Abstract
The aberrant activation of Wnt signaling is a key process in colorectal tumorigenesis. Canonical Wnt signaling controls transcription of target genes via β‐catenin and T‐cell factor/lymphoid enhancer factor family transcription factor complex. Arm protein lost in epithelial cancers, on chromosome X 1 (ALEX1) is a novel member of the Armadillo family which has two Armadillo repeats as opposed to more than six repeats in the classical Armadillo family members. Here we examine cis‐regulatory elements and trans‐acting factors involved in the transcriptional regulation of the ALEX1 gene. Site‐directed mutations of a cyclic AMP response element (CRE) and an E‐box impaired the basal activity of human ALEX1 promoter in colorectal and pancreatic cancer cell lines. Moreover, overexpression of CRE‐binding protein (CREB) increased the ALEX1 promoter activity in these cell lines, whereas knockdown of CREB expression decreased the expression level of ALEX1 mRNA. Interestingly, luciferase reporter analysis and quantitative real‐time RT‐PCR demonstrated that the ALEX1 promoter was up‐regulated in a CRE‐dependent manner by continuous activation of Wnt/β‐catenin signaling induced by a glycogen synthase kinase‐3 inhibitor and overexpression of β‐catenin. These results indicate that the CRE and E‐box sites are essential cis‐regulatory elements for ALEX1 promoter activity, and ALEX1 expression is regulated by CREB and Wntk/β‐catenin signaling. (Cancer Sci 2010)
Wnt signaling is important for embryonic development, stem cell maintenance, and tumorigenesis.( 1 , 2 ) Two Armadillo (Arm) family members, β‐catenin and adenomatous polyposis coli (APC), are essential components of the Wnt signaling pathway. β‐Catenin is mainly localized to the plasma membrane in normal colon mucosa, whereas the cytoplasmic β‐catenin is phosphorylated by the degradation complex composed of casein kinase Ι, glycogen synthase kinase‐3β (GSK‐3β), APC, and AXIN, and subsequently degraded by the ubiquitin–proteasome pathway. In over 80% of colorectal cancers, β‐catenin aberrantly accumulates in the cytoplasmic and nuclear regions due to inactivating mutations in APC, forming a complex with the T‐cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors, leading to activation of the Wnt signaling pathway.( 3 , 4 ) To elucidate the role of the Wnt signaling in tumor development and stem cell maintenance, numerous candidate target genes of the β‐catenin/TCF have been identified.( 5 , 6 , 7 )
The cyclic AMP response element (CRE)‐binding protein/activating transcription factor (CREB/ATF) belongs to the basic region/leucine zipper transcription factor family.( 8 ) Traditionally, it has been believed that CREB dimers bind to CRE sites under basal conditions, but they are inactive; and that activation of CREB is mediated by phosphorylation at a specific serine residue. This phosphorylation promotes the association of CREB with the transcriptional co‐activator CREB‐binding protein and its paralogue p300, leading to activation of its target genes through histone modification and recruitment of an active transcription complex.( 8 ) It is noteworthy that CREB synergistically or predominantly activates some of its target genes through β‐catenin via CRE sites.( 9 , 10 , 11 )
Arm protein lost in epithelial cancers, on chromosome X (ALEX; also known as Armadillo repeat containing, X‐linked [ARMCX]) is a novel member of the Arm family which has one or two Arm repeats as opposed to more than six repeats in the classical Arm family members.( 12 ) The ALEX family, consisting of at least three variants (ALEX1, ALEX2, and ALEX3), is closely localized on chromosome Xq21.33–q22.2.( 12 ) Human ALEX1 is composed of four exons with the coding region residing entirely in a single exon and encodes a predicted protein of 453 amino acids which is highly conserved between humans and mice (95% amino acid similarity).( 13 ) ALEX1 mRNA is widely expressed in human tissues but is dramatically reduced or even undetectable in several human carcinoma cell lines and tissues, suggesting that ALEX1 may play a role as a tumor suppressor.( 12 ) However, the regulatory mechanism of the ALEX1 gene in normal and cancer cells remains largely unknown. Here we examined the transcriptional regulation of the ALEX1 gene in human cancer cell lines and found that ALEX1 expression is regulated by CREB and Wnt/β‐catenin signaling.
Materials and Methods
Cells and culture conditions. Human colon cancer cell lines HCT116 and SW480 were obtained from DS Pharma Biomedical (Osaka, Japan) and the Cell Resource Center for Biomedical Research, Tohoku University (Sendai, Japan), respectively. Human pancreatic cancer cell line PANC‐1 was provided by the RIKEN Cell Bank (Tsukuba, Japan). HCT116 and SW480 cells were cultured in DMEM (Invitrogen, Tokyo, Japan) and PANC‐1 cells were cultured in RPMI‐1640 (Invitrogen) supplemented with 10% heat‐inactivated FBS (Invitrogen) in a 5% CO2 atmosphere at 37°C.
Plasmid constructs and transfection. The putative promoter region of the human ALEX1 gene from −1933 to +487 was amplified by PCR and subcloned into the pCR‐Blunt II‐TOPO plasmid (Invitrogen). An approximately 2400‐bp DNA fragment containing the ALEX1 promoter sequence was inserted into the EcoRV and HindIII site of the pGL4.10 plasmid (Promega, Tokyo, Japan). The luciferase reporter plasmids driven by deleted mutant types of ALEX1 promoter were generated by the same PCR‐based method. Point mutations at potential TCF/LEF‐binding element (TBE(s)), CRE, and E‐box sites were introduced by PCR‐based site‐directed mutagenesis with PrimeSTAR polymerase (Takara Bio, Shiga, Japan).
Full‐length open reading frames for human ALEX1 and CTNNB1 genes (accession nos NM_016608 and NM_001904, respectively) were amplified by PCR and inserted into the XhoI site of pCAGIPuro plasmid (kindly provided by Dr. H. Niwa, RIKEN), designated as pCAGIPuro/ALEX1 and pCAGIPuro/β‐catenin, respectively. The plasmid containing the complete sequence of human ALEX1 gene and the cDNA derived from HCT116 cells which expresses both wild‐type and Ser45‐deleted β‐catenin were used as templates for the amplification of ALEX1 and CTNNB1 genes, respectively.( 12 ) The pCAGIPuro/EGFP plasmid (kindly provided by Dr. H. Niwa, RIKEN) encoding enhanced green fluorescence protein (EGFP) was used as a control.
Human TCF4 (also known as TCF7L2), ΔN‐TCF4, and CREB (CREBΔ isoform; accession no. NM 004379) genes were amplified by PCR and inserted into the EcoRI site of pCAGIPuro plasmid. Two dominant‐negative mutants of the CREBΔ gene, CREBR287L (KCREB) and CREBS119A (CREBM1 and mCREB) in which Arg287 and Ser119 is converted to Leu and Ala, respectively, were generated by PCR‐based site‐directed mutagenesis.
Plasmid transfections were performed by Lipofectamine 2000 or Lipofectamine LTX (Invitrogen) according to the manufacturer’s instructions. Primer sequences used for cloning and mutagenesis are available in Table S1 (Supporting information).
Luciferase reporter assay. The luciferase reporter assay was performed using the Dual‐Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. A total of 5000 cells were plated in a 96‐well plate. After overnight culture, cells were transfected with 100 ng of the expression plasmid of Firefly reporter gene driven by human ALEX1 promoter and 50 ng of Renilla luciferase control plasmid (pGL4.74; Promega) using Lipofectamine LTX and cultured for 48 h. TOPflash and FOPflash reporter plasmids (Millipore, Tokyo, Japan), which contain six wild‐type and six mutated TBE sites, respectively, were used to evaluate β‐catenin/TCF‐dependent transcriptional activity. To activate Wnt/β‐catenin signaling, cells were treated for 48 h with 6‐bromoindirubin‐3′‐oxime (BIO) (Merck, Tokyo, Japan), at a final concentration of 5 μM, 4 h post transfection. As a control, the cells were treated with deionized distilled water and 5 μM 1‐mehyl‐6‐bromoindirubin‐3′‐oxime (MeBIO) (Merck), respectively. Firefly and Renilla luciferases were measured by using the 1420 Multilabel Counter ARVO MX. The experiment was performed in triplicate and repeated at least three times.
Quantitative real‐time RT‐PCR. Total RNA was extracted by RNeasy mini kit (Qiagen, Tokyo, Japan) according to the manufacturer’s instructions. First‐strand cDNA was synthesized from 1 μg of total RNA using Transcriptor Reverse Transcriptase (Roche Diagnostics, Tokyo, Japan) in a total volume of 20‐μL reaction mixtures. Quantitative real‐time RT‐PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems, Tokyo, Japan) and the MX3000P Real‐time PCR System (Stratagene, La Jolla, CA, USA) according to the manufacturers’ instructions. Primer sequences are available in Table S1 (Supporting information).
siRNA transfection. PANC‐1 cells were transfected with the ON‐TARGETplus Non‐Targeting Control siRNA or siRNAs targeting CREB using DharmaFECT1 siRNA Transfection Reagent (Thermo Fisher Scientific, Lafayette, CO, USA), and cultured for 48 h. Quantitative real‐time RT‐PCR was performed using the Rotor‐Gene SYBR Green PCR kit (Qiagen) and the Rotor‐Gene Q PCR system (Qiagen). The following two ON‐TARGETplus siRNAs targeting CREB were used: siRNA1, GAGAGAGGUCCGUCUAAUG; and siRNA2, GCCCAGCCAUCAGUUAUUC.
Chromatin immunoprecipitation (ChIP). ChIP assay was carried out in accordance with a previous report with several modification.( 10 ) Immunoprecipitation was carried out with Dynabeads Protein G (Invitrogen) and polyclonal anti‐CREB antibody (#9197; Cell Singling Technology, Danvers, MA, USA). The primer pair used to amplify ALEX1 and cyclin D1 promoter sequences containing the putative CRE, and ALEX1 3′‐distal region sequence were as follows: ALEX1 promoter, 5′‐GCTGCTGATGGGAGTGGTA‐3′ and 5′‐CGGACCAAAC‐GAAGACTAGG‐3′; cyclin D1 promoter, 5′‐CTCCCGCTCCC‐ATTCTCT‐3′ and 5′‐ACTCTGCTGCTCGCTGCTAC‐3′; and ALEX1 3′‐distal, 5′‐ATGTGCAGCATGAGTCCAAG‐3′ and 5′‐ATTCCCATGGCCACCAGTA‐3′.
Indirect immunofluorescence. The cells were washed in PBS, fixed in 100% methanol at −20°C for 5 min and in 4% paraformaldehyde in PBS at room temperature for 30 min, and permeabilized with 0.2% Triton X‐100 in PBS at room temperature for 15 min. Permeabilized cells were washed three times with PBS, treated with 3% bovine serum albumin in PBS for 30 min, followed by incubation with antihuman β‐catenin antibody at 1:250 dilution at room temperature for 1 h. After washing three times in PBS, the cells were incubated with Rhodamine‐conjugated antimouse IgG antibody (Chemicon) at 1:100 in PBS at room temperature for 1 h. Finally, the cells were mounted in Vectashield Mounting Medium with DAPI (4′,6′‐diamidino‐2‐phenylindole; Vector Laboratories, Burlingame, CA, USA). The images were obtained using a Axiovert 200M fluorescent microscope (Carl Zeiss MicroImaging, Tokyo, Japan).
Statistical analysis. Statistics were performed using the Mann–Whitney U‐test for the expression analysis and luciferase reporter assay. A P‐value less than 0.05 was considered to be statistically significant.
Nucleotide sequence accession numbers. The sequences of human TCF4 gene and human ALEX1 promoter cloned in this study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers AB440195 and AB440194, respectively.
Results
CREB regulates human ALEX1 promoter activity. To identify candidates for cis‐regulatory elements involved in the regulation of ALEX1 gene expression, we carried out a computational search for transcription factor binding sites, and found a CREB/ATF binding site, CRE, and a basic helix‐loop‐helix transcription factors‐binding site, E‐box, at the proximal region of the transcription start site of human ALEX1 gene (Fig. 1a), which are highly conserved between humans and mice. In addition, six consensus TCF/LEF‐binding element (TBE) sites (site nos 8, 9, 10, 11, 12, and 13) and seven imperfect TBE sites (site nos 1, 2, 3, 4, 5, 6, and 7) are located in the upstream and genomic regions of human ALEX1 gene, respectively (Fig. 1a). Luciferase reporter analysis showed that human ALEX1 promoter (−1933 to +487), which included a CRE, an E‐box, and eight TBE sites, was active in HCT116, SW480, and PANC‐1 cells. The site‐directed mutations of each CRE and E‐box dramatically impaired the luciferase activities in HCT116, SW480, and PANC‐1 cells, whereas the mutations of eight TBE sites slightly up‐regulated the ALEX1 promoter activity (Fig. 1b). In addition, co‐transfection with a wild‐type CREB expression vector induced approximately 4.3‐, 2.0‐, and 2.1‐fold increase in ALEX1 promoter activities in HCT116, SW480, and PANC‐1 cells, respectively (Fig. 1c), whereas knockdown of CREB expression using two specific siRNAs decreased the ALEX1 mRNA level in PANC‐1 cells (Fig. 1d). These data indicate that both the CRE and E‐box sites are essential cis‐regulatory elements for basal promoter activity of the ALEX1 gene, and CREB up‐regulates the ALEX1 promoter activity.
Figure 1.
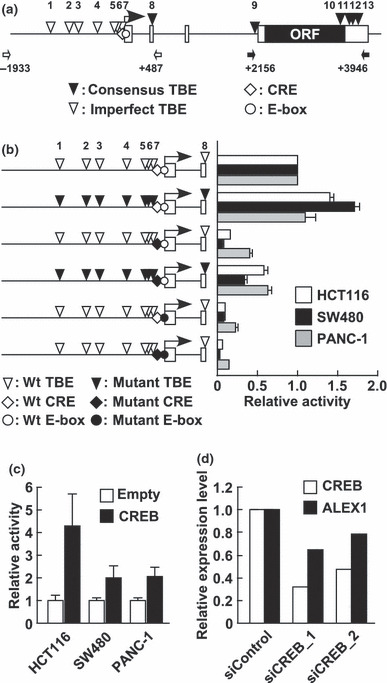
The cyclic AMP response element (CRE) site and CRE‐binding protein (CREB) are involved in human Arm protein lost in epithelial cancers, on chromosome X 1 (ALEX1) regulation. (a) A schematic representation of the genomic structure of the ALEX1 gene on human chromosome X. Open and filled boxes represent the exons of the ALEX1 gene and the ORF encoding the ALEX1 protein, respectively. The bent arrow indicates the transcription start site of the ALEX1 gene. The positions of consensus (CTTTGA/TA/T and A/TA/TCAAAG) and imperfect TCF/LEF‐binding element (TBE)s are indicated by filled and open reverse triangles, respectively, and numbered. An open diamond and circle represent the putative CRE (TGACGTG) and E‐box (CACGTG) site, respectively. The open and closed arrows indicate the primers used for promoter and intron 3‐exon 4 regions of the ALEX1 gene, respectively. (b) In the left diagram, open reverse triangles represent wild‐type TBE sites, and filled reverse triangles represent mutated TBE sites. Open and filled diamonds represent the wild‐type and mutant type of CRE, respectively. Open and filled circles represent the wild‐type and mutant type of E‐box, respectively. The right bar graph represents the relative luciferase activities in HCT116, SW480, and PANC‐1 cells transfected with the reporter plasmid driven by wild‐type and a series of site‐directed mutant type of the ALEX1 promoter. Error bars indicate the SD. (c) Luciferase reporter assay of HCT116, SW480, and PANC‐1 cells co‐transfected with wild‐type ALEX1 promoter‐driven reporter plasmid and the expression plasmid of the indicated genes. Error bars indicate the SD. (d) Quantitative real‐time RT‐PCR analysis of endogenous CREB and ALEX1 mRNA expression in PANC‐1 cells transfected with control siRNA and siRNAs targeting CREB. Relative expression level of CREB and ALEX1 mRNA for each sample was normalized against the expression level of GAPDH mRNA. The average of two independent experiments is shown.
Continuous activation of β‐catenin up‐regulates ALEX1 expression. The consensus TBE sequence recognized by the β‐catenin/TCF complex locates in the proximal promoter regions of several known target genes of canonical Wnt signaling( 6 , 7 , 14 ). To determine whether ALEX1 gene expression is regulated by Wnt/β‐catenin signaling, we examined the expression level and promoter activity of the ALEX1 gene in GSK‐3 inhibitor‐treated PANC‐1 cells. PANC‐1 cells exhibited low levels of β‐catenin expression and β‐catenin/TCF transcriptional activity (2, 3), and detectable level of endogenous ALEX1 mRNA by RT‐PCR. Activation of the β‐catenin/TCF target promoter (TOPflash) and cytoplasmic and nuclear accumulation of β‐catenin were markedly induced by GSK‐3 inhibitor BIO, but not by MeBIO (a kinase‐inactive analog of BIO) (Fig. 2a,b). The luciferase activity of ALEX1 promoter‐driven reporter plasmid was also increased in BIO‐treated PANC‐1 cells 24 h and 48 h post treatment in comparison to that in solvent‐treated control and MeBIO‐treated PANC‐1 cells (Fig. 2c). In addition, quantitative real‐time RT‐PCR revealed an increase of endogenous ALEX1 mRNA by BIO‐treatment (Fig. 2d). Lithium chloride, which is known to function as a GSK‐3 inhibitor, also induced an increase of ALEX1 mRNA (data not shown). These results demonstrate that activation of Wnt/β‐catenin signaling by GSK‐3 inhibition up‐regulates the promoter activity and gene expression of ALEX1.
Figure 2.
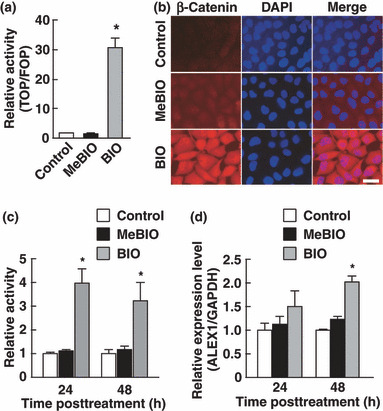
Glycogen synthase kinase‐3 (GSK‐3) inhibitor induces human Arm protein lost in epithelial cancers, on chromosome X 1 (ALEX1) expression. Luciferase reporter assay of PANC‐1 cells treated with/without a GSK‐3 inhibitor, 6‐bromoindirubin‐3′‐oxime (BIO), or an inactive analog of BIO, 1‐mehyl‐6‐bromoindirubin‐3′‐oxime (MeBIO). The bar graph represents the mean ratios between the luciferase activities from TOPflash and FOPflash reporter (TOP/FOP ratio) (a), and relative activities driven by wild‐type ALEX1 promoter (c). (b) Indirect immunofluorescense analysis for β‐catenin with PANC‐1 cells treated with control solvent, MeBIO, and BIO. Nucleus was stained with DAPI. (d) Quantitative real‐time RT‐PCR analysis of endogenous ALEX1 mRNA expressed in PANC‐1 cells treated with BIO or MeBIO for 24 and 48 h. Relative expression level of ALEX1 mRNA for each sample was normalized against the expression level of GAPDH mRNA. Error bars indicate the SD. *P < 0.05 compared to control and MeBIO‐treated cells.
Figure 3.
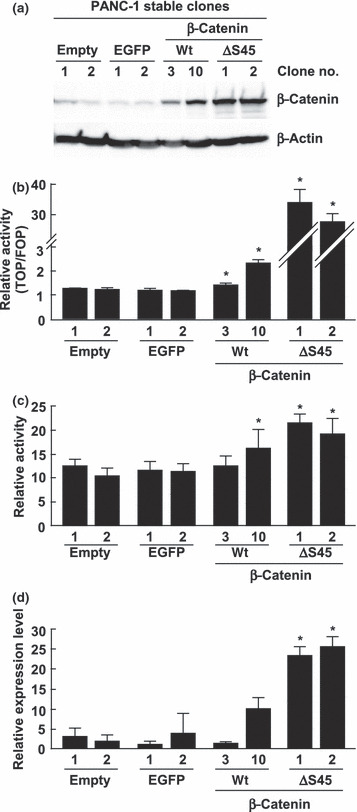
Overexpression of the constitutive‐active mutant type of β‐catenin induces human Arm protein lost in epithelial cancers, on chromosome X 1 (ALEX1) expression. (a) Western blot analysis for β‐catenin was performed with the cell lysate from stably transfected PANC‐1/empty, PANC‐1/EGFP, PANC‐1/β‐catenin‐wt, and PANC‐1/β‐catenin‐ΔS45 clones. β‐Actin protein served as an internal control. (b) Luciferase reporter assay of stably transfected PANC‐1 clones transfected with reporter constructs. The bar graph represents the mean ratios between the luciferase activities from TOPflash and FOPflash reporter (TOP/FOP ratio). The error bars represent the SD. *P < 0.05 compared to each control clone (empty nos 1 and 2, and EGFP nos 1 and 2). (c) Luciferase reporter assay of each stably transfected PANC‐1 clone transfected with wild‐type ALEX1 promoter‐driven reporter plasmid. The bar graph represents the relative luciferase activities, and the error bars represent the SD. *P < 0.05 compared to each control clone (empty nos 1 and 2, and EGFP nos 1 and 2). (d) Quantitative real‐time RT‐PCR analysis of endogenous ALEX1 mRNA of stably transfected PANC‐1 clone. The relative level of ALEX1 mRNA expression for each sample was normalized against the expression level of GAPDH mRNA. *P < 0.05 compared to each control clone (empty nos 1 and 2, and EGFP nos 1 and 2).
Furthermore, to validate whether cytoplasmic and nuclear accumulation of β‐catenin using genetic mutant also activates the ALEX1 gene, we generated the stable clones of PANC‐1 cells expressing the degradation‐resistant β‐catenin mutant with a deletion of Ser45, which is a phosphorylation site for GSK‐3β priming, designated as PANC‐1/β‐catenin‐ΔS45. As a result, two PANC‐1/β‐catenin‐ΔS45 clones showed higher expression levels of β‐catenin protein and activation of the target promoter of β‐catenin/TCF compared with those in the control PANC‐1/empty and PANC‐1/EGFP clones (Fig. 3a,b). Quantitative real‐time RT‐PCR and luciferase reporter analysis revealed significant elevations of endogenous mRNA expression and promoter activity of ALEX1 in the PANC‐1/β‐catenin‐ΔS45 clones (Fig. 3c,d), indicating that ALEX1 gene is regulated by Wnt/β‐catenin signaling.
Regulation of ALEX1 gene expression by β‐catenin is CRE dependent. Since comparable induction by BIO was obtained using the ALEX1 promoter‐driven reporter plasmids with/without intron 3‐exon 4 region containing five TBE sites (+2156 to +3946; Fig. 4a), we used the ALEX1 promoter‐driven reporter plasmid without intron 3‐exon 4 region to elucidate a regulatory region required for β‐catenin stimulation. A series of 5′ deletions and site‐directed mutations of TBE sites exhibited little impairment in response to Wnt/β‐catenin signaling induced by BIO (Fig. 4b). Furthermore, the activation of the ALEX1 promoter was suppressed by the overexpression of the N‐terminal deletion mutant of TCF4 (ΔN‐TCF4) known to block the transcriptional activity of β‐catenin due to lack of the β‐catenin binding domain (Fig. 4c). Previous studies have reported that transcription factors, including CREB, are capable of recruiting β‐catenin;( 10 , 15 , 16 , 17 ) thus, we evaluated the effect of the CRE site on β‐catenin‐mediated transactivation of the ALEX1 promoter by luciferase reporter assay. Interestingly, the mutation of the CRE site resulted in almost complete loss of induction by BIO (Fig. 4b). These data suggest that the CRE site plays an important role in mediating β‐catenin‐dependent transcriptional activation of the ALEX1 promoter, whereas the TBE sites play only a minor role although TCF4 may contribute to this activation.
Figure 4.
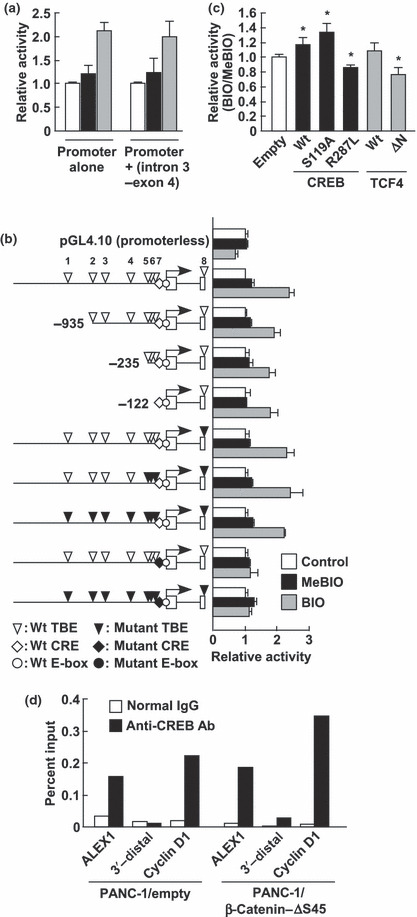
Cyclic AMP response element (CRE)‐site and CRE‐binding protein (CREB)‐mediated activation of human Arm protein lost in epithelial cancers, on chromosome X1 (ALEX1) promoter by the glycogen synthase kinase‐3 (GSK‐3) inhibitor. (a) Luciferase reporter assay of PANC‐1 cells transfected with wild‐type ALEX1 promoter‐driven reporter plasmid with/without the intron 3‐exon 4 region containing five TBE sites (+2156 to +3946). The bar graph represents the relative luciferase activities. (b) The left diagram is shown as described in the legend for Figure 1(b). The right bar graph represents the relative luciferase activities in control, 1‐mehyl‐6‐bromoindirubin‐3′‐oxime (MeBIO)‐, and 6‐bromoindirubin‐3′‐oxime (BIO)‐treated PANC‐1 cells transfected with the reporter plasmid driven by wild‐type, a series of 5′ deleted type, or a series of the site‐directed mutant type of human ALEX1 promoter. The error bars indicate the SD. (c) Luciferase reporter assay of PANC‐1 cells co‐transfected with wild‐type ALEX1 promoter‐driven reporter plasmid and expression plasmid of the indicated genes. The bar graph represents the mean ratios between the luciferase activities in PANC‐1 cells treated with BIO and MeBIO (BIO/MeBIO ratio), and the error bars represent the SD. *P < 0.05 compared to control (empty). (d) Chromatin immunoprecipitation followed by quantitative real‐time PCR was carried out with normal rabbit IgG or anti‐CREB antibody using the PANC‐1/empty clone 1 and PANC‐1/β‐catenin‐ΔS45 clone 1. Sequences of cyclin D1 promoter and 3′‐distal region of the ALEX1 gene were used as a positive and negative control for CREB binding, respectively.
The transcriptional relevance of CREB in regulating the ALEX1 expression in the PANC‐1/empty and the PANC‐1/β‐catenin‐ΔS45 clones was further confirmed by ChIP assay (Fig. 4d). To further investigate whether CREB mediates activation of the ALEX1 promoter by β‐catenin, wild‐type CREB and two dominant‐negative mutants, CREBR287L and CREBS119A, were overexpressed in BIO‐treated PANC‐1 cells. The luciferase reporter analysis for the ALEX1 promoter showed that the response to β‐catenin was enhanced by the wild‐type CREB but was suppressed by a DNA‐binding defective mutant CREBR287L (Fig. 4c), indicating that CREB, at least in part, mediates the transcriptional activation of the ALEX1 gene by Wnt/β‐catenin signaling. Surprisingly, a Ser119 phosphorylation‐defective mutant CREBS119A enhanced the activation of the ALEX1 promoter by BIO, suggesting that BIO‐induced activation of the ALEX1 promoter is independent of the phosphorylation of CREB at Ser119 which is important for transcriptional activation of several CREB target genes.
Discussion
Arm family members exert diverse functions through interactions of their Arm repeat domain with several binding partners. ALEX1 and its homologs are not classical members of the Arm family and represent a new family of proteins with less than six Arm repeats. Here we revealed the cis‐regulatory elements and trans‐acting factors involved in the transcriptional regulation of the ALEX1 gene. First, we demonstrated that potential CRE and E‐box sites were essential cis‐regulatory elements for basal transcription of the human ALEX1 gene. A CRE and an E‐box are known to be occupied by the CREB/ATF family and the basic helix‐loop‐helix family of transcription factors, respectively. Since overexpression and knockdown of CREB altered the ALEX1 promoter activity, CREB may play an important role in regulating ALEX1 gene expression. In addition, ChIP assay also supported the role of CREB in transcriptional activation of the ALEX1 gene via binding to the proximal promoter region including the CRE site. However, the response of the ALEX1 promoter to CREB induction was not diminished by the CRE mutation (Fig. S1, Supporting information). Several genes have been found to be regulated by CREB via multiple CRE sites, and point mutations in one CRE site alone are not enough to impair its promoter activity. Luciferase reporter analysis using a series of 5′ deletions of the ALEX1 promoter‐driven vector suggests that at least the proximal promoter region (−122 to +487) contains a CREB response sequence (Fig. S1, Supporting information). In the case of the ALEX1 gene, CREB might regulate via multiple CRE sites which are uncovered in this paper. Further studies are needed to identify transcription factors regulating ALEX1 gene expression through the E‐box site. Second, we showed that Wnt/β‐catenin signaling up‐regulated ALEX1 gene expression and transcriptional activation of the ALEX1 gene by β‐catenin mediated by the CRE site. It is believed that β‐catenin binds DNA indirectly through the interaction with a member of the TCF/LEF family because β‐catenin does not possess a DNA‐binding domain.( 18 ) However, a genome‐wide analysis of β‐catenin occupancy revealed a lack of consensus TBE motif in 16% of target genes,( 19 ) suggesting that β‐catenin binds to this group of targets indirectly through a nonconsensus TBE motif or an unrelated factor‐binding site. In fact, the transcription factors FOXO, Pit1, and Prop1 recruit β‐catenin to their target gene promoters.( 15 , 16 , 17 ) Furthermore, recent studies have provided evidence supporting the importance of CRE sites in Wnt signaling, since interaction with CREB is required for β‐catenin to activate the cyclin D1 and cyclooxygenase‐2 promoter.( 10 , 20 ) In this report, a GSK‐3 inhibitor and β‐catenin up‐regulated ALEX1 gene expression (2, 3). Importantly, BIO‐induced activation of the ALEX1 promoter was little affected by the mutations of TBE sites, but almost completely abolished by the mutation of the CRE site (Fig. 4a). Moreover, overexpression of CREB enhanced BIO‐induced ALEX1 promoter activation (Fig. 4b). Taken together, ALEX1 is thought to be regulated by Wnt/β‐catenin signaling; however, the regulation of ALEX1 gene expression by β‐catenin may be mediated by the CRE but not TBE sites. Third, the CREB‐mediated regulation of ALEX1 gene expression by β‐catenin was CREB phosphorylation independent. The CREBR287L mutant, which blocks binding of wild‐type CREB to CRE sites through heterodimer formation,( 21 ) was observed to reduce the response to β‐catenin (Fig. 4c). However, the overexpression of the CREBS119A mutant, which underwent a point mutation at a potential phosphorylation site by several kinases such as a cAMP‐dependent protein kinase, was observed to accelerate responses to β‐catenin at similar levels of induction as those by overexpression of wild‐type CREB (Fig. 4c). The phosphorylation of Ser119 is essential for the transcriptional activation of CREB, but not for DNA binding.( 8 ) Therefore, the binding of CREB to the CRE sites is considered to be a prerequisite for the transcriptional activation of the ALEX1 gene by β‐catenin, and CREB phosphorylation may be dispensable for this activation.
The dysregulation of the Wnt signaling can contribute to tumor development mainly by activation of target gene expression such as c‐myc and cyclin D1, whereas Wnt antagonists such as secreted frizzled‐related proteins, dickkopf‐1, and Wnt inhibitor factor‐1 are also up‐regulated by Wnt signaling, leading to the negative feedback regulation of Wnt signaling. The present data indicate that ALEX1 is regulated by Wnt/β‐catenin signaling. In addition, overexpression of ALEX1 suppresses the colony formation of colorectal cancer cell lines and is frequently down‐regulated in human colorectal cancer (manuscript in preparation). Thus, it is speculated that ALEX1 might act as a negative regulator of cell proliferation promoted by aberrant activation of Wnt/β‐catenin signaling.
In summary, the current results indicate that ALEX1 is a potential target gene of CREB, and Wnt/β‐catenin signaling regulates ALEX1 gene expression. The CRE is important for basal promoter activity and the transcriptional activation of ALEX1 gene response to β‐catenin. This is the first study to investigate the transcriptional regulation of the ALEX family, and these findings may provide new insights into the molecular mechanism underlying the CREB function and Wnt/β‐catenin signaling.
Supporting information
Table S1.Primer sequences used in this study.
Fig. S1.The proximal regulatory region of the Arm protein lost in epithelial cancers, on chromosome X 1 (ALEX1) gene responds to cyclic AMP response element binding protein (CREB) induction. Luciferase reporter assay of PANC‐1 cells co‐transfected with the reporter plasmid driven by a series of deletion and site‐directed mutant types of the ALEX1 promoter, and the CREB expression plasmid or control empty plasmid. Error bars indicate the SD.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Acknowledgments
The authors thank Hitoshi Niwa of the RIKEN Center for Developmental Biology for the generous gift of the pCAGIPuro and pCAGIPuro/EGFP plasmids. The authors also thank members of the Division of Functional Genomics and Systems Medicine for helpful discussion and advice. This work was partly supported by a Grant‐in‐Aid for Development of New Technology from The Promotion and Mutual Aid Corporation for Private Schools of Japan (2008).
References
- 1. Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005; 434: 843–50. [DOI] [PubMed] [Google Scholar]
- 2. Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res 2007; 13: 4042–5. [DOI] [PubMed] [Google Scholar]
- 3. Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol 2000; 18: 1967–79. [DOI] [PubMed] [Google Scholar]
- 4. Wong SC, Lo ES, Lee KC, Chan JK, Hsiao WL. Prognostic and diagnostic significance of β‐catenin nuclear immunostaining in colorectal cancer. Clin Cancer Res 2004; 10: 1401–8. [DOI] [PubMed] [Google Scholar]
- 5. Shtutman M, Zhurinsky J, Simcha I et al. The cyclin D1 gene is a target of the b‐catenin/LEF‐1 pathway. Proc Natl Acad Sci USA 1999; 96: 5522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He TC, Sparks AB, Rago C et al. Identification of c‐MYC as a target of the APC pathway. Science 1998; 281: 1509–12. [DOI] [PubMed] [Google Scholar]
- 7. Tetsu O, McCormick F. β‐Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999; 398: 422–6. [DOI] [PubMed] [Google Scholar]
- 8. Mayr B, Montminy M. Transcriptional regulation by the phosphorylation‐dependent factor CREB. Nat Rev Mol Cell Biol 2001; 2: 599–609. [DOI] [PubMed] [Google Scholar]
- 9. Xu L, Corcoran RB, Welsh JW, Pennica D, Levine AJ. WISP‐1 is a Wnt‐1‐ and b‐catenin‐responsive oncogene. Genes Dev 2000; 14: 585–95. [PMC free article] [PubMed] [Google Scholar]
- 10. Pradeep A, Sharma C, Sathyanarayana P et al. Gastrin‐mediated activation of cyclin D1 transcription involves β‐catenin and CREB pathways in gastric cancer cells. Oncogene 2004; 23: 3689–99. [DOI] [PubMed] [Google Scholar]
- 11. Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF‐1‐induced DNA bending and multiple protein‐protein interactions. Genes Dev 1995; 9: 995–1008. [DOI] [PubMed] [Google Scholar]
- 12. Kurochkin IV, Yonemitsu N, Funahashi SI, Nomura H. ALEX1, a novel human armadillo repeat protein that is expressed differentially in normal tissues and carcinomas. Biochem Biophys Res Commun 2001; 280: 340–7. [DOI] [PubMed] [Google Scholar]
- 13. Smith CA, McClive PJ, Sinclair AH. Temporal and spatial expression profile of the novel armadillo‐related gene, Alex2, during testicular differentiation in the mouse embryo. Dev Dyn 2005; 233: 188–93. [DOI] [PubMed] [Google Scholar]
- 14. Easwaran V, Lee SH, Inge L et al. β‐Catenin regulates vascular endothelial growth factor expression in colon cancer. Cancer Res 2003; 63: 3145–53. [PubMed] [Google Scholar]
- 15. Kioussi C, Briata P, Baek SH et al. Identification of a Wnt/Dvl/β‐Catenin ‐‐> Pitx2 pathway mediating cell‐type‐specific proliferation during development. Cell 2002; 111: 673–85. [DOI] [PubMed] [Google Scholar]
- 16. Olson LE, Tollkuhn J, Scafoglio C et al. Homeodomain‐mediated β‐catenin‐dependent switching events dictate cell‐lineage determination. Cell 2006; 125: 593–605. [DOI] [PubMed] [Google Scholar]
- 17. Essers MA, De Vries‐Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between β‐catenin and FOXO in oxidative stress signaling. Science 2005; 308: 1181–4. [DOI] [PubMed] [Google Scholar]
- 18. Xu W, Kimelman D. Mechanistic insights from structural studies of β‐catenin and its binding partners. J Cell Sci 2007; 120: 3337–44. [DOI] [PubMed] [Google Scholar]
- 19. Yochum GS, McWeeney S, Rajaraman V, Cleland R, Peters S, Goodman RH. Serial analysis of chromatin occupancy identifies β‐catenin target genes in colorectal carcinoma cells. Proc Natl Acad Sci USA 2007; 104: 3324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H, Wen S, Bunnett NW, Leduc R, Hollenberg MD, MacNaughton WK. Proteinase‐activated receptor‐2 induces cyclooxygenase‐2 expression through β‐catenin and cyclic AMP‐response element‐binding protein. J Biol Chem 2008; 283: 809–15. [DOI] [PubMed] [Google Scholar]
- 21. Shaywitz AJ, Greenberg ME. CREB: a stimulus‐induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 1999; 68: 821–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.Primer sequences used in this study.
Fig. S1.The proximal regulatory region of the Arm protein lost in epithelial cancers, on chromosome X 1 (ALEX1) gene responds to cyclic AMP response element binding protein (CREB) induction. Luciferase reporter assay of PANC‐1 cells co‐transfected with the reporter plasmid driven by a series of deletion and site‐directed mutant types of the ALEX1 promoter, and the CREB expression plasmid or control empty plasmid. Error bars indicate the SD.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


