.02 |
| 4 | 0.2% curcumin | 1.08 ± 0.12 | 30.4 ± 5.7 | 0.10 ± 0.02 |
| 5 | NMBA + 0.05% curcumin | 1.08 ± 0.08 | 31.2 ± 5.4 | 0.13 ± 0.03 |
| 6 | NMBA + 0.2% curcumin | 1.04 ± 0.11 | 29.8 ± 3.2 | 0.10 ± 0.01 |
Each test was carried out with liver microsomes pooled from five rats. The results are expressed as means ± SD of between four and eight experiments.
Discussion
It has been reported that CYP1A1, but not CYP2B1, 2E1 or 3A1/2, is detectable in esophageal microsomes from male F344( 15 ) and Wistar( 14 ) rats and CYP2E1 in those from ethanol‐treated rats.( 43 ) However, the present results indicate that CYP2B1, 2E1, 1A1, 1A2 and 3A2 are constitutively detected in esophageal microsomes from male F344 rats. The reasons for the discrepancies are currently unknown, but it is suggested that the differences might be due to experimental conditions, such as the transfer conditions, the antibodies used against rat or human CYP species or the detection method. In any case, this is, to our knowledge, the first demonstration of the presence of CYP1A2, 2B1, 2E1 and 3A2 in the esophagus from uninduced rats; our results support the previous finding that CYP1A1, 1A2, 2B6, 2E1 and 3A4 and CYP1A2 mRNA are detected in esophageal mucosa from human and rat, respectively.( 44 , 45 , 46 , 47 ) In this study it has also been demonstrated that CYP2B1 and 2E1 are constitutively detected in five other extrahepatic microsomes, except for CYP2E1 in the small intestine. Both CYPs are known to be present in the lung, small intestine, kidney( 48 , 49 , 50 ) and large intestine,( 51 ) except for CYP2E1 in the small intestine and CYP2B1 in the large intestine, in rats, and CYP2E1 in the stomach from ethanol‐induced rats.( 43 ) Accordingly, this is the first report of the presence of CYP2B1 protein in the stomach and large intestine and CYP2E1 protein in the stomach from uninduced rats, in agreement with the finding of CYP2B1 mRNA expression in both tissues of rats.( 52 , 53 )
CYP2A3 expressed in a baculovirus system metabolizes NMBA, predominantly by methylene hydroxylation, and CYP2A3 and its mRNA are constitutively expressed in rat esophagus, but their expressions are 15‐ and 1600‐fold less than in the nasal mucosa, respectively.( 16 , 54 ) Therefore, Gopalkrishnan et al. have suggested that there must be an enzyme other than CYP2A3 responsible for activating NMBA in the esophagus, ( 54) taking account of the finding that rat esophageal explants efficiently metabolize NMBA.( 55 ) We have shown that CYP2B1 and 2B2 are equally involved in the mutagenic activation of NMBA by PB‐induced liver, and CYP2B2 in uninduced liver, in rats.( 11 ) In the rat, hepatic CYP1A1/2, CYP2B1/2 and CYP3A2 have been reported to contribute to the mutagenic activation of HCAs, BP and AFB1.( 36 , 56 ) It has been shown that rat CYP2E1 and CYP2B1/2 contribute differently to the mutagenic activation of various N‐nitrosodialkylamines, depending on the length of the alkyl chain and the dose of substrate.( 57 , 58 , 59 , 60 ) CYP2E1 activates NPYR and 1 mg dose of DMN and DEN to mutagens and CYP2B1/2 do BHP, NDMM and 10 mg dose of DMN and DEN. Thus, it seems reasonable that feeding a 0.05% or 0.2% curcumin‐containing diet for 6 weeks and intragastric treatment produce no effect on the mutagenic activation of these carcinogens by liver S9, reflecting no alteration of hepatic CYP2B1/2, 2E1, 3A2 and 1A1/2 in rats.
Differing results have been reported for the effect of curcumin or turmeric on metabolic activities specific to each CYP species in rodent liver, lung and stomach. Feeding of 2% curcumin for 2 weeks causes a modest reduction of hepatic EROD activity in female A/J mice,( 28 ) whereas 10% turmeric (equivalent to 0.5% curcumin) for 4 weeks shows no alteration of hepatic aryl hydrocarbon hydroxylase activity in male Wistar rats.( 25 ) Feeding of 1% turmeric for 15 days produces no alteration of EROD or PROD activities, but exerts a suppressive effect on MROD activity, BP‐induced EROD and MROD activity and PB‐induced PROD activity in rat liver.( 24 ) In conjunction with the present findings, with the mutagenic activation of seven carcinogens by hepatic CYP in curcumin‐treated rats, it is concluded that curcumin and turmeric exert no suppression of metabolic activities specific to each CYP species in uninduced rodent liver, with the exception of MROD activity, but cause clear suppression in rats induced by PB and MC.
In rat liver, UDPGT1A1, 1A6 and 2B1 are major enzymes inducible by clofibrate, MC and PB, respectively.( 61 ) Carcinogenic NOC, including NMBA, are known to be substrates for UDPGT,( 62 , 63 , 64 ) and UDPGT2B1 is suggested to be the probable enzyme responsible for the glucuronidation of DEN and N‐nitrosomethyl‐n‐pentylamine.( 65 ) Feeding of 10% turmeric for 4 weeks clearly elevates 4‐NP UDPGT activity,( 25 ) but feeding of 5% turmeric (equivalent to 0.25% curcumin) does not. Therefore, it is reasonable that no alterations in three kinds of UDPGT activity were observed in rats fed 0.05% or 0.2% curcumin for 6 weeks. Together with the results of UDPGT activities in rats treated with 70 or 270 mg/kg curcumin as a single dose, treatment with a higher dose of curcumin might be needed for induction of UDPGT activities. The present finding on hepatic UDPGT activities suggests that neither 0.05% nor 0.2% curcumin modify NMBA‐induced esophageal carcinogenesis through detoxification by the enzymes under the experimental conditions used. However, curcumin caused a decrease in esophageal and gastric levels of CYP2B1 and 2E1 and the mutagenic activation of NMBA, DEN and DMN by the tissue S9, in agreement with the previous finding that curcumin suppresses the metabolic activity specific to CYP2B1 and 2B2 more strongly in stomach microsomes than in liver microsomes in rats.( 24 ) In contrast, curcumin increased large intestinal CYP2B1 and the mutagenic activation of NMBA and DMN. Together with the findings that high levels of esophageal DNA alkylation are induced by NOC,( 8 , 12 , 13 ) and DNA methylation by NMBA and N‐nitrosomethyl‐n‐butylamine in rats occurs to a higher extent in esophagus than in liver,( 66 ) it suggests that modification of metabolic activation of NOC by the target organ plays a critical role in chemoprevention of NOC‐induced carcinogenesis in rats. It has been reported that curcumin inhibits the expression of c‐Jun and c‐Fos/AP‐1,( 67 ) a transcriptional factor that plays an important role in the expressions of CYP2B1/2( 68 ) and CYP2E1,( 69 ) but not CYP1A,( 70 ) and 3 A, suggesting that the suppression of AP‐1‐induced transcription by curcumin might produce a decrease in CYP2B1 and 2E1 expressions in the esophagus and stomach. However, this is not consistent with the present findings of enhancement in mutagenic activation by large intestinal CYP and no suppression in that by liver CYP. As there are no reports on enhancement by curcumin of CYP2B1 or 2E1 expression in any tissue, the mechanisms underlying modification of tissue‐specific actions by curcumin remain to be elucidated, and further investigations are now required.
In conclusion, it has been demonstrated that dietary feeding or intragastric treatment with curcumin shows a suppressive effect on the mutagenic activation of three NOC, including NMBA, by either esophageal or gastric CYP2B1 and 2E1, but shows a promotive effect on the mutagenic activation of two NOC by large intestinal CYP2B1. Consequently, it suggests that suppression by curcumin of NMBA‐induced esophagus carcinogenesis in F344 rats can be attributed to a decrease in the metabolic activation of NMBA by esophageal CYP2B1 during the initiation phase, without the contribution of metabolic activation or inactivation by glucuronidation in rat liver. The present data also suggest that dietary exposure to curcumin might suppress esophageal carcinogenesis initiated by DEN, N‐nitrosopiperidine and methyl‐n‐amylnitrosamine,( 71 , 72 , 73 ) or gastric carcinogenesis initiated by other carcinogens activated by CYP2B1/2 and 2E1, but that curcumin might enhance large intestinal carcinogenesis induced by these carcinogens, although curcumin is known to suppress colon carcinogenesis initiated with azoxymethane, a direct carcinogen.( 74 ) Together with findings that anti‐inflammatory( 75 ) and antioxidant actions( 76 ) by curcumin are possible inhibitory mechanisms, it is reasonable to assume that curcumin could affect chemically‐induced carcinogenesis through multiple mechanisms.
References
- 1. Lin HJ, Chan WC, Fong YY, Newberne PM. Zinc levels in serum, hair, and tumors from patients with esophageal carcinoma. Nutr Rep Int 1977; 15: 635–43. [Google Scholar]
- 2. Yang CS. Research on esophageal cancer in China: a review. Cancer Res 1980; 40: 2633–44. [PubMed] [Google Scholar]
- 3. National Academy of Sciences. The Health Effects of Nitrate, Nitrite, and N‐Nitroso Compounds. Washington: National Academy Press, 1981. [Google Scholar]
- 4. Van Rensburg SJ. Epidemiologic and dietary evidence for a specific nutritional predisposition to esophageal cancer. J Natl Cancer Inst 1981; 67: 243–51. [PubMed] [Google Scholar]
- 5. Druckrey H, Preussmann R, Ivankovic S, Schmahl D. Organotropic carcinogenic effects of 65 various N‐nitrosocompounds on BD rats. Z Krebsforsch 1967; 69: 103–201. [PubMed] [Google Scholar]
- 6. Lijinsky W, Saavedra JE, Reuber MD, Singer SS. Esophageal carcinogenesis in F344 rats by nitrosomethylethylamines substituted in the ethyl group. J Natl Cancer Inst 1982; 68: 681–4. [PubMed] [Google Scholar]
- 7. Labuc GE, Archer MC. Esophageal and hepatic microsomal metabolism of N‐nitrosomethylbenzylamine and N‐nitrosodimethylamine in the rat. Cancer Res 1982; 42: 3181–6. [PubMed] [Google Scholar]
- 8. Kouros M, Monch W, Reiffer FJ, Dehnen W. The influence of various factors on the methylation of DNA by the oesophageal carcinogen N‐nitrosomethylbenzylamine. I. The importance of alcohol. Carcinogenesis 1983; 4: 1081–4. [DOI] [PubMed] [Google Scholar]
- 9. Wilkinson JT, Morse MA, Kresty LA, Stoner GD. Effect of alkyl chain length on inhibition of N‐nitrosomethylbenzylamine‐induced esophageal tumorigenesis and DNA methylation by isothiocyanates. Carcinogenesis 1995; 16: 1011–5. [DOI] [PubMed] [Google Scholar]
- 10. Aze Y, Toyoda K, Furukawa F, Mitsumori K, Takahashi M. Enhancing effect of ethanol on esophageal tumor development in rats by initiation of diethylnitrosamine. Carcinogenesis 1993; 14: 37–40. [DOI] [PubMed] [Google Scholar]
- 11. Mori Y, Koide A, Kobayashi Y, Morimura K, Kaneko M, Fukushima S. Effect of ethanol treatment on metabolic activation and detoxification of esophagus carcinogenic N‐nitrosamines in rat liver. Mutagenesis 2002; 17: 251–6. [DOI] [PubMed] [Google Scholar]
- 12. Swann PF, Coe AM, Mace R. Ethanol and dimethylnitrosamine and diethylnitrosamine metabolism and disposition in the rat. Possible relevance to the influence of ethanol on human cancer incidence. Carcinogenesis 1984; 5: 1337–43. [DOI] [PubMed] [Google Scholar]
- 13. Von Hofe E, Schmerold I, Lijinsky W, Jeltsch W, Kleihues P. DNA methylation in rat tissues by a series of homologous aliphatic nitrosamines ranging from N‐nitrosodimethylamine to N‐nitrosomethyldodecylamine. Carcinogenesis 1987; 8: 1337–41. [DOI] [PubMed] [Google Scholar]
- 14. Pinto LF, Moraes E, Albano RM et al. Rat oesophageal cytochrome P450 (CYP) monooxygenase system: comparison to the liver and relevance in N‐nitrosodiethylamine carcinogenesis. Carcinogenesis 2001; 22: 1877–83. [DOI] [PubMed] [Google Scholar]
- 15. Ahn D, Putt D, Kresty L, Stoner GD, Fromm D, Hollenberg PF. The effects of dietary ellagic acid on rat hepatic and esophageal mucosal cytochromes P450 and phase II enzymes. Carcinogenesis 1996; 17: 821–8. [DOI] [PubMed] [Google Scholar]
- 16. Gopalakrishnan R, Morse MA, Lu J et al. Expression of cytochrome P450 2A3 in rat esophagus: relevance to N‐nitrosobenzylmethylamine. Carcinogenesis 1999; 20: 885–91. [DOI] [PubMed] [Google Scholar]
- 17. Kelloff GJ, Crowell JA, Steele VE et al. Progress in cancer chemoprevention: development of diet‐derived chemopreventive agents. J Nutr 2000; 130 (2S Suppl): 467S–71S. [DOI] [PubMed] [Google Scholar]
- 18. Ushida J, Sugie S, Kawabata K et al. Chemopreventive effect of curcumin on N‐nitrosomethylbenzylamine‐induced esophageal carcinogenesis in rats. Jpn J Cancer Res 2000; 91: 893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng AL, Hsu CH, Lin JK et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high‐risk or pre‐malignant lesions. Anticancer Res 2001; 21: 2895–900. [PubMed] [Google Scholar]
- 20. Mukundan MA, Chacko MC, Annapurna VV, Krishnaswamy K. Effect of turmeric and curcumin on BP‐DNA adducts. Carcinogenesis 1993; 14: 493–6. [DOI] [PubMed] [Google Scholar]
- 21. Krishnaswamy K, Goud VK, Sesikeran B, Mukundan MA, Krishna TP. Retardation of experimental tumorigenesis and reduction in DNA adducts by turmeric and curcumin. Nutr Cancer 1998; 30: 163–6. [DOI] [PubMed] [Google Scholar]
- 22. Oetari S, Sudibyo M, Commandeur JN, Samhoedi R, Vermeulen NP. Effects of curcumin on cytochrome P450 and glutathione S‐transferase activities in rat liver. Biochem Pharmacol 1996; 51: 39–45. [DOI] [PubMed] [Google Scholar]
- 23. Azuine MA, Bhide SV. Chemopreventive effect of turmeric against stomach and skin tumors induced by chemical carcinogens in Swiss mice. Nutr Cancer 1992; 17: 77–83. [DOI] [PubMed] [Google Scholar]
- 24. Thapliyal R, Maru GB. Inhibition of cytochrome P450 isozymes by curcumins in vitro and in vivo . Food Chem Toxicol 2001; 39: 541–7. [DOI] [PubMed] [Google Scholar]
- 25. Goud VK, Polasa K, Krishnaswamy K. Effect of turmeric on xenobiotic metabolising enzymes. Plant Foods Hum Nutr 1993; 44: 87–92. [DOI] [PubMed] [Google Scholar]
- 26. Van Der Logt EMJ, Roelofs HMJ, Nagengast FM, Peters WHM. Induction of rat hepatic and intestinal UDP‐glucuronosyltransferases by naturally occurring dietary anticarcinogens. Carcinogenesis 2003; 24: 1651–6. [DOI] [PubMed] [Google Scholar]
- 27. Susan M, Rao MN. Induction of glutathione S‐transferase activity by curcumin in mice. Arzneimittelforschung 1992; 42: 962–4. [PubMed] [Google Scholar]
- 28. Singh SV, Hu X, Srivastava SK et al. Mechanism of inhibition of benzo[a]pyrene‐induced forestomach cancer in mice by dietary curcumin. Carcinogenesis 1998; 19: 1357–60. [DOI] [PubMed] [Google Scholar]
- 29. Okada K, Wangpoengtrakul C, Tanaka T, Toyokuni S, Uchida K, Osawa T. Curcumin and especially tetrahydrocurcumin ameliorate oxidative stress‐induced renal injury in mice. J Nutr 2001; 131: 2090–5. [DOI] [PubMed] [Google Scholar]
- 30. Mori Y, Yamazaki H, Toyoshi K et al. Mutagenic activation of carcinogenic N‐nitrosopropylamines by rat liver: evidence for a cytochrome P‐450 dependent reaction. Carcinogenesis 1985; 6: 415–20. [DOI] [PubMed] [Google Scholar]
- 31. Omura T, Sato R. The carbon monoxide binding pigment of liver microsomes. J Biol Chem 1964; 239: 265–75. [PubMed] [Google Scholar]
- 32. Koide A, Fuwa K, Furukawa F, Hirose M, Nishikawa A, Mori Y. Effect of cigarette smoke on the mutagenic activation of environmental carcinogens by rodent liver. Mutat Res 1999; 428: 165–76. [DOI] [PubMed] [Google Scholar]
- 33. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680–5. [DOI] [PubMed] [Google Scholar]
- 34. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 1979; 76: 4350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yahagi T, Nagao M, Seino Y, Matsushima T, Sugimura T. Mutagenicities of N‐nitrosamines on Salmonella . Mutat Res 1977; 48: 121–9. [DOI] [PubMed] [Google Scholar]
- 36. Mori Y, Koide A, Fuwa K, Kobayashi Y. N‐benzylimidazole for preparation of S9 fraction with multi‐induction of metabolizing enzymes in short‐term genotoxicity assays. Mutagenesis 2001; 16: 479–86. [DOI] [PubMed] [Google Scholar]
- 37. Smith C, Payne V, Doolittle DJ, Dbnath AK, Lawlor T, Hansch C. Mutagenic activity of a series of synthetic and naturally occurring HCAs in Salmonella . Mutat Res 1992; 279: 61–73. [DOI] [PubMed] [Google Scholar]
- 38. Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat Res 1983; 113: 173–215. [DOI] [PubMed] [Google Scholar]
- 39. Ueno Y, Kubota K, Ito T, Nakamura Y. Mutagenicity of carcinogenic mycotoxins in Salmonella typhimurium . Cancer Res 1978; 38: 536–42. [PubMed] [Google Scholar]
- 40. Heirwegh KP, Van de Vijver M, Fevery J. Assay and properties of dititonin‐activated bilirubin uridine diphosphate glucuronyltransferase from rat liver. Biochem J 1972; 129: 605–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Isselbacher KJ, Charabas MF, Quinn RC. The solubilization and partial purification of a glucuronyl transferase from rabbit liver microsomes. J Biol Chem 1962; 237: 3033–6. [PubMed] [Google Scholar]
- 42. Matern H, Heinemann H, Matern S. Radioassay of UDP‐glucuronosyltransferase activities toward endogenous substrates using labeled UDP‐glucuronic acid and an organic solvent extraction procedure. Anal Biochem 1994; 219: 182–8. [DOI] [PubMed] [Google Scholar]
- 43. Shimizu M, Lasker JM, Tsutsumi M, Lieber CS. Immunohistochemical localization of ethanol‐inducible P450IIE1 in the rat alimentary tract. Gastroenterology 1990; 99: 1044–53. [DOI] [PubMed] [Google Scholar]
- 44. Lechevrel M, Casson AG, Wolf CR et al. Characterization of cytochrome P450 expression in human oesophageal mucosa. Carcinogenesis 1999; 20: 243–8. [DOI] [PubMed] [Google Scholar]
- 45. Nakajima T, Wang RS, Nimura Y et al. Expression of cytochrome P450s and glutathione S‐transferases in human esophagus with squamous‐cell carcinomas. Carcinogenesis 1996; 17: 1477–81. [DOI] [PubMed] [Google Scholar]
- 46. Hughes SJ, Morse MA, Weghorst CM et al. Cytochromes P450 are expressed in proliferating cells in Barrett's metaplasia. Neoplasia 1999; 1: 145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Traber PG, McDonnell WM, Wang W, Florence R. Expression and regulation of cytochrome P‐450I genes (CYP1A1 and CYP1A2) in the rat alimentary tract. Biochim Biophys Acta 1992; 1171: 167–75. [DOI] [PubMed] [Google Scholar]
- 48. Takemoto K, Yamazaki H, Tanaka Y, Nakajima M, Yokoi T. Catalytic activities of cytochrome P450 enzymes and UDP‐glucuronosyltransferases involved in drug metabolism in rat everted sacs and intestinal microsomes. Xenobiotica 2003; 33: 43–55. [DOI] [PubMed] [Google Scholar]
- 49. Liu H, Bigler SA, Henegar JR, Baliga R. Cytochrome P450 2B1 mediates oxidant injury in puromycin–induced nephrotic syndrome. Kidney Int 2002; 62: 868–76. [DOI] [PubMed] [Google Scholar]
- 50. De Waziers I, Cugnenc PH, Yang CS, Leroux JP, Beaune PH. Cytochrome P450 isoenzymes, epoxide hydrolase and glutathione transferases in rat and human hepatic and extrahepatic tissues. J Pharmacol Exp Ther 1990; 253: 387–94. [PubMed] [Google Scholar]
- 51. Hakkak R, Korourian S, Ronis MJ, Ingelman‐Sundberg M, Badger TM. Effects of diet and ethanol on the expression and localization of cytochromes P450 2E1 and P450 2C7 in the colon of male rats. Biochem Pharmacol 1996; 51: 61–9. [DOI] [PubMed] [Google Scholar]
- 52. Pan J, Hong JY, Ma BL, Ning SM, Paranawithana SR, Yang CS. Transcriptional activation of cytochrome P450 2B1/2 genes in rat liver by diallyl sulfide, a compound derived from garlic. Arch Biochem Biophys 1993; 302: 337–42. [DOI] [PubMed] [Google Scholar]
- 53. Vang O, Jensen H, Autrup H. Induction of cytochrome P‐450IA1, IA2, IIB1, IIB2 and IIE1 by broccoli in rat liver and colon. Chem Biol Interact 1991; 78: 85–96. [DOI] [PubMed] [Google Scholar]
- 54. Gopalakrishnan R, Gupta A, Carlton PS, Morse MA, Stoner GD. Functional role of cytochrome p‐450 2a3 in N‐nitrosomethylbenzylamine metabolism in rat esophagus. J Toxicol Environ Health A 2002; 65: 1077–91. [DOI] [PubMed] [Google Scholar]
- 55. Morse MA, Lu J, Gopalakrishnan R et al. Mechanism of enhancement of esophageal tumorigenesis by 6‐phenylhexyl isothiocyanate. Cancer Lett 1997; 112: 119–25. [DOI] [PubMed] [Google Scholar]
- 56. Degawa M, Ueno H, Miura S, Ohta A, Namiki M. A simple method for assessment of rat cytochrome P‐448 isozymes responsible for the mutagenic activation of carcinogenic chemicals. Mutat Res 1988; 203: 333–8. [DOI] [PubMed] [Google Scholar]
- 57. Yahagi T, Nagao M, Seino Y, Matsushima T, Sugimura T. Mutagenicities of N‐nitrosamines on Salmonella . Mutat Res 1977; 48: 121–9. [DOI] [PubMed] [Google Scholar]
- 58. Yoo JS, Yang CS. Enzyme specificity in the metabolic activation of N‐nitrosodimethylamine to a mutagen for Chinese hamster V79 cells. Cancer Res 1985; 45: 569–74. [PubMed] [Google Scholar]
- 59. Kawanishi T, Ohno Y, Takanaka A et al. N‐nitrosodialkylamine dealkylation in reconstituted systems containing cytochrome P‐450 purified from phenobarbital‐ and beta‐naphthoflavone‐treated rats. Arch Toxicol 1992; 66: 137–42. [DOI] [PubMed] [Google Scholar]
- 60. Shu L, Hollenberg PF. Identification of the cytochrome P450 isozymes involved in the metabolism of N‐nitrosodipropyl‐, N‐nitrosodibutyl‐ and N‐nitroso‐n‐butyl‐n‐propylamine. Carcinogenesis 1996; 17: 839–48. [DOI] [PubMed] [Google Scholar]
- 61. Narayanan R, LeDuc B, Williams DA. Determination of the kinetics of rat UDP‐glucuronosyltransferases (UGTs) in liver and intestine using HPLC. J Pharm Biomed Anal 2000; 22: 527–40. [DOI] [PubMed] [Google Scholar]
- 62. Mori Y, Takahashi H, Yamazaki H et al. Distribution, metabolism and excretion of N‐nitrosobis (2‐hydroxypropyl) amine in Wistar rats. Carcinogenesis 1984; 5: 1443–7. [DOI] [PubMed] [Google Scholar]
- 63. Kokkinakis DM, Scarpelli DG, Subbarao V, Hollenberg PF. Species differences in the metabolism of N‐nitroso (2‐hydroxypropyl) (2‐oxopropyl) amine. Carcinogenesis 1987; 8: 295–303. [DOI] [PubMed] [Google Scholar]
- 64. Wiessler M, Rossnagel G. Alpha‐glucuronides of N‐nitrosomethylbenzylamine. In: Bartsch H, O’Neill I, Schulte‐Hermann R, eds. The Relevance of N‐Nitroso Compounds to Human Cancer, Exposure and Mechanism . Lyon: IARC, 1987; 170–2. [PubMed] [Google Scholar]
- 65. Wiench K, Frei E, Schroth P, Wiessler M. 1‐C‐glucuronidation of N‐nitrosodiethylamine and N‐nitrosomethyl‐n‐pentylamine in vivo and in primary hepatocytes from rats pretreated with inducers. Carcinogenesis 1992; 13: 867–72. [DOI] [PubMed] [Google Scholar]
- 66. Hodgson RM, Schweinsberg F, Wiessler M, Kleihues P. Mechanism of esophageal tumor induction in rats by N‐nitrosomethylbenzylamine and its ring‐methylated analog N‐nitrosomethyl (4‐methylbenzyl) amine. Cancer Res 1982; 42: 2836–40. [PubMed] [Google Scholar]
- 67. Huang TS, Lee SC, Lin JK. Suppression of c‐Jun/AP‐1 activation by an inhibitor of tumor promotion in mouse fibroblast cells. Proc Natl Acad Sci USA 1991; 88: 5292–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roe AL, Blouin RA, Howard G. In vivo phenobarbital treatment increases protein binding to a putative AP‐1 site in the CYP2B2 promoter. Biochem Biophys Res Commun 1996; 228: 110–4. [DOI] [PubMed] [Google Scholar]
- 69. Langouet S, Corcos L, Abdel‐Razzak Z, Loyer P, Ketterer B, Guillouzo A. Up‐regulation of glutathione S‐transferases alpha by interleukin 4 in human hepatocytes in primary culture. Biochem Biophys Res Commun 1995; 216: 793–800. [DOI] [PubMed] [Google Scholar]
- 70. Chung I, Jung K. No role of protected region B of human cytochrome P4501A2 gene (CYP1A2) as an AP‐1 response element. Arch Pharm Res 2002; 25: 375–80. [DOI] [PubMed] [Google Scholar]
- 71. Baker JR, Mason MM, Yerganian G, Weisburger EK, Weisburger JH. Induction of tumors of the stomach and esophagus in inbred Chinese hamsters by oral diethylnitrosamine. Proc Soc Exp Biol Med 1974; 146: 291–3. [DOI] [PubMed] [Google Scholar]
- 72. Ito N, Kamamoto Y, Hiasa Y, Makiura S, Marugami M. Histopathological and ultrastructural studies on esophageal tumors in rats treated with N‐nitrosopiperidine. Gann 1971; 62: 445–51. [PubMed] [Google Scholar]
- 73. Tanaka T, Makita H, Kawabata K et al. Modulation of N‐methyl‐N‐amylnitrosamine‐induced rat oesophageal tumourigenesis by dietary feeding of diosmin and hesperidin, both alone and in combination. Carcinogenesis 1997; 18: 761–9. [DOI] [PubMed] [Google Scholar]
- 74. Izbicki JR, Nagelschmidt M, Dornschneider G, Kusche J, Schmitz R. Proteolytic enzymes as new tumor markers in chemical carcinogenesis of intestinal tumors. Cancer Detect Prev 1987; 10: 31–6. [PubMed] [Google Scholar]
- 75. Rao CV, Rivenson A, Simi B, Reddy BS. Chemoprevention of colon carcinogenesis by dietary curcumin, a naturally occurring plant phenolic compound. Cancer Res 1995; 55: 259–66. [PubMed] [Google Scholar]
- 76. Soudamini KK, Kuttan R. Inhibition of chemical carcinogenesis by curcumin. J Ethnopharmacol 1989; 27: 227–33. [DOI] [PubMed] [Google Scholar]


