Abstract
The human homolog of the Drosophila prune protein (from PRUNE, which encodes h‐prune), which interacts with glycogen synthase kinase 3, promotes cellular motility. H‐prune also interacts with nm23‐H1, a suppressor of cancer metastasis. It has been reported that stimulation of cellular motility by h‐prune is enhanced by its interaction with nm23‐H1 in breast cancer cells. In the present study, we examined the expression of h‐prune and nm23‐H1 during tumor progression in gastric cancer (GC). PRUNE mRNA was overexpressed in 12 (32%) of 38 GC cases by quantitative reverse transcription–polymerase chain reaction. PRUNE mRNA levels correlated significantly with advanced T grade, N grade and tumor stage. Immunohistochemical analysis revealed that 43 (30%) of 143 GC cases were positive for h‐prune, and h‐prune‐positive GC cases showed more advanced T grade, N grade and tumor stage than h‐prune‐negative GC cases. One hundred and twenty‐four (87%) of 143 GC cases were positive for nm23‐H1, and nm23‐H1 was expressed in almost all (42 cases, 98%) h‐prune‐positive GC cases. Many GC cases positive for both h‐prune and nm23‐H1 showed more advanced T grade, N grade and tumor stage than other type GC cases. Patients with h‐prune‐positive GC had a significantly worse survival rate than patients with h‐prune‐negative GC. These findings indicate that overexpression of h‐prune is associated with tumor progression and aggressiveness of GC. nm23‐H1 may enhance motility of cancer cells by interacting with h‐prune. (Cancer Sci 2007; 98: 1198–1205)
According to the World Health Organization, gastric cancer (GC) is the fourth most common malignancy worldwide, with approximately 870 000 new cases occurring yearly. Mortality due to GC is second only to that due to lung cancer.( 1 ) Advances in diagnostic tools and treatment have led to excellent long‐term survival for early GC.( 2 ) However, despite improvements in diagnostic and therapeutic strategies, the prognosis of advanced GC with extensive invasion and metastasis remains poor. Several discrete steps can be discerned in the biological cascade of metastasis: loss of cellular adhesion, increased motility and invasiveness, entry and survival in the circulation, exit into new tissue, and eventual colonization of a distant site.( 3 ) Several molecules associated with invasion and metastasis have been identified,( 4 , 5 ) but these molecules cannot completely explain the mechanism of each step of metastasis. In addition, many genes have been analyzed to understand the molecular basis of GC,( 6 , 7 ) but only a few with frequent alterations have been identified so far.
Previously, we identified the human homolog of Drosophila prune protein (PRUNE, which encodes h‐prune) as a glycogen synthase kinase 3 (GSK‐3)‐binding protein.( 8 ) GSK‐3 inhibitors or small interfering RNA (siRNA) for GSK‐3 and h‐prune inhibit cell motility. H‐prune is localized to focal adhesions, and the siRNA for GSK‐3 or h‐prune delays the disassembly of paxillin. Tyrosine phosphorylation of focal adhesion kinase (FAK) and activation of Rac are suppressed in GSK‐3 or h‐prune knock‐down cells.( 8 ) These results suggest that GSK‐3 and h‐prune act cooperatively to regulate cellular motility. In fact, overexpression of h‐prune has been reported in breast cancers and is also associated with high metastatic potential.( 9 ) In our previous study, overexpression of h‐prune was correlated with T grade (depth of invasion), N grade (degree of lymph node metastasis), and tumor stage in colorectal cancers (CRC).( 8 )
H‐prune has phosphodiesterase (PDE) activity, with a preferential affinity for cAMP over cGMP as substrate.( 9 ) PDE are a diverse superfamily of molecules that catalyze the hydrolysis of 3′,5′‐cyclic nucleotides to their corresponding nucleoside 5′‐monophosphates.( 10 ) H‐prune PDE activity is involved in cellular motility,( 9 ) and h‐prune also interacts with nm23‐H1 (NME1, which encodes nm23‐H1),( 11 ) a known suppressor of cancer metastasis.( 12 ) H‐prune and nm23‐H1 proteins partially colocalize in the cytoplasm.( 11 ) It has been suggested that overexpression of h‐prune inhibits the antimetastasis function of nm23‐H1 during the metastatic process.( 9 , 13 ) Several studies have suggested an association between reduced expression of nm23 mRNA or protein and increasing metastatic activity, resulting in poorer prognosis in breast cancers,( 14 ) malignant melanomas( 15 ) and hepatocellular carcinomas.( 16 ) In contrast, in GC and CRC, overexpression of nm23‐H1 has been reported.( 17 , 18 ) High nm23‐H1 expression has been shown to correlate with tumor progression and poor prognosis of GC.( 19 )
Taken together, the currently available data suggest that analysis of h‐prune is necessary to clarify the association between expression of nm23‐H1 and metastatic potential. Correlation between high nm23‐H1 expression and metastatic potential in GC may be due to overexpression of h‐prune. However, expression of h‐prune has not been investigated in GC. In the present study, we examined the expression and distribution of both h‐prune and nm23‐H1 in human GC by immunohistochemistry and reverse transcription–polymerase chain reaction (RT‐PCR).
Materials and Methods
Tissue samples. In a retrospective study design, 181 primary tumors were collected from patients diagnosed with GC who underwent surgery between 1991 and 2001 at the Department of Surgical Oncology, Hiroshima University Hospital (Hiroshima, Japan). All patients underwent curative resection. Only patients without preoperative radiotherapy or chemotherapy and without clinical evidence for distant metastasis were enrolled in the study.
For quantitative RT‐PCR, 38 GC samples and corresponding non‐neoplastic mucosa samples were used. The samples were obtained during surgery at Hiroshima University Hospital. We confirmed microscopically that the tumor specimens were predominantly cancer tissue (>50% on a nuclear basis). Samples were frozen immediately in liquid nitrogen and stored at –80°C until use.
For immunohistochemical analysis, we used archival formalin‐fixed, paraffin‐embedded tissues from 143 patients who had undergone surgical excision for GC. Fifty‐nine of the 143 patients had early GC, and 84 had advanced GC. Early GC is limited to the mucosa or the mucosa and submucosa, regardless of nodal status.( 2 ) Advanced GC is a tumor that has invaded beyond the muscularis propria. Information on patient prognosis was available for 84 GC cases. Because information on postoperative chemotherapy was not available, postoperative chemotherapeutic backgrounds were not involved in the prognostic analysis.
Tumor staging was carried out according to the tumor–node–metastasis (TNM) stage grouping.( 20 ) Histological classification of GC was carried out according to the Lauren classification system.( 21 ) Because written informed consent was not obtained, for strict privacy protection, identifying information for all samples was removed before analysis; this procedure is in accordance with Ethical Guidelines for Human Genome/Gene Research enacted by the Japanese Government and approved by the Ethical Review Committee of the Hiroshima University School of Medicine.
Quantitative RT‐PCR analysis. Total RNA was extracted with an RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and 1 µg of total RNA was converted to cDNA with a First Strand cDNA Synthesis Kit (Amersham Biosciences, Piscataway, NJ, USA). Quantitation of PRUNE mRNA levels in human tissue samples was done by real‐time fluorescence detection as described previously.( 22 ) PRUNE primer sequences were 5′‐GAA GTC CTG GAA CGC TCC C‐3′ and 5′‐GGT TAG GGT GGG TAC TTG AGG C‐3′. PCR was carried out using a SYBR Green PCR Core Reagents Kit (Applied Biosystems, Foster City, CA, USA). Real‐time detection of the emission intensity of SYBR green bound to double‐stranded DNA was done with an ABI PRISM 7700 Sequence Detection System (Applied Biosystems) as described previously.( 23 ) ACTB‐specific PCR products were amplified from the same RNA samples and served as internal controls.
Immunohistochemistry. A Dako LSAB kit (Dako, Carpenteria, CA, USA) was used for immunohistochemical analysis. In brief, sections were pretreated by microwaving in citrate buffer for 30 min to retrieve antigenicity. After peroxidase activity was blocked with 3% H2O2–methanol for 10 min, sections were incubated with normal goat serum (Dako) for 20 min to block non‐specific antibody binding sites. Sections were incubated with rabbit polyclonal antih‐prune (diluted 1:50, antih‐prune antibody was raised in our laboratory)( 8 ) and rabbit polyclonal antinm23‐H1 (1:20, Santa Cruz Biotechnology, Santa Cruz, CA, USA). The specificity of the h‐prune antibody has been characterized in detail.( 8 ) Sections were incubated with primary antibody for 1 h at 25°C, followed by incubations with biotinylated antirabbit IgG and peroxidase‐labeled streptavidin for 10 min each. Staining was completed with a 10‐min incubation with the substrate–chromogen solution. The sections were counterstained with 0.1% hematoxylin. The percentage of stained cancer cells was evaluated for each antibody.
Statistical methods. Correlations between clinicopathological parameters and h‐prune or nm23‐H1 expression were analyzed by Fisher's exact test. Kaplan–Meier survival curves were constructed for h‐prune‐ or nm23‐H1‐positive and h‐prune‐ or nm23‐H1‐negative patients. Survival rates were compared between h‐prune‐ or nm23‐H1‐positive and h‐prune‐ or nm23‐H1‐negative groups. Differences between survival curves were tested for statistical significance using the log‐rank test.( 24 ) Cox proportional hazards multivariate model was used to examine the association of clinical and pathological factors and the expression of h‐prune with survival. A P‐value of less than 0.05 was considered statistically significant.
Results
mRNA expression of PRUNE in GC. Expression of PRUNE mRNA was analyzed by quantitative RT‐PCR of RNA from 38 GC samples and corresponding non‐neoplastic mucosa samples. We calculated the ratio of PRUNE mRNA levels between GC tissue (T) and corresponding non‐neoplastic mucosa (N). When a T/N ratio > two‐fold was considered to represent overexpression, PRUNE was overexpressed in 12 (32%) of the 38 GC samples. As shown in Fig. 1, PRUNE levels were significantly higher in T3/4 cases than in T1/2 cases (P = 0.0156; Mann–Whitney U‐test). Moreover, PRUNE levels correlated significantly with N status (P = 0.0122; Mann–Whitney U‐test) and tumor stage (P = 0.0127; Mann–Whitney U‐test). There was no clear tendency between PRUNE level and histological type.
Figure 1.
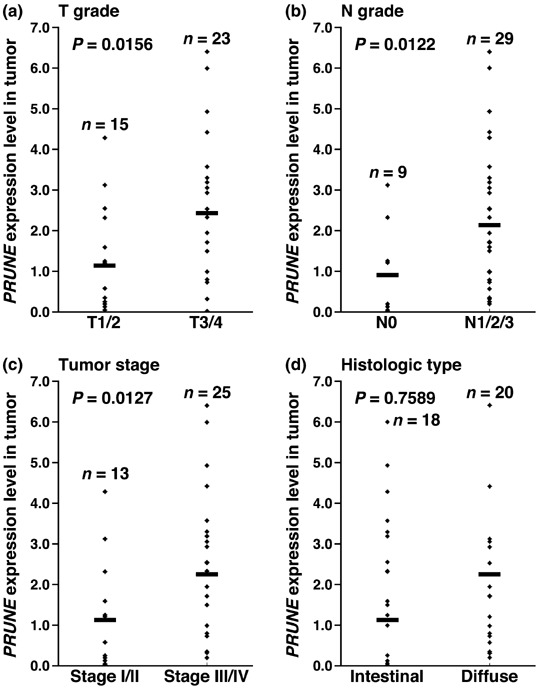
Expression of PRUNE mRNA in gastric cancer tissues. Each point represents the levels of PRUNE mRNA in an individual specimen. Horizontal bar represents the mean PRUNE mRNA expression level. (a) PRUNE mRNA expression levels were significantly higher in T3/4 cases than in T1/2 cases (P = 0.0156; Mann–Whitney U‐test). (b) The PRUNE mRNA expression levels were significantly higher in N1/2/3 cases than in N0 cases (P = 0.0122; Mann–Whitney U‐test). (c) The PRUNE mRNA expression levels were significantly higher in stage III/IV cases than in stage I/II cases (P = 0.0127; Mann–Whitney U‐test). (d) There was no clear association between PRUNE mRNA expression level and histological type.
Immunohistochemical analysis of h‐prune in GC . We next investigated the expression and distribution of h‐prune by immunohistochemistry of 143 GC tissue samples. In corresponding non‐neoplastic gastric mucosa, only weak or no staining of h‐prune was observed in epithelial and stromal cells (Fig. 2a,b). However, GC tissue showed stronger, more extensive staining than corresponding non‐neoplastic mucosa. In 14 (10%) of the 143 GC cases, h‐prune‐positive tumor cells were restricted to the invasive front (Fig. 2a,c,d). In these cases, less than 50% of tumor cells were stained. However, in the majority of GC cases containing h‐prune‐positive tumor cells, more than 50% of the tumor cells showed cytoplasmic staining for h‐prune (43 of 143 GC cases). In these GC cases, h‐prune‐positive tumor cells were observed at the invasive front in addition to the central part of the tumors. The remaining cases showed 10–50% immunopositive tumor cells and h‐prune staining was not observed at the invasive front. H‐prune was detected in the cytoplasm of tumor cells in intestinal‐type (Fig. 2e) and diffuse‐type GC (Fig. 2f,g). We next analyzed the relationship between h‐prune expression and clinicopathological characteristics. Several previous studies have indicated that observation of the invasive front is important in the analysis of tumor cells, because it reflects the invasive potential of those cells. To investigate the significance of restricted h‐prune staining at the invasive front, the relationship between h‐prune staining and clinicopathological characteristics was examined in GC cases in which less than 50% of tumor cells were stained. In these cases, GC cases in which h‐prune‐positive tumor cells were restricted to the invasive front showed more advanced T grade (P = 0.0083, Fisher's exact test) than those in which h‐prune‐positive tumor cells were not restricted to the invasive front (Table 1). GC cases in which h‐prune‐positive tumor cells were restricted to the invasive front did not show advanced N grade or tumor stage (Table 1). In contrast, among all of the cases we studied, GC cases in which more than 50% of tumor cells were stained showed more advanced T grade (P = 0.0158, Fisher's exact test), N grade (P = 0.0035, Fisher's exact test) and tumor stage (P = 0.0466, Fisher's exact test) than those in which less than 50% of tumor cells were stained (Table 1). Therefore, when more than 50% of tumor cells were stained, the immunostaining was considered positive for h‐prune.
Figure 2.
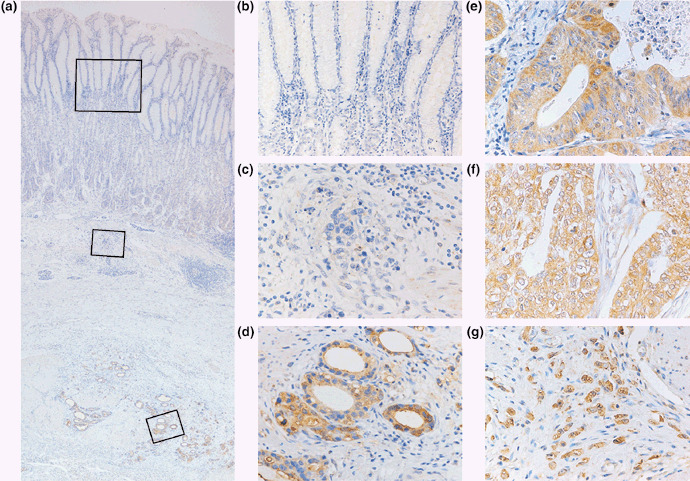
Immunohistochemical analysis of h‐prune in gastric cancer (GC) tissues. (a) GC case in which h‐prune expression was strong at the invasive front (original magnification, ×40). (b,c,d) High magnification images of the fields indicated by boxes in panel (a). (b) In corresponding non‐neoplastic gastric mucosa, only weak or no staining of h‐prune was observed in epithelial and stromal cells (original magnification, ×100). (c) In the superficial layer of GC tissue, only weak or no staining of h‐prune was observed (original magnification, ×400). (d) In the invasive front of GC tissue, strong cytoplasmic staining of h‐prune was observed in GC cells (original magnification, ×400). (e) H‐prune‐positive intestinal‐type GC. Strong cytoplasmic staining of h‐prune was observed in GC cells (original magnification, ×400). (f) H‐prune‐positive diffuse‐type GC. Strong cytoplasmic staining of h‐prune was observed in GC cells (original magnification, ×400). (g) H‐prune‐positive diffuse‐type GC (so‐called scirrhous‐type GC). Strong cytoplasmic staining of h‐prune was observed in GC cells (original magnification, ×400).
Table 1.
Association of h‐prune expression with clinicopathologic features of gastric cancer
| Feature | H‐prune expression (%) | P‐value † | P‐value ‡ | ||
|---|---|---|---|---|---|
| Positive | Invasion front positive | Negative | |||
| Age (years) | |||||
| >65 | 20 (25%) | 6 (7%) | 55 | 0.1504 | 0.1410 |
| ≤65 | 23 (37%) | 8 (13%) | 31 | ||
| Sex | |||||
| Male | 25 (27%) | 10 (11%) | 57 | 0.7703 | 0.3442 |
| Female | 18 (35%) | 4 (8%) | 29 | ||
| T grade § | |||||
| T1 | 11 (19%) | 2 (3%) | 46 | 0.0083 | 0.0158 |
| T2/3/4 | 32 (38%) | 12 (14%) | 40 | ||
| N grade § | |||||
| N0 | 16 (20%) | 6 (8%) | 58 | 0.1305 | 0.0035 |
| N1/2/3 | 27 (43%) | 8 (13%) | 28 | ||
| Stage § | |||||
| Stage I/II | 25 (25%) | 8 (8%) | 68 | 0.0944 | 0.0466 |
| Stage III/IV | 18 (43%) | 6 (14%) | 18 | ||
| Histological type ¶ | |||||
| Intestinal | 22 (26%) | 9 (11%) | 54 | 1.0000 | 0.1988 |
| Diffuse | 21 (36%) | 5 (9%) | 32 | ||
Invasion front positive versus negative. Statistical significance was determined with Fisher's exact test.
‡ Positive versus invasion front‐positive plus‐negative. Statistical significance was determined with Fisher's exact test.
Stage was classified according to the criteria of the International Union Against Cancer TNM classification of malignant tumors. 20
¶ Histology was classified according to the criteria of Lauren.
Association between h‐prune and nm23‐H1 expression. Because it has been suggested that overexpression of h‐prune in breast cancer inhibits the antimetastasis function of nm23‐H1 during metastasis,( 9 , 13 ) we also carried out immunohistochemical analysis of nm23‐H1 in the same 143 GC cases. Consistent with previous reports,( 19 ) strong cytoplasmic staining of nm23‐H1 was observed in tumor cells. In corresponding non‐neoplastic mucosa, weak staining of nm23‐H1 was observed. In the present study, to compare the h‐prune staining pattern with the nm23‐H1 staining pattern, the same cut‐off point for nm23‐H1 and h‐prune immunostaining was set. When more than 50% of tumor cells were stained, the immunostaining was considered positive for nm23‐H1. In total, 124 (87%) of 143 GC cases were positive for nm23‐H1. The h‐prune staining pattern was then compared with the nm23‐H1 staining pattern. Of 43 h‐prune‐positive GC cases, 42 (98%) were positive for nm23‐H1, and in these 42 GC cases there was a tendency for h‐prune and nm23‐H1 to be expressed in the same tumor cells (Fig. 3a,b). All 18 GC cases that were negative for both h‐prune and nm23‐H1 were signet ring cell carcinoma. In contrast, the majority (82 of 143 GC cases, 57%) of cases were positive for nm23‐H1 but not h‐prune (Fig. 3c,d). The frequency of nm23‐H1 expression in h‐prune‐positive GC cases (42 of 43 cases, 98%) was significantly higher than that in h‐prune‐negative GC cases (82 of 100 cases, 82%, P = 0.0134, Fisher's exact test) (Table 2).
Figure 3.
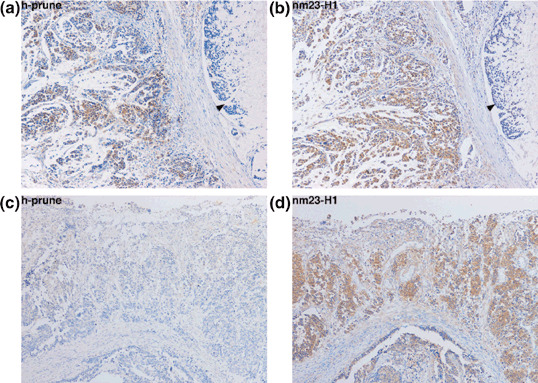
Expression and distribution of h‐prune and nm23‐H1 in gastric cancer (GC) tissues. (a) Immunohistochemistry of h‐prune in h‐prune‐positive GC case. Staining for h‐prune was observed in diffuse‐type GC cells but not in signet ring cell carcinoma components (arrowhead) (original magnification, ×100). (b) Immunohistochemistry of nm23‐H1 in the same GC case as in panel (a). H‐prune and nm23‐H1 tended to be located in the same GC cells (original magnification, ×100). (c) Immunohistochemistry of h‐prune in h‐prune‐negative GC case (original magnification, ×100). (d) Immunohistochemistry of nm23‐H1 in the same GC case as in panel (c). Staining of nm23‐H1 was observed in GC cells (original magnification, ×100).
Table 2.
Distribution of h‐prune and nm23‐H1 expression in gastric cancer
| H‐prune expression | nm23‐H1 expression | P‐value † | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 42 (98%) | 1 | 0.0134 |
| Negative | 82 (82%) | 18 | |
Statistical significance was determined with Fisher's exact test.
We next analyzed the relationship between nm23‐H1 expression and clinicopathological characteristics. Expression of nm23‐H1 was not correlated with age, sex, T grade, N grade or tumor stage (Table 3). As reported previously,( 19 ) expression of nm23‐H1 was observed more frequently in intestinal‐type GC (79 of 85 cases, 93%) than in diffuse‐type GC (45 of 58 cases, 78%, P = 0.0113, Fisher's exact test) (Table 3). It has been reported that in the majority of nm23‐H1‐positive tumors, more than 75% of the tumor cells contained a homogeneous cytoplasmic pattern for nm23‐H1.( 19 ) GC cases containing more than 75% of nm23‐H1‐positive tumor cells were found in 113 of 143 GC cases. GC cases containing more than 75% of nm23‐H1‐positive tumor cells did not show advanced T grade, N grade or tumor stage (data not shown). Among the nm23‐H1‐positive GC cases, h‐prune‐positive GC cases showed advanced T grade (P = 0.0341, Fisher's exact test), N grade (P = 0.0083, Fisher's exact test) and tumor stage (P = 0.0371, Fisher's exact test) more frequently than h‐prune‐negative GC cases (Fig. 4a–c). In contrast, among h‐prune‐negative GC cases there was no statistically significant correlation between nm23‐H1 positivity and T grade, N grade or tumor stage (Fig. 4a–c).
Table 3.
Association of nm23‐H1 expression with clinicopathologic features of gastric cancer
| Feature | nm23‐H1 expression (%) | P‐value † | |
|---|---|---|---|
| Positive | Negative | ||
| Age (years) | |||
| >65 | 70 (86%) | 11 | 1.0000 |
| ≤65 | 54 (87%) | 8 | |
| Sex | |||
| Male | 80 (87%) | 12 | 1.0000 |
| Female | 44 (86%) | 7 | |
| T grade ‡ | |||
| T1 | 49 (83%) | 10 | 0.3220 |
| T2/3/4 | 75 (89%) | 9 | |
| N grade ‡ | |||
| N0 | 68 (85%) | 12 | 0.6219 |
| N1/2/3 | 56 (89%) | 7 | |
| Stage ‡ | |||
| Stage I/II | 88 (87%) | 13 | 0.7926 |
| Stage III/IV | 36 (86%) | 6 | |
| Histological type § | |||
| ntestinal | 79 (93%) | 6 | 0.0113 |
| Diffuse | 45 (78%) | 13 | |
Statistical significance was determined with Fisher's exact test.
Stage was classified according to the criteria of the International Union Against Cancer TNM classification of malignant tumors. 20
§ Histology was classified according to the criteria of Lauren.
Figure 4.
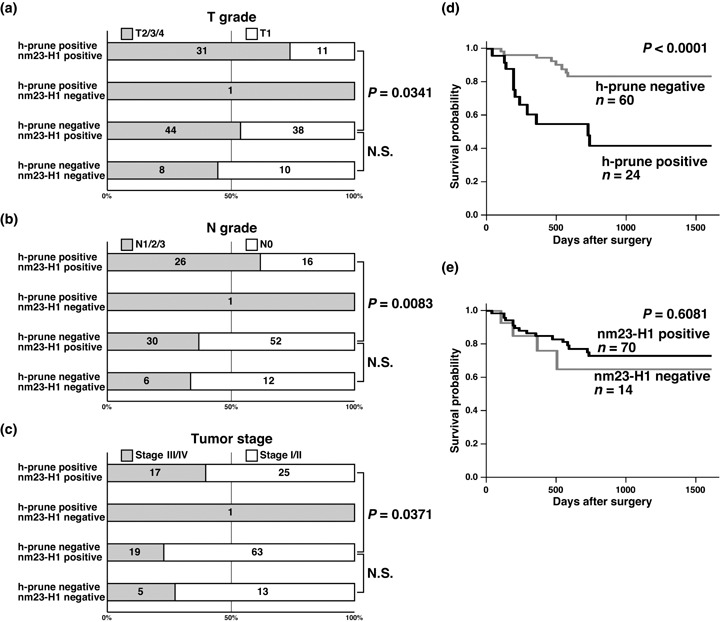
Correlation of h‐prune and nm23‐H1 expression with clinicopathologic features. Many gastric cancer (GC) cases positive for both h‐prune and nm23‐H1 showed advanced (a) T grade, (b) N grade and (c) tumor stage. There were no clear differences between h‐prune‐negative/nm23‐H1‐positive GC cases and h‐prune‐negative/nm23‐H1‐negative GC cases. There was only h‐prune‐positive/nm23‐H1‐negative GC among 143 GC cases. (d) The survival of patients with h‐prune‐positive GC was significantly worse in 84 patients with GC (P < 0.0001, log‐rank test). (e) There was no statistical difference between the survival rate of patients with nm23‐H1‐positive GC and that of patients with nm23‐H1‐negative GC (P = 0.6081, log‐rank test).
We also examined the relationship between survival and expression of h‐prune and nm23‐H1 in GC (n = 84). On univariate analysis, T grade (P < 0.0001, log‐rank test), N grade (P = 0.0474, log‐rank test), tumor stage (P = 0.0001, log‐rank test) and h‐prune expression (P < 0.0001, log‐rank test) (Fig. 4d) were significant prognostic factors of survival in patients with GC, whereas age, sex, histological type and nm23‐H1 expression (Fig. 4e) were not correlated with survival. Next, Cox proportional hazards multivariate model was used to examine the association of clinicopathological factors and the expression of h‐prune with survival. Multivariate analysis indicated that T grade, N grade, tumor stage and h‐prune expression were independent predictors of survival in patients with GC (Table 4).
Table 4.
Multivariate analysis of factors influencing survival
| Factor | Hazard ratio | 95% CI | χ2 | P‐value |
|---|---|---|---|---|
| T grade † | ||||
| T1 | 1 | Reference | 7.42 | 0.0064 |
| T2/3/4 | 18.99 | 2.28–157.86 | ||
| N grade † | ||||
| N0 | 1 | Reference | 5.57 | 0.0182 |
| N1/2/3 | 8.24 | 1.43–47.46 | ||
| Stage † | ||||
| Stage I/II | 1 | Reference | 5.71 | 0.0168 |
| Stage III/IV | 6.42 | 1.39–29.43 | ||
| H‐prune expression | ||||
| Negative | 1 | Reference | 8.18 | 0.0043 |
| Positive | 4.87 | 1.63–14.48 | ||
Stage was classified according to the criteria of the International Union Against Cancer TNM classification of malignant tumors. 20
Discussion
Metastasis fundamentally involves the movement of cells from one site to another. The molecular mechanisms that underlie cell migration involve dynamic cytoskeletal changes, cell–matrix interactions, localized proteolysis, actin–myosin contractions and focal contact disassembly.( 25 ) We reported previously that h‐prune is localized to focal adhesions and that knockdown of h‐prune inhibits cell motility. Suppression of tyrosine phosphorylation of FAK and activation of Rac are involved in inhibition of cell motility.( 8 ) In the present study, overexpression of PRUNE mRNA was observed in 32% of cases by quantitative RT‐PCR analysis, and cytoplasmic staining of h‐prune protein was observed in 30% of cases by immunohistochemistry in GC tissue samples. Expression of h‐prune was correlated with T grade, N grade and tumor stage. In addition, patients with h‐prune‐positive GC had a significantly worse survival rate than patients with h‐prune‐negative GC. Taken together, these results suggest that extensive expression of h‐prune contributes to the malignant behavior of GC, possibly by promoting cancer cell motility. H‐prune may be a good marker of poor survival of GC.
It is generally accepted that cancer progresses as a disease of genetically heterogeneous cell populations. In the present study, there were several cases in which h‐prune‐positive tumor cells were restricted to the invasive front. Among GC cases in which less than 50% of tumor cells were stained, GC cases in which h‐prune‐positive tumor cells were restricted to the invasive front showed more advanced T grade than those in which h‐prune‐positive tumor cells were not restricted to the invasive front; however, these GC cases comprised only 10% of 143 GC cases, and did not show advanced N grade or tumor stage, suggesting that expression of h‐prune may represent tumor invasiveness, especially local invasiveness. Observation of the invasive front is important in the analysis of tumor cells, because it reflects the invasive potential of tumor cells. It has been reported that expression of matrilysin in the invasive front is a promising biomarker predicting nodal metastasis of colorectal cancers.( 26 ) Overexpression of heparanase at the invasive front has been reported in GC, and high expression of heparanase is a strong predictor of poor survival.( 27 ) These results indicate that the proteolytic degradation of extracellular matrix by these molecules is one of the most important mechanisms in tumor progression, and the proteolytic degradation occurs at the invasive front. Because h‐prune promotes cell motility, expression of h‐prune at the invasive front may partly contribute to the malignant behavior of GC, such as local invasiveness.
The mechanism of regulation of h‐prune expression is still unknown. In the present study, overexpression of PRUNE mRNA was observed in 32% by quantitative RT‐PCR analysis, and h‐prune‐positive GC cases were found in 30% by immunohistochemistry. Because GC samples analyzed by quantitative RT‐PCR were different from those analyzed by immunohistochemistry, we could not compare PRUNE mRNA levels with h‐prune protein levels. However, these findings suggest that overexpression of PRUNE occurs at the transcriptional or mRNA level in GC. PRUNE amplification and overexpression have been reported in certain sarcomas and breast cancers.( 13 ) PRUNE is located on chromosome 1q21.3, and gains of 1q21.1‐21.2 and 1q21.3 have been reported in GC.( 28 ) Amplification of the PRUNE gene may be involved in overexpression of h‐prune in GC cases. The main cause of peptic ulceration is Helicobacter pylori, and H. pylori is the strongest risk factor for the development of distal GC.( 29 , 30 ) Because stimulation of matrilysin by H. pylori has been reported,( 31 ) induction of h‐prune by H. pylori should be examined in the near future.
In the present study, immunohistochemical analysis of GC tissues revealed that almost all h‐prune‐positive GC cases (98%) also expressed nm23‐H1. Expression of nm23‐H1 was found in both early‐ and late‐stage GC, whereas expression of h‐prune was detected mainly in late‐stage tumors. In nm23‐H1‐positive GC cases, h‐prune‐positive GC cells may develop in accordance with tumor progression. In GC cases positive for both h‐prune and nm23‐H1, intratumoral distribution of h‐prune‐positive tumor cells and nm23‐H1‐positive tumor cells was similar, and many of these GC cases showed advanced T grade, N grade and tumor stage. Because the PDE activity of h‐prune, which is involved in cellular motility, is enhanced by the interaction with nm23‐H1,( 9 ) expression of both proteins may confer cellular motility to GC cells. Among h‐prune‐negative GC cases, there were no clear differences between nm23‐H1 positivity and clinical characteristics, suggesting that expression of only nm23‐H1 does not influence progression of GC. Although there was no impact of nm23‐H1 expression on patient survival in the present study, it has been reported that high nm23‐H1 expression correlates with tumor progression and poor prognosis.( 9 ) Expression of nm23‐H1 may enhance the PDE activity of h‐prune but may not have antimetastatic activity. All 18 GC cases negative for both h‐prune and nm23‐H1 were signet ring cell carcinoma. Several reports have indicated that the prognosis of patients with signet ring cell carcinoma is relatively favorable in the early stages and poor in advanced stages compared with other histological types of GC.( 32 ) In fact, of 18 signet ring cell carcinoma cases, 12 cases showed stage I and the prognosis of patients with these GC was favorable.
Epigenetic changes, such as DNA methylation of CpG islands, are detected commonly in human cancers. Hypermethylation of CpG islands is associated with silencing of many genes, especially defective tumor‐related genes, and has been proposed as an alternative way to inactivate tumor‐related genes in human cancers.( 33 ) GC show the CpG island methylator phenotype (CIMP).( 34 , 35 ) CIMP‐positive GC also tend to show DNA methylation of the p16INK4a ,( 34 ) hMLH1 ( 6 ) and RIZ ( 6 ) genes, suggesting that CIMP is an important pathway involved in stomach carcinogenesis. Because DNA methylation inhibitors such as 5‐aza‐2′‐deoxycytidine have been shown to increase nm23‐H1 expression in breast cancer cells,( 36 ) h‐prune or nm23‐H1 expression may be associated with CIMP.
Is h‐prune a good therapeutic target for GC? Previous data indicated that dipyridamole, an antiplatelet aggregation drug, inhibits h‐prune PDE activity and cellular motility in a breast cancer cell line.( 9 ) In the SW480 CRC cell line, knockdown of h‐prune decreased cell motility.( 8 ) These results suggest that inhibition of h‐prune may be useful for the prevention or treatment of metastasis. However, because h‐prune is expressed ubiquitously in normal human adult tissues,( 11 ) inhibitors of h‐prune, such as dipyridamole, may have severe adverse effects. However, it is known that components of the blood‐clotting pathway contribute to metastasis by trapping tumor cells in capillaries or by facilitating adhesion of tumor cells to capillary walls. Anti‐platelet aggregation drugs may interfere with this step in the metastatic process. In fact, several studies have indicated that the formation of metastatic tumors could be inhibited by antiplatelet aggregation drugs (reviewed by Hejna et al. (37 )). In GC, combined chemotherapy with dipyridamole with adriamycin and 5‐fluorouracil appears to be safe and may be useful clinically for treatment of GC.( 38 ) Taken together with our present results, h‐prune inhibitors, such as dipyridamole, may be effective for the prevention or treatment of h‐prune‐positive GC without severe adverse effects. The PDE superfamily is large and complex, containing 11 highly related and structurally related gene families and over 60 distinct isoforms. Each of the PDE families contains one to four genes, and many genes generate multiple isoforms.( 39 ) Development of specific h‐prune inhibitors is important because dipyridamole also inhibits the activities of PDE5, PDE6, PDE7, PDE8, PDE10 and PDE11.( 39 )
In conclusion, we found that h‐prune is overexpressed in GC and that this overexpression correlates with tumor progression and poor survival in patients with GC. Although the significance of overexpression of nm23‐H1 is still unclear, nm23‐H1 may contribute to tumor cell motility by upregulating the function of h‐prune. Although the precise mechanism by which h‐prune regulates cellular motility remains unclear, the efficacy of a combination chemotherapy with 5‐fluorouracil, cisplatin and dipyridamole( 40 ) should be investigated in h‐prune‐positive GC.
Acknowledgments
This work was supported, in part, by Grants‐in‐Aid for Cancer Research from the Ministry of Education, Culture, Science, Sports, and Technology of Japan, and from the Ministry of Health, Labor, and Welfare of Japan. We thank Masayoshi Takatani and Masayuki Ikeda for excellent technical assistance and advice. This work was carried out with the kind cooperation of the Research Center for Molecular Medicine, Faculty of Medicine, Hiroshima University. We thank the Analysis Center of Life Science, Hiroshima University for the use of their facilities.
References
- 1. Ohgaki H, Matsukura N. Stomach cancer. In: Stewart BW, Kleihues P, eds. World Cancer Report. Lyon: IARC Press, 2003: 194–7. [Google Scholar]
- 2. Hohenberger P, Gretschel S. Gastric cancer. Lancet 2003; 362: 305–15. [DOI] [PubMed] [Google Scholar]
- 3. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006; 127: 679–95. [DOI] [PubMed] [Google Scholar]
- 4. Oue N, Hamai Y, Mitani Y et al . Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res 2004; 64: 2397–405. [DOI] [PubMed] [Google Scholar]
- 5. Kurayoshi M, Oue N, Yamamoto H et al . Expression of Wnt‐5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res 2006; 66: 10 439–48. [DOI] [PubMed] [Google Scholar]
- 6. Oue N, Mitani Y, Motoshita J et al . Accumulation of DNA methylation is associated with tumor stage in gastric cancer. Cancer 2006; 106: 1250–9. [DOI] [PubMed] [Google Scholar]
- 7. Shutoh M, Oue N, Aung PP et al . DNA methylation of genes linked with retinoid signaling in gastric carcinoma: expression of the retinoid acid receptor beta, cellular retinol‐binding protein 1, and tazarotene‐induced gene 1 genes is associated with DNA methylation. Cancer 2005; 104: 1609–19. [DOI] [PubMed] [Google Scholar]
- 8. Kobayashi T, Hino S, Oue N et al . Glycogen synthase kinase 3 and h‐prune regulate cell migration by modulating focal adhesions. Mol Cell Biol 2006; 26: 898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Angelo A, Garzia L, Andre A et al . Prune cAMP phosphodiesterase binds nm23‐H1 and promotes cancer metastasis. Cancer Cell 2004; 5: 137–49. [DOI] [PubMed] [Google Scholar]
- 10. Beavo JA, Brunton LL. Cyclic nucleotide research – still expanding after half a century. Nat Rev Mol Cell Biol 2002; 3: 710–18. [DOI] [PubMed] [Google Scholar]
- 11. Reymond A, Volorio S, Merla G et al . Evidence for interaction between human PRUNE and nm23‐H1 NDPKinase. Oncogene 1999; 18: 7244–52. [DOI] [PubMed] [Google Scholar]
- 12. Steeg PS, Bevilacqua G, Kopper L et al . Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 1988; 80: 200–4. [DOI] [PubMed] [Google Scholar]
- 13. Forus A, D’Angelo A, Henriksen J et al . Amplification and overexpression of PRUNE in human sarcomas and breast carcinomas – a possible mechanism for altering the nm23‐H1 activity. Oncogene 2001; 20: 6881–90. [DOI] [PubMed] [Google Scholar]
- 14. Bevilacqua G, Sobel ME, Liotta LA, Steeg PS. Association of low nm23 RNA levels in human primary infiltrating ductal breast carcinomas with lymph node involvement and other histopathological indicators of high metastatic potential. Cancer Res 1989; 49: 5185–90. [PubMed] [Google Scholar]
- 15. Florenes VA, Aamdal S, Myklebost O, Maelandsmo GM, Bruland OS, Fodstad O. Levels of nm23 messenger RNA in metastatic malignant melanomas: inverse correlation to disease progression. Cancer Res 1992; 52: 6088–91. [PubMed] [Google Scholar]
- 16. Yamaguchi A, Urano T, Goi T et al . Expression of human nm23‐H1 and nm23‐H2 proteins in hepatocellular carcinoma. Cancer 1994; 73: 2280–4. [DOI] [PubMed] [Google Scholar]
- 17. Nakayama H, Yasui W, Yokozaki H, Tahara E. Reduced expression of nm23 is associated with metastasis of human gastric carcinomas. Jpn J Cancer Res 1993; 84: 184–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haut M, Steeg PS, Willson JK, Markowitz SD. Induction of nm23 gene expression in human colonic neoplasms and equal expression in colon tumors of high and low metastatic potential. J Natl Cancer Inst 1991; 83: 712–16. [DOI] [PubMed] [Google Scholar]
- 19. Muller W, Schneiders A, Hommel G, Gabbert HE. Expression of nm23 in gastric carcinoma: association with tumor progression and poor prognosis. Cancer 1998; 83: 2481–7. [DOI] [PubMed] [Google Scholar]
- 20. Sobin LH, Wittekind CH, eds. TNM Classification of Malignant Tumors, 6th edn. New York: John Wiley & Sons, 2002. [Google Scholar]
- 21. Lauren P. The two histological main types of gastric carcinoma: diffuse and so‐called intestinal‐type carcinoma. An attempt at a histo‐clinical classification. Acta Pathol Microbiol Scand 1965; 64: 31–49. [DOI] [PubMed] [Google Scholar]
- 22. Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT‐PCR. Genome Res 1996; 6: 995–1001. [DOI] [PubMed] [Google Scholar]
- 23. Kondo T, Oue N, Yoshida K et al . Expression of POT1 is associated with tumor stage and telomere length in gastric carcinoma. Cancer Res 2004; 64: 523–9. [DOI] [PubMed] [Google Scholar]
- 24. Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep 1966; 50: 163–70. [PubMed] [Google Scholar]
- 25. Friedl P, Wolf K. Tumour‐cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003; 3: 362–74. [DOI] [PubMed] [Google Scholar]
- 26. Kurokawa S, Arimura Y, Yamamoto H et al . Tumour matrilysin expression predicts metastatic potential of stage I (pT1) colon and rectal cancers. Gut 2005; 54: 1751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takaoka M, Naomoto Y, Ohkawa T et al . Heparanase expression correlates with invasion and poor prognosis in gastric cancers. Laboratory Invest 2003; 83: 613–22. [DOI] [PubMed] [Google Scholar]
- 28. Vauhkonen H, Vauhkonen M, Sajantila A, Sipponen P, Knuutila S. DNA copy number aberrations in intestinal‐type gastric cancer revealed by array‐based comparative genomic hybridization. Cancer Genet Cytogenet 2006; 167: 150–4. [DOI] [PubMed] [Google Scholar]
- 29. Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez‐Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med 1991; 325: 1132–6. [DOI] [PubMed] [Google Scholar]
- 30. Nomura A, Stemmermann GN, Chyou PH, Perez‐Perez GI, Blaser MJ. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann Intern Med 1994; 120: 977–81. [DOI] [PubMed] [Google Scholar]
- 31. Bebb JR, Letley DP, Thomas RJ et al . Helicobacter pylori upregulates matrilysin (MMP‐7) in epithelial cells in vivo and in vitro in a Cag dependent manner. Gut 2003; 52: 1408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kunisaki C, Shimada H, Nomura M, Matsuda G, Otsuka Y, Akiyama H. Therapeutic strategy for signet ring cell carcinoma of the stomach. Br J Surg 2004; 91: 1319–24. [DOI] [PubMed] [Google Scholar]
- 33. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3: 415–28. [DOI] [PubMed] [Google Scholar]
- 34. Toyota M, Ahuja N, Suzuki H et al . Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res 1999; 59: 5438–42. [PubMed] [Google Scholar]
- 35. Ushijima T, Okochi‐Takada E. Aberrant methylations in cancer cells: Where do they come from? Cancer Sci 2005; 96: 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hartsough MT, Clare SE, Mair M et al . Elevation of breast carcinoma Nm23‐H1 metastasis suppressor gene expression and reduced motility by DNA methylation inhibition. Cancer Res 2001; 61: 2320–7. [PubMed] [Google Scholar]
- 37. Hejna M, Raderer M, Zielinski CC. Inhibition of metastases by anticoagulants. J Natl Cancer Inst 1999; 91: 22–36. [DOI] [PubMed] [Google Scholar]
- 38. Sakaguchi Y, Maehara Y, Emi Y, Kusumoto T, Kohnoe S, Sugimachi K. Dipyridamole combination chemotherapy can be used safely in treating gastric cancer patients. Anticancer Drugs 1991; 2: 139–43. [DOI] [PubMed] [Google Scholar]
- 39. Cheng J, Grande JP. Cyclic nucleotide phosphodiesterase (PDE) inhibitors: novel therapeutic agents for progressive renal disease. Exp Biol Med (Maywood) 2007; 232: 38–51. [PubMed] [Google Scholar]
- 40. Kohnoe S, Maehara Y, Takahashi I, Emi Y, Baba H, Sugimachi K. Treatment of advanced gastric cancer with 5‐fluorouracil and cisplatin in combination with dipyridamole. Int J Oncol 1998; 13: 1203–6. [DOI] [PubMed] [Google Scholar]


