Abstract
Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) are human gammaherpesviruses associated with numerous malignancies. Primary effusion lymphoma or body cavity-based lymphoma is a distinct clinicopathological entity that, in the majority of cases, manifests coinfection with KSHV and EBV. In previous analyses, we have characterized the EBV in the BC-1 and BC-2 cell lines as potential intertypic recombinants of the EBV types 1 and 2. In order to examine the infectious and transforming capacities of KSHV and the intertypic EBV recombinants from the BC-1 and BC-2 cell lines, viral replication was induced in these cell lines and fresh human primary B lymphocytes were infected with progeny virus. The transformed clones were analyzed by PCR and Western blotting. All analyzed clones were infected with the intertypic progeny EBV but had no detectable signal for progeny KSHV. Additionally, primary B lymphocytes incubated with viral supernatant containing KSHV alone showed an unsustained initial proliferation, but prolonged growth or immortalization of these cells in vitro was not observed. We also show that the EBV recombinants from BC-1 were less efficient than the EBV recombinants from BC-2 in the ability to maintain the transformed phenotype of the infected human B lymphocytes. From these findings, we conclude that the BC-1 and BC-2 intertypic EBV recombinants can immortalize human primary B lymphocytes, albeit at different levels of efficiency. However, the KSHV induced from BC-1 and BC-2 alone cannot transform primary B cells, nor can it coinfect EBV-positive B lymphocytes under our experimental conditions with B lymphocytes from EBV-seropositive individuals. These results are distinct from those in one previous report and suggest a possible requirement for other factors to establish coinfection with both viral agents.
Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) are the only two known human gammaherpesviruses. EBV (human herpesvirus 4 [HHV-4]) is present in the vast majority of individuals and establishes latent asymptomatic infection in B lymphocytes (23). EBV has been associated with several different human malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and various immunoblastic lymphomas (for a review, see reference 32). KSHV (HHV-8) is a recently discovered gammaherpesvirus belonging to the genus Rhadinovirus (6, 12). It has sequence similarity to herpesvirus saimiri, murine herpesvirus 68, and EBV, all of which have tumorigenic capacity (31). KSHV has been found in all epidemiological forms of Kaposi's sarcoma (21). In addition, KSHV has been detected in primary effusion lymphomas (PELs) (10, 30), in multicentric Castleman's disease (17, 46; A. Gessain, A. Sudaka, J. Briere, N. Fouchard, M. A. Nicola, B. Rio, M. Arborio, X. Troussard, J. Audoin, J. Diebold, et al., Letter, Blood 87:414–416) and in dendritic cells of patients with multiple myeloma (36). Despite the discovery of KSHV's association with many different pathological entities, it is still unclear whether the virus plays a causal role in the onset or manifestation of these diseases.
Primary effusion lymphomas, also known as body cavity-based lymphomas (BCBLs), are a subset of non-Hodgkin's lymphomas with distinctive clinical and biological features (30). PELs are found mostly in patients with AIDS and grow mainly in the body cavities without contiguous tumor masses. Among the distinctive characteristics of PELs are an intimate association with KSHV (110, 32, 331; A. Carbone, U. Tirelli, A. Gloghini, C. Pastore, E. Vaccher, and G. Gaidano, Letter, Eur. J. Cancer 32A:555–556; D. S. Karcher and S. Alkan, Letter, N. Engl. J. Med. 333:797–799) and the presence of EBV in the vast majority of cases (30). In contrast to PELs, only a minority of the solid Kaposi's sarcoma tumors are positive for EBV (16). A growing interest in the analysis of KSHV and EBV coinfection and in the elucidation of a potential synergy between the two latently infected genomes provides considerable motivation for the close characterization of PELs. The initial establishment of the KSHV and EBV coinfected PEL cell lines, BC-1 and BC-2, by Cesarman and colleagues has provided important reagents for the analysis of the KSHV and EBV genomes and any interactions occurring between the coinfecting viruses (11).
Two distinct types of EBV have been isolated and characterized. Originally designated A and B, these two different virus types are now referred to as types 1 and 2, in keeping with the herpes simplex system of nomenclature (37). Even though type 1 and type 2 EBV (EBV-1 and -2) are largely the same throughout most of the genome, the viral genomes can be typed based on known genomic markers. The EBNA loci can be typed at the nucleotide level by PCR (43) or at the protein level by the type-specific reactivity of EBNA epitopes with human sera (41, 42, 45). Virus isolation studies of EBV derived from healthy patients have demonstrated that EBV-1 is most prevalent in these individuals and is the only virus type present in at least 90% of the examined cases (18, 50, 51). However, virus isolation studies of certain T-cell-immunocompromised, human immunodeficiency virus-positive cohorts have shown that EBV-2 exists in much greater proportion in these groups (7, 44, 50, 52, 53). The predominance of a single transforming virus strain, most commonly EBV-1 rather than EBV-2, has been demonstrated in healthy individuals; however, increasing evidence suggests that multiple EBV infections are common within immunocompromised hosts (22, 44, 51, 53). Previous studies have documented the existence of intertypic recombinants of EBV-1 and -2 in both a healthy adult (8) and T-cell-immunocompromised individuals (52).
In earlier studies of BC-1 and BC-2, we have characterized the EBV infecting these cell lines as intertypic recombinants of EBV-1 and -2 (1). The discovery of such intertypic recombinants existing in transformed lymphoblastoid cell lines (LCLs) prompts one to ask whether these intertypic EBV recombinants are responsible for the transformed phenotype and whether they can efficiently transform primary B lymphocytes. Previous work has also shown the ability of these viruses to transform B cells (27). However, the question of whether the KSHV contained in BC-1 and BC-2 can infect and/or immortalize human primary B cells as coinfected or singly infected B lymphocytes is important in understanding the initiation and maintenance of the transformed phenotype. In this work, we have shown that the EBV recombinants in BC-1 and BC-2 can infect and immortalize fresh T-cell-depleted peripheral blood mononuclear cells (PBMC); however, these viruses vary in the efficiency with which they transform B lymphocytes. We have also shown that KSHV from BC-1 and BC-2 alone cannot infect and immortalize human primary B cells. Furthermore, KSHV from BC-1 and BC-2 cannot be established as a coinfection with EBV in immortalized B lymphocytes in our system. Previous work by Haas and colleagues showed immortalization of human primary B lymphocytes by KSHV and coinfection with EBV by using PBMC from EBV-seropositive individuals (24). Further investigations into the ability of KSHV and EBV to coinfect human B lymphocytes will help explain the variations in results from our studies and from previously reported studies.
MATERIALS AND METHODS
Antibodies and cell cultures.
Latent EBNA proteins were analyzed with antibodies derived from a patient serum previously characterized for identification of each of the EBNA proteins (38). Serum samples were adsorbed against the EBV-negative Burkitt's lymphoma cell line BJAB lysate at 4°C for 48 h. Adsorbed serum was then used as a 1:50 dilution in phosphate-buffered saline (PBS) with 1 mM sodium azide. The hybridoma cell lines PE2, A10, and S12 against EBNA2, EBNA3C, and LMP1, respectively, were cultured, and the supernatant was used as a 1:2 dilution in PBS with 1 mM sodium azide. Antiserum recognizing the EBV lytic antigens was obtained from patient samples which had been previously characterized (40). The serum was typically diluted 1:50 in PBS after absorption with cell lysate from the EBV-negative cell line BJAB.
BJAB is an EBV-negative cell line obtained from Elliott Kieff. The B95-8 cell line harbors EBV-1, and the Jijoye cell line is infected with EBV-2 (28, 29). The P3HR-1 cell line is derived from its Jijoye parent and also contains EBV-2; however, the P3HR-1 genome has regions in the EBNALP and EBNA2 genes deleted (34). BC-1, BC-2, and BC-3 were purchased from the American Type Culture Collection. BCBL-1 was obtained from the AIDS Reference and Reagent Program (35). BC-1 and BC-2 are coinfected with EBV and KSHV (20). BC-3 and BCBL-1 are infected with KSHV alone (3, 35). The cell lines were maintained in complete medium, which consisted of RPMI 1640 supplemented with 15 to 20% inactivated fetal bovine serum, 2 mM glutamine, and 10 μg of gentamicin per ml. All cell lines were monitored and routinely fed with fresh medium every 3 to 4 days. BC-1, BC-2, BC-3, and BCBL-1 grew best when maintained at a density of at least 400,000 to 500,000 cells per ml.
Primary B-lymphocyte infections.
Lytic replication was induced in the BC-1, BC-2, BC-3, BCBL-1, B95-8, and Jijoye cell lines with a combination of tetradecanoyl phorbol acetate (TPA) and sodium butyrate. We have established a specific titration of 20 ng of TPA/ml and 2.5 mM sodium butyrate for inducing lytic replication of BC-1, BC-2, BC-3, and BCBL-1. The resultant virus supernatant was passed through a 0.45-μm-pore-size filter and used to infect primary human B lymphocytes. The virus was incubated for 2 h at 37°C with 5 × 107 T-cell-depleted human PBMC. The PBMC were obtained from adult EBV-seropositive donors, and T cells were removed with 2-aminoethyl isothiouronium bromide (Sigma)-treated sheep erythrocytes. Infected primary B cells were resuspended in complete medium at a concentration of 3.3 × 105 per ml and distributed at 150 μl (5 × 104 cells) per well in a 96-microwell plate (40, 48). The cultures were fed 14 days postplating with 100 μl of fresh complete medium. LCLs were macroscopically visible 1 to 4 weeks after plating. The cell lines were expanded in culture for further genetic and biochemical analyses.
PCR analyses. (i) PCR primers.
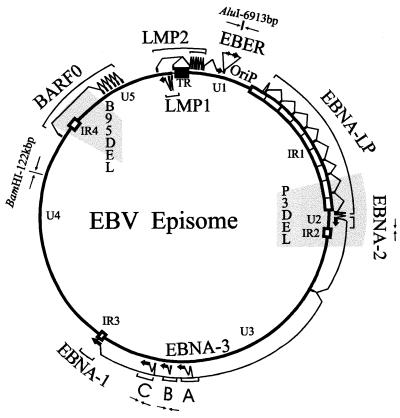
Primers corresponding to five different markers in the EBV genome were used in the PCR typing analyses (Fig. 1). The primer combinations which amplify distinctive EBV-1 and -2 fragments of the EBNA2, EBNA3B, and EBNA3C genes have been previously described (43, 47, 48). The BamHI-C-AluI primers and the Bam122 primers have also been described (49). The BamHI-C-AluI primers amplify a 157-bp fragment for both EBV-1 and -2; however, only the EBV-1 PCR product can be digested with AluI into fragments of 52 and 105 bp. The Bam122 primers produce a PCR product for both type 1 and type 2 genomes. This PCR product contains a BamHI site in the type 1, B95-8 DNA but not in type 2, P3HR-1 DNA. PCR analysis for the presence of the KSHV viral genome in the progeny LCLs was done with primers which amplify a 233-bp fragment (KS330). The sequences of these oligonucleotides have been previously reported (12).
FIG. 1.
Schematic of the EBV genome showing the locations of the three genomic markers used in PCR typing analyses of the BC-1 and BC-2 progeny EBV. The genomic markers are an AluI restriction site at 6,913 bp in the region adjacent to the EBV early RNA region, type-specific polymorphisms within the EBNA3B gene, and a BamHI restriction site located at approximately 122 kbp. The locations of the P3HR-1 and B95-8 deletions are also shown shaded in gray.
(ii) Preparation of viral DNA for PCR analysis.
The supernatant from the induced cell lines used in primary B-lymphocyte infections was prepared and analyzed for the presence of virus. Supernatant (0.5 ml) was centrifuged at 15,000 rpm (International Equipment Co.) for 30 min to pellet the virus particles. The pellet was resuspended in 60 μl of 0.2× PBS and heated at 95°C for 30 min. The solution was then incubated with proteinase K (0.75 ng/μl) at 55°C for 1 h, followed by inactivation of the enzyme for another hour. Six microliters of each preparation of viral DNA was used for PCR analysis.
(iii) PCR and restriction endonuclease digestion.
Crude DNA extracts were prepared for PCR by proteinase K treatment of harvested cells. Cells from 0.5 ml of a well-growing culture were resuspended in 250 μl of 0.2× PBS. This solution was then incubated with proteinase K (0.75 ng/μl) at 55°C for 1 h, followed by inactivation of the enzyme at 95°C for another hour. Five microliters of these DNA solutions was then added to the reaction components to give a 25-μl reaction mixture. The PCR was carried out in 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.5 mM (each) deoxynucleoside triphosphate, 0.1 μM (each) primer, and 0.75 U of Taq polymerase. DNA was amplified in an MJ Research, Inc., thermal cycler machine. A total of 40 cycles were run, each consisting of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C. PCR-amplified DNA was analyzed by electrophoresis in a 1% (wt/vol) ME-agarose, 2% (wt/vol) Nusieve agarose gel and stained with ethidium bromide. For restriction endonuclease digestion, the entire PCR mixture was precipitated in 100% ethanol and resuspended in 10 μl of Tris-EDTA buffer. The DNA was then digested overnight in a 30-μl reaction mixture, based on the manufacturer's suggested protocol.
Immunoblot analysis.
Harvested cells were dispersed mechanically and dissolved in sodium dodecyl sulfate (SDS) lysis buffer. In order to compensate for low levels of expression of viral proteins in BC-1 and BC-2 parental and progeny cell lines, the number of harvested cells from these lines was increased twofold (1,000,000 cells) compared to the number harvested from the B95-8, P3HR-1, and BJAB cell lines (500,000 cells). The cell lysates were fractionated by SDS–8% polyacrylamide gel electrophoresis, and the proteins were transferred to 0.45-μm Bio-Rad nitrocellulose membranes. The membranes were analyzed according to the manufacturer's protocol and incubated in the human serum dilutions at 4°C overnight. Protein A-horseradish peroxidase secondary antibody was used at a 1:7,500 dilution, and anti-mouse immunoglobulin-horseradish peroxidase secondary antibody was used at a 1:2,500 dilution. Proteins were detected with chemiluminescence reagents (Amersham). Images were scanned and prepared for publication with Corel Draw.
RESULTS
Intertypic EBV recombinants from BC-1 and BC-2 immortalize human primary B lymphocytes in vitro.
Previous typing analyses of the EBV genomes in the BC-1 and BC-2 cell lines demonstrated that these viruses are intertypic recombinants of EBV1 and -2 (1). In order to detect the presence of this intertypic recombinant EBV in the progeny BC-1 and BC-2 cell lines, we chose six cell lines derived from each of the BC-1 and BC-2 infections and analyzed representative genomic markers by PCR typing assays. The first marker we examined is located adjacent to the EBV early RNAs. An AluI site is located at 6,913 bp in the prototypic type 1 B95-8 genome but not in the prototypic type 2 P3HR-1 genome. This restriction site is also absent in the BC-1 and BC-2 viral genomes. To further identify the EBV in the transformed BC-1 and BC-2 progeny cell lines, we analyzed for the known type 1 and type 2 polymorphisms in the EBNA2, -3B, and -3C genes. With primers specific for these loci, we used PCR analysis to demonstrate differences in the amplification products for the type 1 and type 2 alleles. The BC-1 viral genome shows type 2 characteristics at these loci, whereas the BC-2 viral genome shows type 1 polymorphisms at these markers. Using primers that amplify from nucleotides 122206 to 122475, we also analyzed the BC-1 and BC-2 progeny EBV genomes for the presence of a BamHI restriction site within this amplification fragment. DNA from the B95-8 cell line and the BC-1 and BC-2 parental cell lines has a BamHI restriction site within this PCR product, while in P3HR-1 DNA this restriction site is absent (4, 19). The data from these typing experiments suggest that the EBV infecting the BC-1 and BC-2 progeny cell lines are the intertypic recombinants derived from the parental cell lines. Table 1 provides a summary of the PCR analyses of the progeny cell lines chosen in these series of experiments.
TABLE 1.
Classification of EBV genome types at five different locia
| Cell line | Genomic marker
|
|||
|---|---|---|---|---|
| AluI (6,913 bp) | EBNA2 | EBNA3B, -3C | BamHI (122 kbp) | |
| BC-1 parent | T2 | T2 | T2, T2 | T1 |
| BC1.1 | T2 | T2 | T2, T2 | T1 |
| BC1.2 | T2 | T2 | T2, T2 | T1 |
| BC1.4 | T2 | T2 | T2, T2 | T1 |
| BC1.13 | T2 | T2 | T2, T2 | T1 |
| BC1.16 | T2 | T2 | T2, T2 | T1 |
| BC1.22 | T2 | T2 | T2, T2 | T1 |
| B95-8 | T1 | T1 | T1, T1 | T1 |
| Jijoye or P3HR-1 | T2 | T2 | T2, T2 | T2 |
| BC-2 parent | T2 | T1 | T1, T1 | T1 |
| BC2.5 | T2 | T1 | T1, T1 | T1 |
| BC2.7 | T2 | T1 | T1, T1 | T1 |
| BC2.9 | T2 | T1 | T1, T1 | T1 |
| BC2.12 | T2 | T1 | T1, T1 | T1 |
| BC2.14 | T2 | T1 | T1, T1 | T1 |
| BC2.18 | T2 | T1 | T1, T1 | T1 |
| B95-8 | T1 | T1 | T1, T1 | T1 |
| Jijoye or P3HR-1 | T2 | T2 | T2, T2 | T2 |
Summary of data acquired from typing analyses of BC-1 and BC-2 parent and progeny viruses. Previous studies have shown that the EBV harbored in the BC-1 and BC-2 cell lines are intertypic recombinants of the prototypic type 1 (T1) and type 2 (T2) viral genomes (1). The EBNA2, -3B, and -3C markers were analyzed with previously described type-specific primers (43, 49). The genomic types at the AluI (6,913-bp) and BamHI (122-kbp) markers were determined by restriction endonuclease digestion of PCR-amplified products. The AluI (6,913-bp) and the BamHI (122-kbp) markers have been previously shown to segregate according to genomic type at each locus (2, 49).
PCR analysis does not detect KSHV DNA in BC-1 and BC-2 progeny cell lines.
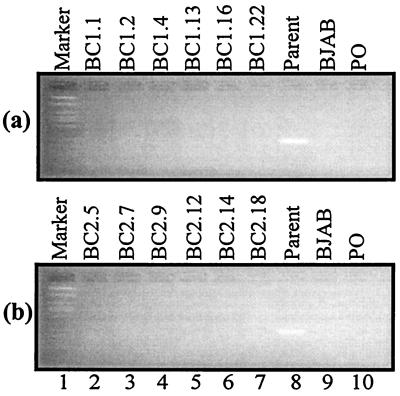
Using KS330 primers that amplify a distinctive 233-bp fragment from the KSHV genome, we performed PCR analysis for the presence of KSHV in the BC-1 and BC-2 progeny cell lines. Our analysis clearly indicates the presence of the KSHV genome in the parental BC-1 and BC-2 cell lines (Fig. 2, lane 8). However, PCRs did not yield any amplification products for the progeny BC-1 and BC-2 (Fig. 2, lanes 2 to 7). We analyzed progeny cell lines from three separate experiments for both BC-1 and BC-2. More than 500 LCLs were screened for the presence of KSHV DNA. No signal was seen from PCR of progeny LCL DNAs; however, our positive controls were clearly detecting KSHV DNA. Thus, we conclude that KSHV is not present in these progeny cell lines.
FIG. 2.
PCR analysis for the presence of KSHV in the BC-1 and BC-2 progeny cell lines by using KS330 primers, which amplify a distinct 233-bp fragment. (a) BC-1 analyses; (b) BC-2 analyses. The appearance of a KS330 amplification product signifies infection with KSHV. BC-1 and BC-2 parents show the presence of KSHV (lanes 8), but the BC-1 and BC-2 progeny cell lines do not appear to be infected with KSHV (lanes 2 to 7). BJAB (lane 9) is an EBV-negative and KSHV-negative cell line. PO denotes a control reaction run in the absence of any source of template DNA.
KSHV is not capable of immortalizing T-cell-depleted human primary B lymphocytes in vitro.
BC-3 and BCBL-1 are BCBL cell lines that are singly infected with KSHV. In order to examine the infection of primary B lymphocytes by KSHV, we harvested virus after the induction of viral replication with TPA and sodium butyrate. We incubated T-cell-depleted PBMCs from EBV-seropositive individuals with BC-3 and BCBL-1 supernatants containing KSHV. In EBV-seropositive individuals, approximately 1 in 105 to 106 of their peripheral-blood B lymphocytes is EBV infected (23). Previous infection studies have shown that the outgrowth of spontaneous LCLs in vitro due to EBV from seropositive individuals is rare (49). After incubation with BC-3 and BCBL-1 supernatants, the cells were plated in 96-microwell plates and observed for evidence of proliferation and immortalization. Table 2 shows the results of three separate experiments. By 2 weeks after plating, initial proliferation and clumping were observed in all wells with a change in pH, as detected by a color change in the medium. After 2 weeks, however, B-cell proliferation did not persist, and no evidence of prolonged growth transformation and immortalization of these cells was observed. Similar initial unsustained proliferation has been seen previously when primary human B lymphocytes were incubated with medium containing TPA and butyrate (E. S. Robertson, unpublished observations). We suggest that the initial proliferation observed in this experiment is not caused by KSHV infection and subsequent mediation of transformation. Rather, we propose that such initial proliferation results from the TPA and butyrate in the media. However, it is possible that KSHV infects primary B cells and triggers proliferation of these cells but this proliferation is not maintained due to the absence of other critical factors.
TABLE 2.
Infection of primary human B lymphocytes by KSHVa
| Expt | Virus | No. of wells positive for B-cell proliferation
|
|||
|---|---|---|---|---|---|
| 1 wk | 2 wk | 4 wk | 12 wk | ||
| 1 | BC-3 | 0 | 96a | 0 | 0 |
| BCBL-1 | 0 | 96a | 0 | 0 | |
| 2 | BC-3 | 0 | 96a | 0 | 0 |
| BCBL-1 | 0 | 96a | 0 | 0 | |
| 3 | BC-3 | 0 | 96a | 0 | 0 |
| BCBL-1 | 0 | 96a | 0 | 0 | |
Initial unsustained proliferation observed in all wells with change in pH as detected by color change in medium and clumping of cells. Similar initial proliferation has been seen when primary human B lymphocytes are incubated with medium containing TPA and butyrate, usually used to induce lytic replication (E. S. Robertson, unpublished observations). We suggest that this observation may be a result of TPA and butyrate which were present in the medium during infection.
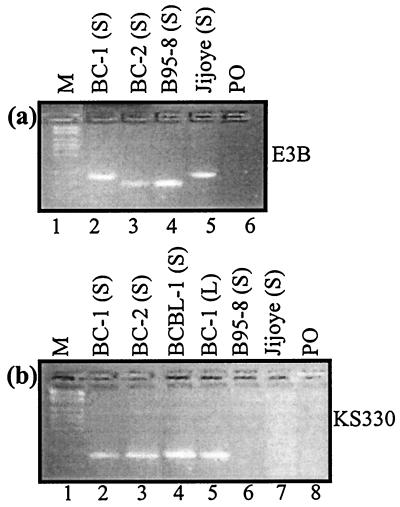
One important question raised in these experiments was the possibility that the KSHV was not easily induced by TPA and sodium butyrate. Additionally, we wanted to approximate the relative levels of virion particles present in the supernatant. Semiquantitative analysis with KS330 primers (12) for PCR of viral DNA indicated that similar levels of viral DNA were present (Fig. 3B). Moreover, the induction of KSHV was similar for the EBV-positive (BC-1 and BC-2) and EBV-negative (BCBL-1) cell lines. Therefore, KSHV was induced and present at relatively equivalent levels for infection of primary B lymphocytes.
FIG. 3.
PCR analysis for the presence of virus in the supernatants of cells induced for production of EBV and KSHV. The source of the template DNA is indicated in parentheses in the label for each lane. (S), virus supernatant; (L), cell lysate. (A) Analysis with type-specific primers for EBNA3B; (B) 233-bp products generated with the KSHV-specific primers. BC-1 and BC-2 are cell lines which are coinfected with EBV and KSHV. BCBL-1 is infected with only KSHV. B95-8 and Jijoye are the prototypic EBV-1 and -2 strains, respectively.
LCLs derived from BC-1 and BC-2 expressed essential latent antigens.
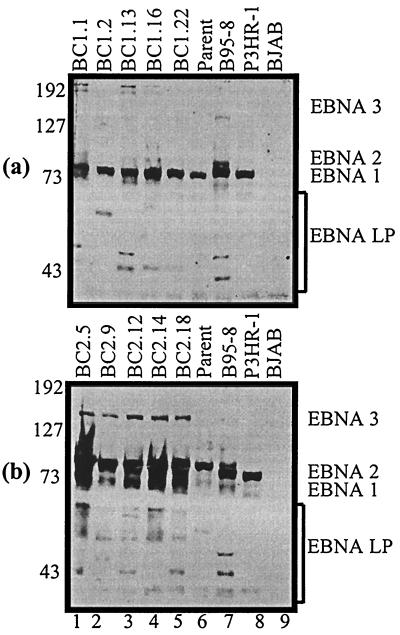
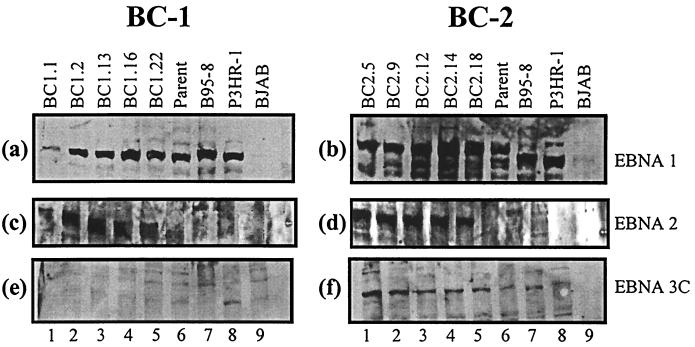
Using adsorbed, specific EBNA human serum capable of detecting EBNA1, EBNALP, EBNA2, and EBNA3A, -3B, and -3C (40), we analyzed membranes blotted with fractionated cell lysates from BC-1 and BC-2 parents and progeny (Fig. 4). EBNA1 is expressed in all the cell lines analyzed. The intensity of the EBNA1 signal was lower in BC-1 (Fig. 4a) than in BC-2 (Fig. 4b). The notable size variability of the EBNA1 protein from BC-2 can be attributed to different numbers of IR3 repeats located within the EBNA1 coding region. Because EBNA2 was not readily distinguishable in the BC-1 and BC-2 cell lines, we postulated that the EBNA2 band was comigrating with the EBNA1 band. To test this hypothesis, we blotted separately for the EBNA1 and EBNA2 proteins. A human serum that recognizes only the EBV EBNA1 antigen was used to detect EBNA1 expression. Immunoblot analysis indicates that EBNA 1 is expressed in all progeny cell lines (Fig. 5a and b). Numerous breakdown products were seen to be migrating faster on the blot in the BC-2 analysis. EBNA2 expression was determined by blotting with supernatant from a hybridoma cell line specific for detection of the EBNA2 protein (Fig. 5c and d). This analysis clearly indicated that EBNA2 was expressed in both the BC-1 and BC-2 progeny cell lines and was comigrating with the EBNA1 protein. However, as expected, the BC-1 and BC-2 parental cell lines did not express detectable levels of EBNA2 (Fig. 5c and d, lanes 6) (9). Expression of the EBNA3C protein was clearly seen in the BC-2 progeny but was almost undetectable in the BC-1 progeny (Fig. 5e and f). Since the monoclonal antibodies were made to type 1 EBNA3C antigens, the absence of or lowered EBNA3C detection in BC-1 cell lines may represent the type specificity of the antibodies used and not the actual expression patterns (26). Some signals for EBNA3C were seen in the parental lanes in BC-1 and BC-2; however, this is probably due to the nonspecific signals seen in most of the lanes after prolonged exposure. EBNALP can typically be seen as a ladder of bands from 17 to 73 kDa due to the multiple splicing of the BamHI W repeats that span the open reading frame. A number of bands are seen in this region in the progeny cell lines, which indicates expression of EBNALP in all progeny lines derived from BC-1 and BC-2 (Fig. 4).
FIG. 4.
Western blot analysis for latent EBNA expression by using human serum specific for EBNA recognition. (a) BC-1 analyses; (b) BC-2 analyses. Latent EBNA proteins are labeled. EBNA1 and EBNA2 appear to be comigrating on SDS–8% polyacrylamide gel electrophoresis. Size differences in the EBNA1 protein are due to different numbers of the IR3 repeats located in the coding region. B95-8 and P3HR-1 are the type 1 and type 2 controls, respectively. BJAB is an EBV-negative cell line. Molecular masses (in kilodaltons) are shown on the left of each gel.
FIG. 5.
Western blot analysis with antibodies specific for EBNA1, -2, and -3. Western blots of BC-1 (a) and BC-2 (b) with human serum specific for EBNA1 recognition are shown. Cross-reactive antibodies to EBNA1 recognized both type 1 and type 2 variants of the protein. All BC-1 and BC-2 parents and progeny express the EBNA1 protein. Western blots with monoclonal antibodies specific for EBNA2 are shown for BC-1 (c) and BC-2 (d). The progeny cell lines appear to be expressing EBNA2 (lanes 1 to 5), but the parent cell lines do not (lane 6). These results confirm that EBNA2 was comigrating with EBNA1 in our experiments. Western blots with monoclonal antibodies specific for recognition of the EBNA3C protein are shown for BC-1 (e) and BC-2 (f), respectively. Since the monoclonal antibodies were made in response to type 1 antigen, apparent expression differences between BC-1 and BC-2 may be attributable to type-specific differences in antigen recognition.
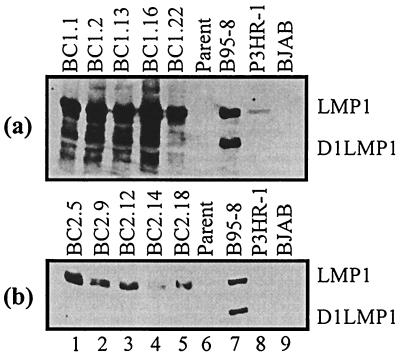
Expression of the essential EBV latent protein LMP1 was analyzed in the progeny cell lines by Western blotting with supernatant from the S12 hybridoma cell line that specifically detects LMP1 (Fig. 6) (25). All progeny LCLs show reasonably high levels of LMP1 expression. A predominant lower band typical of the D1LMP1 protein can be seen in the B95-8 control lanes. This D1LMP1 signal and other smaller bands were observed in BC-1 progeny. This may indicate relatively high levels of spontaneous lytic replication. Additionally, the BC-1 progeny cell lines express LMP1 at markedly higher levels than do the BC-2 progeny. Moreover, these same LCLs expressed high levels of the D1LMP1 variant. However, this form of LMP1 was not detectable in the analyzed BC-2 progeny LCLs.
FIG. 6.
Western blot of BC-1 (a) and BC-2 (b) parent and progeny with a hybridoma cell line specific for the LMP1 S12. The predominant lower band in the B95-8 lane represents the D1LMP1 protein. Most BC-1 progeny LCLs show expression of multiple smaller variants of LMP1, including D1LMP1. BC-2 progeny showed only one major predominant wild-type LMP1 signal.
Early antigen expression is detected in the BC-1 and BC-2 progeny LCLs.
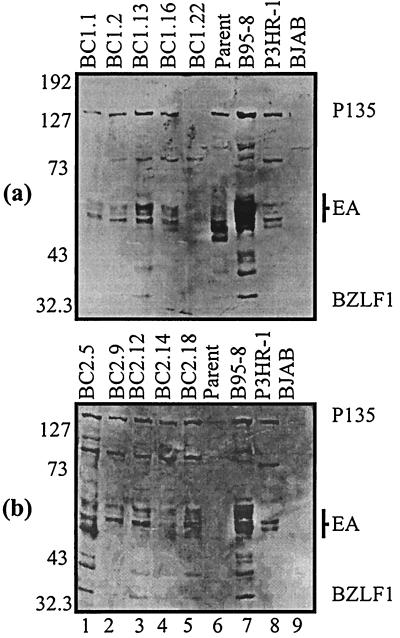
For detection of the EBV early antigens we used a human serum capable of detecting a number of lytic antigens, including BALF2, the single-stranded DNA binding protein, the EA-D (early antigen diffused) complex, and BZLF1 (14, 23). The EA-D antigens range from 43 to 56 kDa. The BALF2 protein and BZLF1 gene product are usually seen in EBV-infected cells undergoing active replication (13, 39). The BC-1 parent expresses EA-D antigens as well as BALF2 (Fig. 7a), whereas the BC-2 parent does not express detectable levels of early antigens (Fig. 7b). Progeny cell lines from both BC-1 and BC-2 appear to be expressing low levels of EA-D antigens. A larger band at approximately 135 kDa indicated expression of the BALF2 single-stranded DNA binding protein in all progeny cell lines (Fig. 7). Only one BC-1 progeny cell line (BC-1.22) was tightly latent, as indicated by undetectable levels of lytic antigen expression (Fig. 7a, lane 5). Another band located at approximately 80 kDa is present in many of the progeny BC-1 and BC-2 cell lines. This band corresponds to the EBNA1 protein that was recognized by the human serum used to detect the early antigens.
FIG. 7.
Western blot for detection of the EBV early antigens with a human serum capable of detecting early antigens from the EA-D complex ranging from 43 to 56 kDa. The same serum also detects the single-stranded DNA binding protein (at approximately 135 kDa) encoded from the BALF2 ORF. BC-1 (a) and BC-2 (b) analyses are shown. Only one progeny cell line from BC-1 had undetectable levels of lytic antigen expression. All other LCLs indicated some level of expression. Molecular masses (in kilodaltons) are shown on the left of each gel.
BC-1 and BC-2 intertypic EBV recombinants transform primary B cells with various efficiencies.
After induction of EBV lytic replication in the BC-1 and BC-2 parental cell lines, T-cell-depleted PBMC were infected with filtered virus supernatant. The infected B lymphocytes were then plated out in a 96-well plate and screened for outgrowth of transformed B lymphocytes. Outgrowth was designated positive if visible clumping of transformed cells and notable color changes in the medium were observed. The experiment was performed three times for verification of the results, and the data are listed in Table 3. In each experiment, BC-2 progeny virus transformed B cells and led to macroscopic outgrowth at a much higher rate than the virus induced from the BC-1, B95-8, and Jijoye cell lines. In experiment 1, for example, BC-2 gave 14 wells positive for B-cell outgrowth at week 1. BC-1 and Jijoye gave no positive wells at week 1, and B95-8 gave only 8 wells positive for B-cell outgrowth. By week 4 of experiment 1, BC-1 showed 62 wells positive for B-cell outgrowth and BC-2 had 76 wells with visible outgrowth of transformed B cells. At week 4, B95-8 and Jijoye had 68 and 71 positive wells, respectively. The number of wells demonstrating visible B-cell transformation continued to increase for all cell lines until week 8, with the total number of positive wells for each cell line in experiment 1 increasing by 23 to 35%. By week 24, a striking difference in prolonged outgrowth was observed. As expected, the total number of positive wells had decreased due to the eventual death of cells that were not truly immortalized. However, BC-1 showed a much more drastic decline in total number of positive wells than BC-2, B95-8, or Jijoye. By week 24, 71% of the BC-1 wells that were once positive for outgrowth of B cells had lost the transformed phenotype and failed to maintain outgrowth. BC-2 demonstrated the greatest number of stable transformants, with only a 25% reduction in the total number of positive wells as measured from week 8. B95-8 and Jijoye underwent reductions of 41 and 55%, respectively. By week 48, only two wells from BC-1 remained positive for outgrowth. BC-2, on the other hand, did not lose any positive wells over the last 24-week period. B95-8 and Jijoye demonstrated 20 and 10% reductions in cells with the transformed phenotype, respectively. The data from experiments 2 and 3 corroborate the data from experiment 1, as seen in Table 3.
TABLE 3.
Comparison of transformation efficiencies of BC-1 and BC-2 intertypic recombinants to that of wild-type EBVa
| Expt | Virus | No. of wells positive for B-cell outgrowth
|
||||
|---|---|---|---|---|---|---|
| 1 wk | 4 wk | 8 wk | 24 wk | 48 wk | ||
| 1 | BC-1 | 0 | 62 | 84 | 24 | 2 |
| BC-2 | 14 | 76 | 96 | 72 | 72 | |
| B95-8 | 8 | 68 | 92 | 54 | 43 | |
| Jijoye | 0 | 71 | 87 | 39 | 35 | |
| 2 | BC-1 | 0 | 57 | 92 | 12 | 6 |
| BC-2 | 17 | 70 | 96 | 84 | 84 | |
| B95-8 | 9 | 67 | 96 | 61 | 60 | |
| Jijoye | 0 | 59 | 84 | 43 | 41 | |
| 3 | BC-1 | 0 | 48 | 80 | 15 | 4 |
| BC-2 | 11 | 73 | 96 | 84 | 75 | |
| B95-8 | ND | ND | ND | ND | ND | |
| Jijoye | ND | ND | ND | ND | ND | |
T-cell-depleted PBMC were infected with virus induced from each of the cell lines indicated. The infected B lymphocytes were distributed in 96-well plates with approximately 50,000 cells per well. Outgrowth was considered positive when visible clumping of transformed cells and notable color changes in the medium were observed. The experiment was repeated three times. ND, not determined. Distinct differences in the transformation competencies of the BC-1 and BC-2 intertypic recombinant EBV are evident in all three experiments.
To verify that the experiments were controlled for levels of virion particles, we analyzed the virus supernatant for EBV produced upon induction of BC-1 and BC-2 by TPA and sodium butyrate. We used the same volume of virus supernatant to maintain a similar exposure to the TPA and butyrate present in the virus supernatant. Similar to the cases of the B95-8 and Jijoye cell lines, EBV was readily induced in BC-1 and BC-2. Moreover, BC-2, which has a more potent transforming potential, had less virus present in the supernatant (Fig. 2A). Hence, while one could postulate that the observed results in Table 3 could be attributed to relatively higher levels of competent virus in BC-2, Fig. 2A indicates that this was not the case.
To determine if these results may also be due to the type of EBNA2 and EBNA3 loci in BC-1, we compared BC-1 to Jijoye, the prototypic type 2 virus used in our experiment. Although the amount of LCLs obtained from infections with Jijoye was lower than the number derived from the type 1, B95-8 virus, it was substantially higher than that seen for BC-1 infections. Hence, the lower transforming potential of BC-1 EBV cannot be attributed solely to its type 2 EBNA2 and EBNA3 genomic loci.
These results indicate that the BC-2 intertypic recombinant EBV is a more potent transforming agent than either the B95-8 and Jijoye controls or the BC-1 EBV. In addition to initiating transformation more effectively, BC-2 EBV also maintained the transformed phenotype at significantly higher levels throughout the experiment. Additionally, BC-1 was unable to maintain long-term outgrowth of LCLs as efficiently as BC-2, suggesting possible genomic abnormalities. The fact that BC-1 and BC-2 are intertypic recombinants of EBV-1 and -2 may provide one explanation. However, the type 2 cell line Jijoye is capable of transforming B lymphocytes with relatively high efficiency. Therefore, we would suggest that there is a greater probability for genomic abnormalities in the BC-1 recombinant.
DISCUSSION
The PCR data presented in this work clearly show that the progeny BC-1 and BC-2 EBV are intertypic recombinants that were derived from the parental cell lines. PCR evidence also demonstrates that no coinfection with KSHV occurred in over 500 LCLs analyzed in our experiments. Western blot analyses of the EBV proteins expressed in the progeny cell lines indicate an expression pattern that is consistent with the latent transformed state. Unlike the parental cell lines, which demonstrate a latency II (Lat II) pattern of expression (9), the progeny cell lines appear to demonstrate a Lat III pattern that is characteristic of transformed LCLs (37). In Lat III, a full pattern of latent gene expression is observed, including expression of the Cp- and Wp-driven set of EBNA transcripts and the BamHI N-derived mRNAs encoding several virus latent membrane proteins (for a review, see reference 32). The fact that the progeny cell lines are expressing the EBNA2 protein, essential for EBV-induced B-cell transformation, and the LMP1 oncogene argues strongly for the ability of the intertypic recombinant viruses to drive cell proliferation.
The discovery of intertypic EBV recombinants in the transformed BC-1 and BC-2 cell lines prompted some crucial questions about the transforming efficiencies of these viruses. Assays to determine the transformation efficiencies of the BC-1 and BC-2 intertypic EBV recombinants indicate that the BC-2 EBV is a more potent transforming virus than the type 1 B95-8 or the type 2 Jijoye. On the other hand, BC-1 EBV was shown to be the least potent of the analyzed transforming viruses. BC-2 EBV was also shown to be most successful in maintaining the transformed phenotype over time, while BC-1 EBV appeared to be least capable of maintaining transformation in cells. B cells transformed by BC-1 EBV showed a significant decrease in the transformed phenotype when observed over a 48-week period following initial infection. It is possible that the recombination events had resulted in aberrant genomes of BC-1 and BC-2 and that the BC-1 genome is rearranged in a manner deleterious to the long-term survival of LCLs in vitro. However, this does not suggest that the BC-1 intertypic EBV has no role in inducing PELs in vivo. It is possible that the BC-1 recombinant virus provides critical functions in patients infected with KSHV to drive cell proliferation in the immunocompromised individuals (20). The BC-1 transformed phenotype may be maintained by the synergistic actions of EBV and KSHV infecting these cells in vivo.
Previous studies of human immunodeficiency virus-positive T-cell immunocompromised cohorts have indicated a greater prevalence of type 2 EBV in these individuals as well as an increased frequency of multiple EBV infections (50, 53). The discovery of intertypic EBV recombinants in these patients (52) has suggested that the impaired immune surveillance and higher titers of EBV in immunocompromised individuals may foster the development of novel recombinant strains. The demonstration of the BC-2 intertypic EBV recombinants as distinctly more potent at transforming PBMC in vitro is an important finding that may provide evidence for intertypic recombination as an evolutionary mechanism for the emergence of more potent and efficient transforming strains of EBV. The selective advantage of a virus with enhanced transforming capacity could lead to altered tropisms for host cells and an increase in the virulence of these strains of EBV. Type 1-type 2 EBV chimeras may also exhibit novel mechanisms of immune evasion. Changes in patterns of gene expression or alterations in programs of EBV latency may enable these recombinant variants to elude established immune responses to EBV infection. New combinations of EBV-1 and -2 antigens could help these emerging intertypic strains to elude the established host immune response, which most often involves antibody recognition of the widely prevalent type 1 antigens.
Since the recent discovery of KSHV, much study has focused on the potential transforming properties of this virus. The virus appears to have much potential to trigger malignancy, including many candidates for transforming genes (for a review, see reference 5). One recent report proposes that KSHV is capable of transforming primary human endothelial cells (15). This is the first report declaring that KSHV can transform any cell type; however, it should be noted that the virus was lost over time in the majority of cells (15). Our data suggest that KSHV alone cannot transform human primary B lymphocytes. One recently published study by Kliche and colleagues posits that KSHV can maintain persistent infection of EBV-positive B lymphocytes and can be serially passaged from KSHV+ EBV+ LCLs to fresh PBMC (24). Furthermore, this group presents data from infection of PBMC from EBV+ donors with BCBL-1 supernatants and suggests that KSHV infection can induce LCL outgrowth of primary B lymphocytes. They also state that KSHV+ EBV+ immortalized cell lines have been derived from infections of PBMC from EBV-seropositive donors with supernatants from the BCBL-1 cell lines. Based on numerous infection studies, we have no evidence that KSHV can induce immortalization of T-cell-depleted human primary B lymphocytes. We also have no data to suggest that infection of PBMC from EBV-seropositive patients with supernatants from induced BCBL-1 or BC-3 cell lines can produce KSHV+ or KSHV+ EBV+ immortalized cell lines. Since Kliche and colleagues observed that infection of PBMC with BC-1 supernatant did not result in the outgrowth of EBV+ LCLs but instead resulted in the outgrowth of LCLs infected with both EBV and KSHV, they propose that this observation may indicate a selection for infection with both viruses. However, our data suggest that the KSHV from the KSHV+ EBV+ BC-1 and BC-2 cell lines can potentially infect primary B cells but cannot maintain a sustained coinfection with EBV in human primary B lymphocytes, thus indicating no selection for coinfection. The contradiction in the observed results from these two studies may indicate the necessity of important cofactors for KSHV infection that were present in one study and absent in the other. Hence, we propose that important differences in the experimental setup and in the isolation methods used in preparing the PBMC for infection in our experiments and in those of Kliche and colleagues may account for the observed differences.
The study of PEL-derived cell lines continues to produce new insights into their unique biology. Of central importance to this biology is the potential synergistic relationship between KSHV and EBV. Much evidence suggests that cells infected with both viruses may have a growth advantage (24). Our data suggest that coinfection of KSHV with EBV may alter the gene expression patterns of EBV. For example, all progeny cell lines express both LMP1 and EBNA2; however, neither parental cell line expresses EBNA2. While a model remains to be established, differences between EBV expression patterns of KSHV-positive parental and KSHV-negative progeny cell lines may indicate that the functions of some KSHV proteins can supplement or possibly even substitute for the functions of certain EBV proteins. Further study of the BC-1 and BC-2 cell lines will continue to elucidate any potential synergism between these two latently infected herpesviruses.
ACKNOWLEDGMENTS
We thank Murray A. Cotter II, Elliott Kieff, and Gary Nabel for their critical comments and discussions prior to submission of this work. BZLF1 and EBNA3C (A10) monoclonal antibodies were a gift from Martin Rowe. S12 monoclonal antibody was kindly provided by David Thorley-Lawson. R3 monoclonal antibody was a gift from Gary Pearson. Elliott Kieff provided PE2 antibody. BCBL-1 was provided by the NIH AIDS Research Reference and Reagent Program.
E.S.R. is a Scholar of the Leukemia Society of America. A.J.A. is supported through an undergraduate research fellowship from the American Society for Microbiology. This work was supported by grants from the Leukemia Society of America, the American Heart Association (AHA9650467N), the National Cancer Institute (CA072150-01), and internal grants from the University of Michigan Comprehensive Cancer Center and the Basic Science Research Partnership Fund to E.S.R.
REFERENCES
- 1.Aguirre, A. J., and E. S. Robertson. Characterization of intertypic recombinants of the Epstein-Barr virus from the body cavity-based lymphomas cell lines BC-1 and BC-2. Virology, in press. [DOI] [PubMed]
- 2.Arrand J R, Young L S, Tugwood J D. Two families of sequences in the small RNA-encoding region of Epstein-Barr virus (EBV) correlate with EBV types A and B. J Virol. 1989;63:983–986. doi: 10.1128/jvi.63.2.983-986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 4.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff C. Kaposi's sarcoma-associated herpesvirus; the 2nd human gammaherpesvirus. Epstein-Barr Virus Rep. 1998;5:3–9. [Google Scholar]
- 6.Boshoff C, Weiss R A. Kaposi's sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 7.Buisson M, Morand P, Genoulaz O, Bourgeat M J, Micoud M, Seigneurin J M. Changes in the dominant Epstein-Barr virus type during human immunodeficiency virus infection. J Gen Virol. 1994;75:431–437. doi: 10.1099/0022-1317-75-2-431. [DOI] [PubMed] [Google Scholar]
- 8.Burrows J M, Khanna R, Sculley T B, Alpers M P, Moss D J, Burrows S R. Identification of a naturally occurring recombinant Epstein-Barr virus isolate from New Guinea that encodes both type 1 and type 2 nuclear antigen sequences. J Virol. 1996;70:4829–4833. doi: 10.1128/jvi.70.7.4829-4833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callahan J, Pai S, Cotter M, Robertson E S. Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: potential switches in latent gene expression due to coinfection. Virology. 1999;262:18–30. doi: 10.1006/viro.1999.9876. [DOI] [PubMed] [Google Scholar]
- 10.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 11.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 12.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 13.Decaussin G, Leclerc V, Ooka T. The lytic cycle of Epstein-Barr virus in the nonproducer Raji line can be rescued by the expression of a 135-kilodalton protein encoded by the BALF2 open reading frame. J Virol. 1995;69:7309–7314. doi: 10.1128/jvi.69.11.7309-7314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrell P. Epstein-Barr virus. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- 15.Flore O, Rafii S, Ely S, O'Leary J J, Hyjek E M, Cesarman E. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature. 1998;394:588–592. doi: 10.1038/29093. [DOI] [PubMed] [Google Scholar]
- 16.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 17.Gessain A. Human herpesvirus 8 and associated diseases: Kaposi's sarcoma, body cavity based lymphoma and multicentric Castleman disease: clinical and molecular epidemiology. Bull Acad Natl Med. 1997;181:1023–1034. . (In French.) [PubMed] [Google Scholar]
- 18.Gratama J W, Oosterveer M A, Weimar W, Sintnicolaas K, Sizoo W, Bolhuis R L, Ernberg I. Detection of multiple ‘Ebnotypes’ in individual Epstein-Barr virus carriers following lymphocyte transformation by virus derived from peripheral blood and oropharynx. J Gen Virol. 1994;75:85–94. doi: 10.1099/0022-1317-75-1-85. [DOI] [PubMed] [Google Scholar]
- 19.Heller M, Dambaugh T, Kieff E. Epstein-Barr virus DNA. IX. Variation among viral DNAs from producer and nonproducer infected cells. J Virol. 1981;38:632–648. doi: 10.1128/jvi.38.2.632-648.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horenstein M G, Nador R G, Chadburn A, Hyjek E M, Inghirami G, Knowles D M, Cesarman E. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. Blood. 1997;90:1186–1191. [PubMed] [Google Scholar]
- 21.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 22.Katz B Z, Andiman W A, Eastman R, Martin K, Miller G. Infection with two genotypes of Epstein-Barr virus in an infant with AIDS and lymphoma of the central nervous system. J Infect Dis. 1986;153:601–604. doi: 10.1093/infdis/153.3.601. [DOI] [PubMed] [Google Scholar]
- 23.Kieff E. Epstein-Barr Virus and its replication. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 24.Kliche S, Kremmer E, Hammerschmidt W, Koszinowski U, Haas J. Persistent infection of Epstein-Barr virus-positive B lymphocytes by human herpesvirus 8. J Virol. 1998;72:8143–8149. doi: 10.1128/jvi.72.10.8143-8149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann K P, Staunton D, Thorley-Lawson D A. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maunders M J, Petti L, Rowe M. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J Gen Virol. 1994;75:769–778. doi: 10.1099/0022-1317-75-4-769. [DOI] [PubMed] [Google Scholar]
- 27.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller G, Robinson J, Heston L, Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci USA. 1974;71:4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nador R G, Cesarman E, Chadburn A, Dawson D B, Ansari M Q, Sald J, Knowles D M. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpes virus. Blood. 1996;88:645–656. [PubMed] [Google Scholar]
- 31.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otsuki T, Kumar S, Ensoli B, Kingma D W, Yano T, Stetler-Stevenson M, Jaffe E S, Raffeld M. Detection of HHV-8/KSHV DNA sequences in AIDS-associated extranodal lymphoid malignancies. Leukemia. 1996;10:1358–1362. [PubMed] [Google Scholar]
- 33.Pastore C, Gloghini A, Volpe G, Nomdedeu J, Leonardo E, Mazza U, Saglio G, Carbone A, Gaidano G. Distribution of Kaposi's sarcoma herpesvirus sequences among lymphoid malignancies in Italy and Spain. Br J Haematol. 1995;91:918–920. doi: 10.1111/j.1365-2141.1995.tb05410.x. [DOI] [PubMed] [Google Scholar]
- 34.Rabson M, Gradoville L, Heston L, Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982;44:834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 36.Rettig M B, Ma H J, Vescio R A, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said J W, Berenson J R. Kaposi's sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science. 1997;276:1851–1854. doi: 10.1126/science.276.5320.1851. [DOI] [PubMed] [Google Scholar]
- 37.Rickinson A, Kieff E. Epstein-Barr Virus. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 38.Robertson E, Kieff E. Reducing the complexity of the transforming Epstein-Barr virus genome to 64 kilobase pairs. J Virol. 1995;69:983–993. doi: 10.1128/jvi.69.2.983-993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson E S, Ooka T, Kieff E D. Epstein-Barr virus vectors for gene delivery to B lymphocytes. Proc Natl Acad Sci USA. 1996;93:11334–11340. doi: 10.1073/pnas.93.21.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson E S, Tomkinson B, Kieff E. An Epstein-Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowe D T, Clarke J R. The type-specific epitopes of the Epstein-Barr virus nuclear antigen 2 are near the carboxy terminus of the protein. J Gen Virol. 1989;70:1217–1229. doi: 10.1099/0022-1317-70-5-1217. [DOI] [PubMed] [Google Scholar]
- 42.Rowe M, Young L S, Cadwallader K, Petti L, Kieff E, Rickinson A B. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol. 1989;63:1031–1039. doi: 10.1128/jvi.63.3.1031-1039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sculley T B, Apolloni A, Hurren L, Moss D J, Cooper D A. Coinfection with A- and B-type Epstein-Barr virus in human immunodeficiency virus-positive subjects. J Infect Dis. 1990;162:643–648. doi: 10.1093/infdis/162.3.642. [DOI] [PubMed] [Google Scholar]
- 45.Sculley T B, Apolloni A, Stumm R, Moss D J, Mueller-Lantczh N, Misko I S, Cooper D A. Expression of Epstein-Barr virus nuclear antigens 3, 4, and 6 are altered in cell lines containing B-type virus. Virology. 1989;171:401–408. doi: 10.1016/0042-6822(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 46.Tissier F, de Pinieux G, Thiounn N, Merran S, Gessain A, Tulliez M, Vieillefond A. Castleman's disease and chromophobe carcinoma of the kidney. An incidental association? Ann Pathol. 1998;18:429–431. . (In French.) [PubMed] [Google Scholar]
- 47.Tomkinson B, Kieff E. Second-site homologous recombination in Epstein-Barr virus: insertion of type 1 EBNA 3 genes in place of type 2 has no effect on in vitro infection. J Virol. 1992;66:780–789. doi: 10.1128/jvi.66.2.780-789.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. Epstein-Barr virus recombinants from overlapping cosmid fragments. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao Q Y, Croom-Carter D S, Tierney R J, Habeshaw G, Wilde J T, Hill F G, Conlon C, Rickinson A B. Epidemiology of infection with Epstein-Barr virus types 1 and 2: lessons from the study of a T-cell-immunocompromised hemophilic cohort. J Virol. 1998;72:4352–4363. doi: 10.1128/jvi.72.5.4352-4363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Q Y, Rowe M, Martin B, Young L S, Rickinson A B. The Epstein-Barr virus carrier state: dominance of a single growth-transforming isolate in the blood and in the oropharynx of healthy virus carriers. J Gen Virol. 1991;72:1579–1590. doi: 10.1099/0022-1317-72-7-1579. [DOI] [PubMed] [Google Scholar]
- 52.Yao Q Y, Tierney R J, Croom-Carter D, Cooper G M, Ellis C J, Rowe M, Rickinson A B. Isolation of intertypic recombinants of Epstein-Barr virus from T-cell-immunocompromised individuals. J Virol. 1996;70:4895–4903. doi: 10.1128/jvi.70.8.4895-4903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao Q Y, Tierney R J, Croom-Carter D, Dukers D, Cooper G M, Ellis C J, Rowe M, Rickinson A B. Frequency of multiple Epstein-Barr virus infections in T-cell-immunocompromised individuals. J Virol. 1996;70:4884–4894. doi: 10.1128/jvi.70.8.4884-4894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]