Abstract
To identify the molecular background of esophageal cancer, we conducted a proteomics study using an antibody microarray consisting of 725 antibodies and surgical specimens from three cases. The microarray analysis identified 24 proteins with aberrant expression in esophageal cancer compared with the corresponding normal mucosa. The overexpression of 14 of the 24 proteins was validated by western blotting analysis of the same samples. These 14 proteins were examined by immunohistochemistry, in which nine proteins showed consistent results with those obtained by western blotting. Among the nine proteins, seven were localized in tumor cells, and two in infiltrating cells. The former included proteins associated with mitotic checkpoint control and the nuclear factor (NF)‐κB pathway. Although mitotic checkpoint gene products (budding uninhibited by benzidazoles 1 homolog beta (BubR1) and mitotic arrest deficient‐like 1 (Mad2)) have previously been reported to be involved in esophageal cancer, the association of NF‐κB‐activating kinase, caspase 10, and activator protein‐1 with esophageal cancer has not been previously reported. These proteins play a key role in the NF‐κB pathway, and NF‐κB is a signal transduction factor that has emerged as an important modulator of altered gene programs and malignant phenotype in the development of cancer. The association of these proteins with esophageal cancer may indicate that mitotic checkpoint gene products and NF‐κB play an important part in the carcinogenesis of esophageal cancer. (Cancer Sci 2009; 100: 1612–1622)
Esophageal cancer is the eighth most common cancer( 1 ) and the sixth leading cause of cancer death worldwide.( 2 ) Despite the use of modern surgical techniques in combination with radiotherapy and chemotherapy, early recurrence is common and the overall 5‐year survival rate remains below 40%.( 3 , 4 , 5 ) ESCC develops through a multistep process from dysplasia through carcinoma in situ to invasive carcinoma, and the acquisition of molecular alterations tightly corresponds to the dysplasia–carcinoma sequence.( 6 ) However, the mechanisms of esophageal cancer progression remain largely obscure. The characterization of molecular alterations inherently linked to ESCC development and an in‐depth understanding of the molecular mechanisms underlying carcinogenesis and growth control may therefore provide information relevant to early tumor detection, refined prognosis, and development of novel targeted therapeutics.
As the proteome is a functional translation of the genome, directly regulating cancer phenotypes, its study may further our understanding of esophageal cancer. Esophageal cancer research has made significant progress due to proteomics studies on two fronts to date. First, tissue proteomics have identified numerous intracellular proteins that had not been previously reported to be implicated in esophageal cancer. For instance, the use of two‐dimensional polyacrylamide gel electrophoresis followed by mass spectrometry and database search led to the identification of the aberrant expression of proteins correlating with certain clinicopathological parameters in esophageal cancer.( 7 ) Second, the use of proteomics tools has led to the discovery of a novel candidate plasma biomarker for early diagnosis.( 8 ) This is important as the majority of esophageal cancer cases has advanced locally or has developed distant metastases by the time of diagnosis, rendering the cancer surgically inoperable,( 9 , 10 ) and the existing plasma tumor markers (e.g. squamous cell carcinoma‐related antigen (SCC), cinoembryonic antigen (CEA), and cytokeratin‐19 fragments (CYFRA21‐1)) have obvious limitations in terms of sensitivity and specificity in detecting patients with localized and resectable esophageal cancer.( 11 )
The major proteomics tools such as two‐dimensional gel electrophoresis and mass spectrometry essentially have two major drawbacks. First, these techniques tend to uncover the proteins in the order of their expression level; the proteins with higher expression levels are more frequently observed than proteins with low abundance such as transcription factors, receptors, and growth factors. Second, the present approaches do not allow the systematic profiling of protein expression. The proteins are currently identified when their physical characteristics, such as the molecular weight and charge, fit the conditions of the proteomic methods, and such protein characteristics do not necessarily correlate with protein functions. As a consequence, pathway comprehensive approaches such as those identifying all proteins in certain molecular pathways cannot be carried out using the present proteomics tools. To partially address these drawbacks, multidimensional separation by liquid chromatography and gel electrophoresis were used to reduce sample complexity and increase the number of observable proteins. However, further improvements are still required to overcome the limitations of cancer proteomics.
The recently developed antibody‐based proteomics may address the aforementioned limitations of cancer proteomics. A large‐scale library of antibodies with high specificity and sensitivity may be used to detect the proteins with low expression levels, and thus enable pathway comprehensive analysis. For instance, a large‐scale immunoblotting analysis with 900 well‐characterized antibodies resulted in the identification of 102 deregulated proteins in pancreatic cancer cells.( 12 ) An antibody proteomics project was launched to detect all proteins encoded by the human genome.( 13 ) In addition to the characterization of antibodies already developed, the development of techniques for hundreds or thousands of antibodies will be the challenge in antibody‐based proteomics.
In this paper, we examined the proteins with aberrant expression levels in esophageal cancer tissues and normal tissues using an antibody microarray. The antibody microarray is commercially available and enables comprehensive high‐throughput studies. We validated the antibody microarray study results by western blotting and immunohistochemistry. Through the comparison between the normal mucosa and tumor tissues, we discussed the reproducibility of the antibody microarray results, the degree of inconsistency between the different methods, and the functional contribution of the identified members of the NF‐κB pathway to esophageal cancer progression.
Materials and Methods
Patients. Tumor tissues (1T, 2T and 3T) and their adjacent normal mucosal tissues (1N, 2N and 3N) were collected from three cases of ESCC, which were surgically resected in 2007 and 2008 at the National Cancer Center Hospital. The resected tissues were snap frozen in liquid nitrogen and stored at –80°C until use. All cases were newly diagnosed as squamous cell carcinoma of the esophagus (ESCC). The patients did not receive anticancer treatment prior to surgery. All patients were male and in the seventh decade of their lives, and all tumors were located in the lower thoracic segment of the esophagus and were classified as T3N1 according to the International Union against Cancer TNM classification.( 14 ) Only one case with distant lymph node metastasis was classified as M1.( 14 ) This study was approved by the ethics committee of the National Cancer Center and written informed consent was obtained from the patients.
Antibody microarrays. The Panorama Antibody Microarray XPRESS Profilter725, XP725 was purchased from Sigma‐Aldrich (St Louis, MO, USA). Seven hundred and twenty‐five antibodies were spotted on each glass slide in 32 subarrays, each containing duplicate spots for 23 antibodies, as well as duplicate positive control spots for Cy3 and Cy5 (monoclonal antibodies labeled with Cy3 and Cy5), and duplicate negative controls, resulting in 1536 spots per slide. These antibodies represent families of proteins known to be involved in a variety of different biological pathways. Detailed information on each antibody can be obtained in the antibody specific datasheet found on the Sigma‐Aldrich website at http://www.sigma.com/arrays.
Protein extraction. Frozen samples were crushed to powder with a CryoPress (Microtech Nichion, Shizuoka, Japan) under cooling with liquid nitrogen. The frozen powder was then treated with urea lysis buffer (7 mol/L urea, 2 mol/L thiourea, 3% CHAPS, and 1% Triton X‐100). After centrifugation at 17 400 g for 10 min, the supernatant was used as the source of cellular proteins for protein expression studies. The protein concentrations ranged between 3.46 and 8.45 mg/mL.
Protein labeling and microarray assays. Briefly, 100 µg (77 µL) of protein extracted from each tumor and normal sample were labeled with Cy5 (Cy5 Mono Reactive Dye; GE Healthcare Biosciences, Uppsala, Sweden), using a scaled‐down version of the protocol provided by the manufacturer. The protein extracts were mixed with 20 µL of 0.5 M sodium bicarbonate, pH 8.5. The labeling reaction was carried out for 30 min at room temperature. Labeled samples were purified from free excess dye using the Sigma Spin columns supplied with the antibody microarray kit. The 100 mg labeled protein samples (1 mg/ml) were applied onto the column, and centrifuged first for 2 min and then for 4 min at 800 g . The protein concentration of the recovered samples was adjusted to 1 mg/mL in 100 µL. Before hybridization, the dye‐to‐protein molar ratio was adjusted to 2.93–4.84 mol of dye/mol of protein according to the manufacturer's instructions. Labeled protein extracts were incubated on Panorama Ab Microarray slides (Sigma‐Aldrich) for 30 min with moderate shaking. The slides were washed three times in PBS, in Tween 20 for 5 min, and immersed in water for 2 min. Finally, the slides were air dried and scanned with a laser scanner at the appropriate wave length (Typhoon Trio; GE Healthcare Biosciences).
Data analysis and identification of alterations. Microarray images were analyzed with the Array Vision software version 8.0 (GE Healthcare Biosciences). Fluorescence intensity measurements from each array element were compared with local background, and background subtraction was carried out. The reported value represents the average of all the pixels remaining in the spot, after first removing the pixels with density values that exceeded a threshold value set at four times the absolute deviation value above the median. This measure removes the influence of image artifacts, such as dust particles, on density estimation. The proteins showing ≥2.5‐fold difference in expression in more than two cases of three were deemed significant.
Western blotting. Five µg of each protein sample was separated by SDS‐PAGE on a 10–12.5% polyacrylamide gradient gel and subsequently blotted on a nitrocellulose membrane. Immunoblot analysis was carried out using the antibodies listed in Table 1 and horseradish peroxidase‐conjugated secondary antibodies (1:1000; GE Healthcare Biosciences). Antibody–antigen complexes were visualized with an ECL system (GE Healthcare Biosciences) using LAS 3000 (Fuji Film, Tokyo, Japan). The intensity of the protein bands was quantified and the relative intensity of the examined proteins was calculated on the basis of the intensity of the actin bands on the same membrane.
Table 1.
List of the proteins found to be overexpressed in the microarray analysis and examined by western blotting and immunohistochemistry
| No. | Antibody commercial name | Company | Product no. | Dilution | |
|---|---|---|---|---|---|
| Western blotting | Immunohistochemistry | ||||
| 1 | Acetylated protein | Sigma | A5463 | 1: 100 | – |
| 2 | Actin | Sigma | A5060 | 1: 1000 | – |
| 3 | AP‐1 | Sigma | A5968 | 1: 200 | 1: 5000 |
| 4 | AP‐2a | Sigma | A0844 | 1: 100 | 1: 500 |
| 5 | BAD | BD Biosciences | 610391 | 1: 500 | 1: 500 |
| 6 | BUBR1 | BD Biosciences | 612502 | 1: 500 | 1: 500 |
| 7 | Calbindin‐D | Sigma | C7354 | 1: 250 | – |
| 8 | Caspase 10 | Sigma | C8351 | 1: 250 | 1: 500 |
| 9 | Cdk1 | Sigma | C4973 | 1: 250 | 1: 500 |
| 10 | phosph‐β‐catenin | Sigma | C4231 | 1: 250 | 1: 5000 |
| 11 | Cytokeratin 18 | Sigma | C1399 | 1: 500 | – |
| 12 | HABH3 | Sigma | A8353 | 1: 250 | 1: 500 |
| 13 | hnRNP‐U | Sigma | R6278 | 1: 500 | 1: 500 |
| 14 | Mad 2 | Sigma | M8694 | 1: 250 | 1: 5000 |
| 15 | NAK | Sigma | N2661 | 1: 250 | 1: 50 |
| 16 | NG2 | Sigma | N8912 | 1: 250 | 1: 50 |
| 17 | b‐NOS | Sigma | N2280 | 1: 500 | 1: 200 |
| 18 | NGFb | Sigma | N3279 | 1: 100 | – |
| 19 | RAB 9 | ABR‐Affinity Bioreagents | MA3‐067 | 1: 250 | 1: 200 |
| 20 | Protein phosphatase 2A alpha | Sigma | P8998 | 1: 250 | – |
| 21 | Protein Tyrosine Phosphatase | Sigma | P9109 | 1: 250 | – |
| 22 | phospho‐Pyk2 | Sigma | P7114 | 1: 250 | – |
| 23 | SKK2 | Abnova | H00005606‐M02 | 1: 100 | – |
| 24 | Tropomyosin (Sarcimatic) | Sigma | T9283 | 1: 100 | – |
Immunohistochemistry and tissue microarray. The cellular localization of the proteins was examined immunohistochemically on methanol‐fixed, paraffin‐embedded tissues using the Dako REAL EnVision Detection System (DAKO, Glostrup, Denmark) following the manufacturer's instructions. In brief, the sections were deparaffinized, dehydrated, and blocked with 0.3% H2O2 in methanol for 30 min to remove endogenous peroxidase activity. The sections were autoclaved in 10 mM citrate buffer (pH 6.0) at 121°C for 10 min. The primary antibodies and dilutions used are shown in Table 1.
We obtained two commercial tissue microarrays (ES801; US Biomax, Rockville, MD, USA; and A218(II) AccuMax array; ISU ABXIS, Seoul, Korea) including 40 and 30 esophageal cancer cases with matched normal adjacent tissue, respectively, and used them to examine the expression of the identified proteins and their relationship with the corresponding clinicopathological data. The sex, age, and histological grade were available for all 70 cases on the tissue microarrays, whereas the tumor stage, lymph node metastasis status, distant metastasis status, and tumor size were only available for the 30 cases on the A218(II) array.
mRNA expression analysis. Total RNA was isolated from six ESCC tissues using an Isogen kit (Nippon Gene, Toyama, Japan). The integrity of the RNA was determined on the Agilent 2100 Bioanalyser using the RNA 6000 Nano Kit (Agilent Technologies, Palo Alto, CA, USA). RNA expression analysis was carried out using the GeneChip 3’ IVT expression kit (Affymetrix, Santa Clara, CA, USA) according to the supplier's protocol. Labeled targets were hybridized to the GeneChip Human Genome U133 Plus 2.0 Array. Gene expression analysis was carried out using the Affymetrix GeneChip Command Console Software. Experimental data were quantile normalized in Expression Console using the RMA algorithm for probe set summarization. The maximal signal was adopted for subsequent analysis when there were multiple probes corresponding to the same protein.
Statistical analysis. The numerical data were imported into the Expressionist software (Genedata, Basel, Switzerland) for scatter plotting and calculation of the correlation coefficient. The correlation between protein expression and clinicopathological features was evaluated using the Fisher exact test for categorical variables and the Mann–Whitney U‐test for continuous variables. P‐value differences of <0.05 were considered to be significant. Statistical analyses were carried out using the SPSS 11.0 statistical package (SPSS, Chicago, IL, USA).
Results
Antibody microarray performance. The protein contents of surgical specimens from three patients diagnosed with ESCC were extracted. The scanned image of an antibody microarray for a representative tumor extract (1T) is shown in Figure 1. The shape of the spots, the dynamic range of the intensity response, and the signal‐to‐noise ratio, as well as the positive and negative controls were routinely checked. The reproducibility of the results was monitored by comparing duplicate spots on the same slide. The scatter plot demonstrated that the intensity values of 63.1–94.1% of the antibody spots was scattered within a twofold difference, and that the correlation coefficient was 0.7172–0.9332 in all six samples examined (Fig. 2). Although spot intensity showed high reproducibility for spots with higher intensity in the microarray slide, it showed poor reproducibility for spots with lower intensity, even when adjacent duplicated spots on the same slide were compared.
Figure 1.
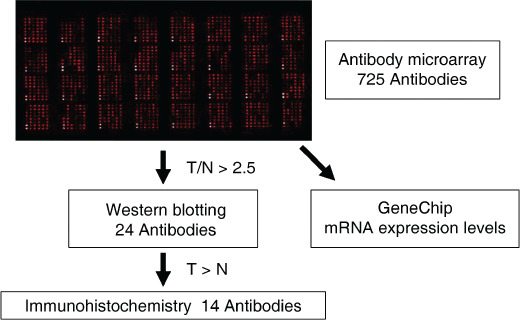
Scheme showing the experimental flow: the antibody microarray analysis results were validated using western blotting and immunohistochemistry. N, normal; T, tumor.
Figure 2.
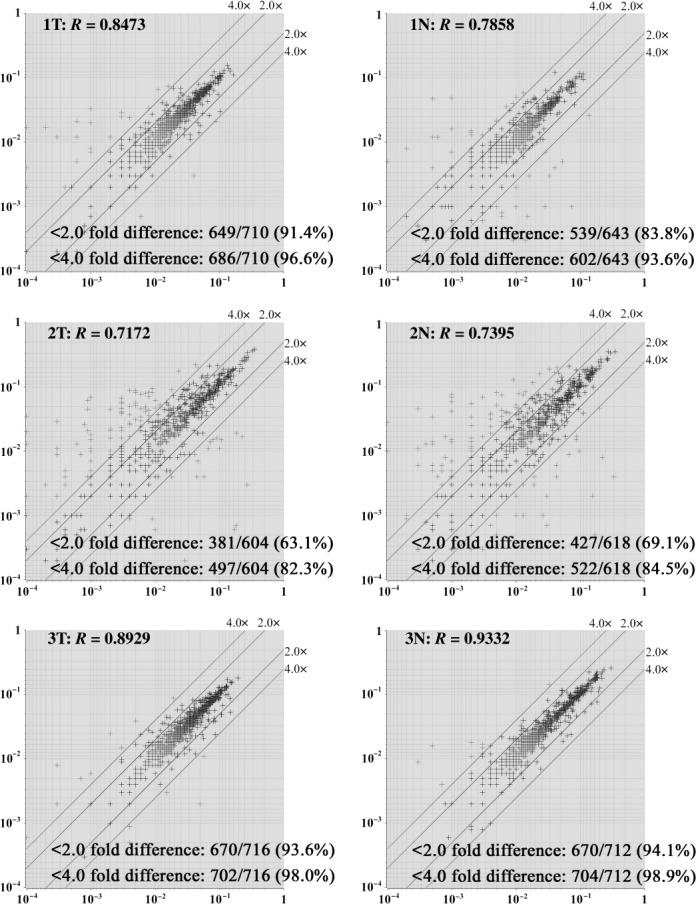
Reproducibility of the antibody microarray analysis. The scatter plot shows that the intensity values of 63.1–94.1% of the antibodies contained in the microarrays used were scattered within a twofold difference, and that the correlation coefficient (R) was 0.7172–0.9332. X‐axis, intensity of one spot of duplicate spots; Y‐axis, intensity of other spot of duplicate spots.
Differential protein expression analysis. We initially searched for proteins with upregulated expression in esophageal cancer compared to the corresponding normal reference mucosa. Twenty‐four proteins, including two non‐unique proteins (whole acetylated proteins and protein tyrosine phosphatases), showed a notable upregulation (more than 2.5‐fold change in at least two samples) in the tumor compared to the normal tissues (Table 2). No proteins showed notable downregulation in the tumor tissues compared with their normal reference mucosa. We classified the identified proteins based on their biological function according to Gene Ontology web site and literature searching by the author. Proteins associated with signal transduction were the most frequently identified, followed by those involved in apoptosis, cell cycle, and the cytoskeleton (Table 2). The locus of the identified proteins was variable, and absence of constant chromosomal localization was noted (Table 2).
Table 2.
Proteins found to be upregulated in esophageal squamous cell carcinoma tissues in the antibody microarrays
| No. | Commercial antibody name | Official protein name | Gene symbol | Entrez gene ID | Biological function | Locus | Cellular localization |
|---|---|---|---|---|---|---|---|
| 1 | Acetylated protein | – | – | – | – | – | – |
| 2 | Actin | Actin, alpha 1, skeletal muscle | ACTA1 | 58 | Cytoskeleton | 1q42.13 | Cytoplasm |
| 3 | AP‐1 | jun oncogene | JUN | 3725 | Cell cycle | 1p32‐p31 | Cytoplasm, nucleus |
| 4 | AP‐2a | Transcription factor AP‐2 alpha | TFAP2A | 7020 | Transcription | 6p24 | Nucleus |
| 5 | BAD | Bcl‐associated death promoter | BAD | 572 | Apoptosis | 11q13.1 | Cytoplasm, membrane, mitochondrion |
| 6 | BUBR1 | BUB 1 budding uninhibited by benzidazoles 1 homolog beta | BUB1B | 701 | Cell cycle | 15q15 | Cytoplasm, nucleus |
| 7 | Calbindin‐D | Calbindin 1 | CALB1 | 793 | Calcium‐associated | 8q21.3–q22.1 | Cytoplasm, nucleus |
| 8 | Caspase 10 | Caspase 10, apoptosis‐related cysteine peptidase | CASP10 | 843 | Apoptosis | 2q33–q34 | Cytoplasm |
| 9 | Cdk1 | Cell division cycle 2, G1 to S and G2 to M | CDC2 | 983 | Cell cycle | 10q21.1 | Cytoplasm, nucleus |
| 10 | phosph‐beta‐catenin | Catenin (cadherin associated protein), beta 1 | CTNNB1 | 1499 | Signal transduction | 3p21 | Cytoplasm, nucleus, membrane |
| 11 | Cytokeratin 18 | Keratin 18 | KRT18 | 3875 | Cytoskeleton | 12q13 | Cytoplasm |
| 12 | HABH3 | alkB, alkylation repair homolog 3 | ALKBH3 | 221 120 | Oxidation reduction | 11p11.2 | Cytoplasm, nucleus |
| 13 | hnRNP‐U | Heterogeneous nuclear ribonucleoprotein U | HNRPU | 3192 | RNA splicing | 1q44 | Nucleus, cell surface |
| 14 | Mad 2 | MAD2 mitotic arrest deficient‐like 1 | MAD2L1 | 4085 | Cell cycle | 4q27 | Cytoplasm, nucleus |
| 15 | NAK | TANK‐binding kinase 1 | TBK1 | 29 110 | Immune response | 12q14.1 | Cytoplasm |
| 16 | NG2 | Chondroitin sulfate proteoglycan 4 | CSPG4 | 1464 | Signal transduction | 15q24.2 | Membrane, cell surface |
| 17 | b‐NOS | Nitric oxide synthase 1 (neuronal) | NOS1 | 4842 | Oxidation reduction | 12q24.2–q24.31 | Cytoplasm |
| 18 | NGFb | Nerve growth factor | NGFB | 4803 | Signal transduction | 1p13.1 | Extracellular region, Golgi |
| 19 | RAB 9 | RAB9, member RAS oncogene family | RAB9 | 9367 | Signal transduction | Xp22.2 | Golgi, lysosome, membrane |
| 20 | Protein phosphatase 2A alpha | Protein phosphatase 2, catalytic subunit, alpha isoform | PPP2CA | 5515 | RNA splicing | 5q31.1 | Cytoplasm, nucleus, mitochondrion, membrane |
| 21 | Protein tyrosine phosphatase | – | – | – | – | – | – |
| 22 | phospho‐Pyk2 | PTK2 protein tyrosine kinase 2 beta | PTK2B | 2185 | Apoptosis | 8p21.1 | Cytoplasm, membrane |
| 23 | SKK2 | Mitogen‐activated protein kinase kinase 3 | MAP2K3 | 5606 | Signal transduction | 17q11.2 | Protein complex |
| 24 | Tropomyosin (Sarcimatic) | Tropomyosin 1 (alpha) | TPM1 | 7168 | Cytoskeleton | 15q22.1 | Cytoplasm, nucleus |
Western blotting analysis. The 24 proteins identified in the microarray analysis were validated using western blotting. Fourteen of the 24 proteins showed overexpression in the tumor tissues, the results being consistent with the results of microarray analysis. Six proteins showed discordant results; five proteins showed inverse expression compared to the microarray results, whereas actin was found to be consistently expressed in the tumor and normal tissues in western blotting (Table 3; Fig. 3). Four proteins were undetectable in western blotting.
Table 3.
Summary of the results of western blotting and immunohistochemistry
| No. | Antibody commercial name | WB | R (WB array) | Ihc localization |
|---|---|---|---|---|
| 1 | Acetylated protein | Undetectable | ND | ND |
| 2 | Actin | T = N | ND | ND |
| 3 | AP‐1 | T > N | † | Cytoplasm, nucleus |
| 4 | AP‐2a | T > N | † | Inflamatory cells |
| 5 | BAD | T > N | 0.351 | Inflamatory cells |
| 6 | BUBR1 | T > N | –0.178 | Cytoplasm |
| 7 | Calbindin‐D | T < N | ND | ND |
| 8 | Caspase 10 | T > N | –0.572 | Cytoplasm |
| 9 | Cdk1 | T > N | 0.583 | Nucleus |
| 10 | phosph‐beta‐catenin | T > N | –0.699 | Cytoplasm |
| 11 | Cytokeratin 18 | T < N | ND | ND |
| 12 | HABH3 | T > N | –0.981 | Nucleus |
| 13 | hnRNP‐U | T > N | –0.458 | Nucleus |
| 14 | Mad 2 | T > N | 0.934 | Cytoplasm |
| 15 | NAK | T > N | 0.763 | Cytoplasm |
| 16 | NG2 | T > N | –0.933 | Undetectable |
| 17 | b‐NOS | T > N | 0.708 | Undetectable |
| 18 | NGFb | Undetectable | ND | ND |
| 19 | RAB 9 | T > N | 0.805 | Cytoplasm |
| 20 | Protein phosphatase 2A alpha | T < N | ND | ND |
| 21 | Protein tyrosine phosphatase | T < N | ND | ND |
| 22 | phospho‐Pyk2 | Undetectable | ND | ND |
| 23 | SKK2 | Undetectable | ND | ND |
| 24 | Tropomyosin (Sarcimatic) | T < N | ND | ND |
Not calculated due to missing array data. IHC, immunohistochemistry; ND, not done; R, correlation coefficiency of the western blotting compared to the microarray results; WB, western blotting.
Figure 3.
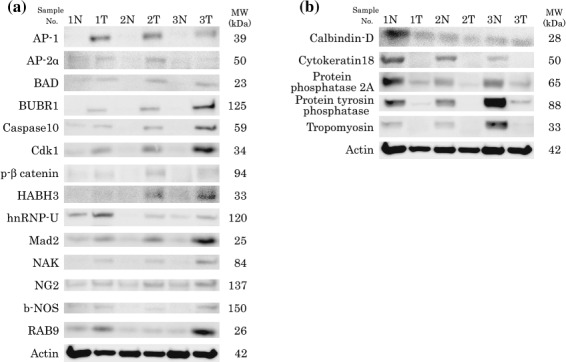
Western blotting results. (a) Fourteen of the 24 proteins identified in the microarray analysis as being overexpressed in tumor tissues showed concordant results in western blotting analysis. (b) Five proteins showed discordant results.
The scattergram (Fig. 4) demonstrates the correlation between the intensity of the duplicate antibody signals of the 24 proteins, in relation to the consistency between western blotting and antibody microarray experiments. The intensity values of the proteins found to be upregulated in tumor tissues in the microarrays showed high reproducibility, irrespective of whether these microarray results were discordant (R = 0.7666) or concordant (R = 0.6921) with western blotting results, or whether these proteins were not detected in western blotting (R = 0.6317). We concluded that reproducibility of the antibody microarray experiments may not guarantee concordant results between western blotting and antibody microarray experiments.
Figure 4.
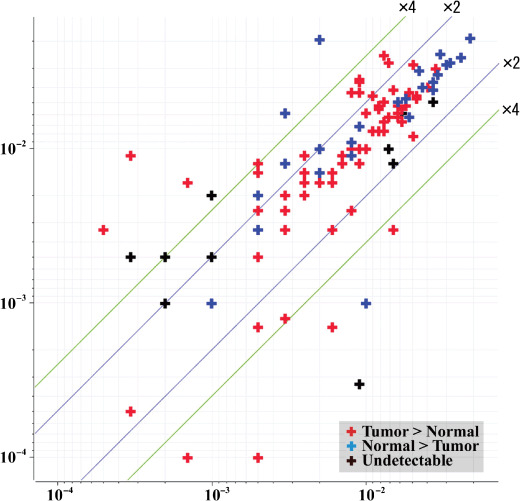
The scattergram demonstrates the correlation between the intensity of duplicate antibody signals of the 24 proteins found to be overexpressed in the microarray analysis. The intensity values of the proteins found to be overexpressed in tumor tissues in the microarrays showed high reproducibility, irrespective of whether these microarray results were discordant (R = 0.7666) or concordant (R = 0.6921) with western blotting results, or whether these proteins were not detected by western blotting (R = 0.6317). X‐axis, intensity of one spot of duplicate spots; Y‐axis, intensity of other spot of duplicate spots.
Correlation between microarray and western blotting data. The quantitative correlation between the microarray and western blotting data was examined. Each tumor tissue's data were normalized by the normal counterpart's data. The correlation coefficient for AP‐1 and AP‐2α could not be calculated due to missing microarray values. The 14 proteins overexpressed in tumor tissues in western blotting included proteins with a high correlation coefficient, such as Mad2 (R = 0.934) and NAK (R = 0.763), as well as proteins with a poor correlative relationship, such as hABH3 (R = –0.981) and NG2 (R = –0.933) (Table 3; Fig. 5).
Figure 5.
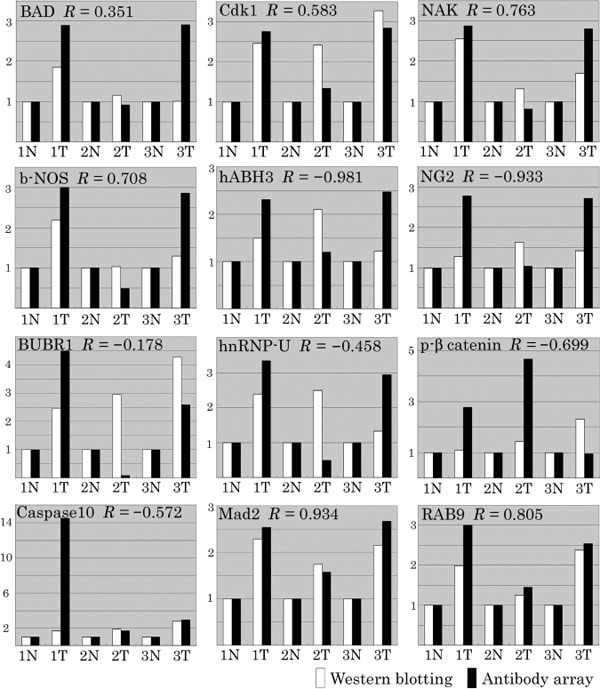
Comparison of the microarray with the western blotting data. The correlation coefficient of the 14 proteins found to be overexpressed in tumor tissues in western blotting was calculated. Normalization of each tumor tissue's data was carried out using the normal counterpart's data.
Immunohistochemistry. We carried out immunohistochemistry to verify the abundance and cellular localization of the 14 proteins with high expression levels in ESCC as revealed by western blotting (Table 3). The expression of six proteins (AP‐1, BUBR1, caspase 10, Cdk1, NAK, and Mad2), either cytoplasmic or nuclear, was stronger in ESCC compared with normal epithelium. Nuclear expression of heterogeneous nuclear ribonucleoprotein U (hn‐RNP‐U) was observed in all tumor and normal tissues. The expression of Bcl‐associated death promoter (BAD) and AP‐2α was higher in infiltrating inflammatory cells compared with the tumor stroma (Fig. 6). The expression of p‐β‐catenin, HABH3, and member RAS oncogene family (RAB) was similar in tumor and normal tissues (data not shown). Nitric oxide synthase 1 (b‐NOS) and NG2 expression were not detected immunohistochemically (data not shown).
Figure 6.
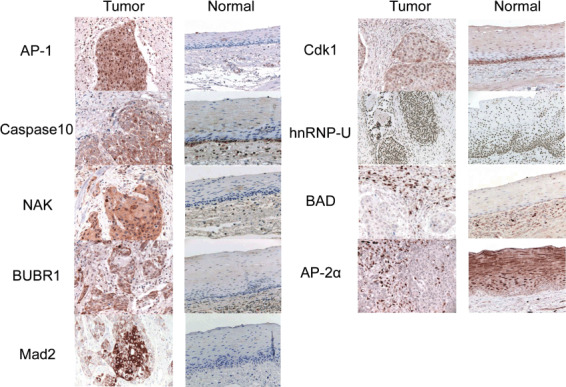
Immunohistochemistry. The expression of six proteins (activator protein [AP]‐1, BUBR1, caspase 10, cyclin‐dependent kinase [Cdk] 1, nuclear factor‐κB‐activating kinase [NAK], and Mad2), either cytoplasmic or nuclear, was stronger in esophageal squamous cell carcinoma compared with normal epithelium. Nuclear expression of hn‐RNP‐U was observed in all tumor and normal tissues. The expression of BAD and AP‐2α was higher in infiltrating inflammatory cells compared with the tumor stroma. BUBR, budding uninhibited by benzidazoles 1 homolog beta; Mad, mitotic arrest deficient‐like 1; hn‐RNP‐U, heterogeneous nuclear ribonucleoprotein U; BAD, Bcl‐associated death promoter.
Of the six proteins with stronger immunohistochemical expression in ESCC, tissue microarray analysis showed that BUBR1, Mad2, AP‐1, caspase 10, NAK, and Cdk1 were overexpressed in 80, 86, 73, 73, 63, and 90% of ESCC patients, respectively, compared with normal tissues. There was no association between BUBR1, AP‐1, caspase 10, or Cdk1 expression with clinicopathological characteristics. Overexpression of Mad2 and NAK was infrequent in tumors histologically categorized as grade 3 or with lymph node metastases, respectively (Table 4).
Table 4.
Tissue microarray results
| Variable | n | BUBR1 | Mad2 | AP‐1 | Caspase 10 | NAK | Cdk1 |
|---|---|---|---|---|---|---|---|
| All cases | 70 | 56 (80%) | 60 (86%) | 51 (73%) | 51 (73%) | 44 (63%) | 63 (90%) |
| Sex | |||||||
| Male | 55 | 44 (80%) | 49 (89%) | 40 (73%) | 40 (73%) | 33 (60%) | 49 (89%) |
| Female | 15 | 12 (80%) | 11 (73%) | 11 (73%) | 11 (73%) | 11 (73%) | 15 (100%) |
| Age (years) | |||||||
| <60 | 38 | 29 (76%) | 32 (84%) | 27 (71%) | 25 (66%) | 20 (53%) | 34 (89%) |
| ≥60 | 32 | 27 (84%) | 28 (88%) | 24 (75%) | 26 (81%) | 24 (75%) | 29 (91%) |
| Histological grade | |||||||
| 1 | 16 | 12 (75%) | 16 (100%)* | 13 (81%) | 9 (56%) | 6 (38%) | 14 (88%) |
| 2 | 42 | 33 (79%) | 39 (93%)* | 31 (74%) | 33 (79%) | 29 (69%) | 39 (93%) |
| 3 | 9 | 8 (89%) | 2 (22%)* | 4 (44%) | 6 (67%) | 7 (78%) | 8 (89%) |
| X | 3 | 3 (100%) | 3 (100%) | 3 (100%) | 3 (100%) | 2 (67%) | 2 (67%) |
| Tumor stage | |||||||
| T1 | 12 | 10 (83%) | 12 (100%) | 9 (75%) | 9 (75%) | 9 (75%) | 11 (92%) |
| T2 | 7 | 5 (71%) | 6 (86%) | 7 (100%) | 6 (86%) | 4 (57%) | 7 (100%) |
| T3 | 10 | 8 (80%) | 8 (80%) | 8 (80%) | 10 (100%) | 5 (50%) | 9 (90%) |
| T4 | 1 | 0 (0%) | 1 (100%) | 1 (100%) | 0 (0%) | 0 (0%) | 1 (100%) |
| Lymph node metastasis | |||||||
| Negative | 17 | 13 (76%) | 15 (88%) | 12 (71%) | 13 (76%) | 14 (82%)* | 16 (94%) |
| Positive | 13 | 10 (77%) | 12 (92%) | 13 (100%) | 12 (92%) | 4 (31%)* | 12 (92%) |
| Distant metastasis | |||||||
| Negative | 25 | 19 (76%) | 23 (92%) | 21 (84%) | 20 (80%) | 15 (60%) | 23 (92%) |
| Positive | 5 | 4 (80%) | 4 (80%) | 4 (80%) | 5 (100%) | 3 (60%) | 5 (100%) |
| Tumor size (mm) | |||||||
| <40 | 14 | 9 (64%) | 12 (86%) | 11 (79%) | 10 (71%) | 10 (71%) | 14 (100%) |
| ≥40 | 16 | 14 (88%) | 15 (94%) | 14 (88%) | 15 (94%) | 8 (50%) | 14 (88%) |
*Statistically significant differences (P < 0.05). BUBR, budding uninhibited by benzidazoles 1 homolog beta; Mad, mitotic arrest deficient‐like 1; AP, activator protein; NAK, TANK‐binding kinase 1; Cdk, cyclin‐dependent kinase.
mRNA expression. The mRNA expression levels of 22 proteins identified by antibody microarray as overexpressed in tumors were examined and compared with western blotting results (Fig. 7). Among the 14 proteins found to be overexpressed in ESCC in western blotting analysis, nine (64%) proteins were also overexpressed at the mRNA level. The expression of two (50%) of four proteins found to be lower in ESCC by western blotting was also lower at the mRNA level. Discordance between the mRNA and western blotting results was observed in approximately 40% of the proteins studied.
Figure 7.
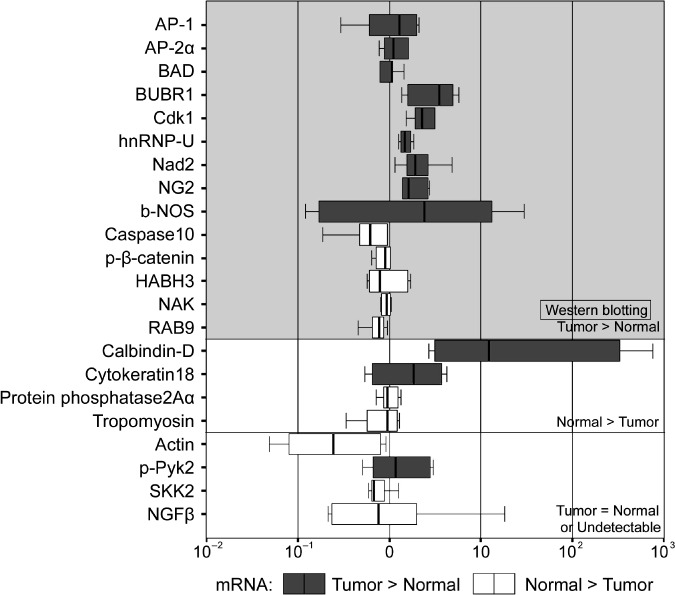
The mRNA expression levels of the 22 identified proteins. Among the 14 proteins overexpressed in esophageal squamous cell carcinoma (ESCC) by western blotting analysis, nine were also found to be overexpressed at the mRNA level. The expression of two of four proteins found to be lower in ESCC in western blotting was also lower at the mRNA level.
Discussion
The study of cancer through the use of antibody microarrays provides valuable information on protein expression levels, isoforms expressed, and post‐translational modifications concerning the particular cancer studied. Furthermore, as many reports have demonstrated discordance between mRNA and protein expression,( 15 , 16 , 17 ) which concerned approximately 40% of proteins in this study, the information provided more closely reflects cancer phenotypes. As well as in separation‐based proteomics such as that using two‐dimensional gel electrophoresis,( 7 , 18 , 19 ) antibody microarrays have been used in the study of colorectal,( 20 ) breast,( 21 , 22 ) pancreatic,( 23 , 24 ) bladder,( 25 ) and prostate cancer,( 26 ) squamous cell carcinoma of the oral cavity,( 27 , 28 ) hepatocellular carcinoma,( 29 ) and gastric cancer.( 30 ) Although several proteomic studies have been reported,( 7 , 8 ) antibody microarrays have not been used to date in esophageal cancer.
Microarray analysis identified 24 proteins that were overexpressed in the tumor tissues compared with the corresponding normal tissues. Examination of these proteins by western blotting and immunohistochemistry validated these results for six of these proteins. Of the six proteins, Mad2 and BubR1 are crucial components of a functional mitotic checkpoint by organizing spindle assembly.( 31 ) Overexpression of the Mad2 gene has been reported in nasopharyngeal,( 32 ) gastric,( 33 ) ovarian,( 34 ) colorectal,( 35 ) hepatocellular,( 36 ) and esophageal cancer,( 37 ) whereas BubR1 is overexpressed in colorectal,( 38 ) gastric,( 39 ) and esophageal cancer.( 37 ) Tanaka et al. reported that Mad2 and BubR1 are coordinately overexpressed in ESCC, and were overexpressed in 89 and 95% of ESCC, respectively, compared with normal tissues.( 37 ) Although they reported no correlation between any clinicopathological data, including the histological grade and the expression of these proteins, we identified a correlation between Mad2 expression and histological grade (Table 4). This correlation has also been reported in gastric cancer( 33 ) and hence is a finding that warrants further investigation. In this study, we detected overexpression of Mad2 and BubR1, which were included in cell cycle regulation, in ESCC in different cohorts. Recently, the relationship between the mitotic checkpoint and drug sensitivity has been the subject of extensive investigations.( 40 ) These proteins may be considered as important determinants of the efficacy of microtubule‐targeting drugs in killing cancer cells.
Cdk1 is a member of the CDK family, which regulates phase transitions and checkpoints within the cell cycle.( 41 , 42 ) Under non‐neoplastic conditions, Cdk1 is localized to the nucleus where it regulates cell cycle progression as a subunit of the M‐phase promoting factor, together with a cyclin B subunit. Within the normal esophageal mucosa, Cdk1 is localized to the parabasal cell layer, where the majority of normal cellular proliferation occurs.( 43 ) In this study, the fact that these three proteins serving crucial roles in M‐phase regulation were concurrently identified may indicate that one or all of them may contribute significantly to the carcinogenesis of ESCC.
The remaining three proteins, AP‐1, caspase 10, and NAK, are associated with NF‐κB, which is a signal transduction factor that has emerged as an important modulator of altered gene programs and malignant phenotype in the development of cancer.( 44 ) In one esophageal adenocarcinoma study, 61% of resected tumors displayed NF‐κB immunoreactivity and 87.5% of the NF‐κB‐positive tumors were Stage IIb or III.( 45 ) Constitutive activation of the NF‐κB and AP‐1 signal transduction pathways have been identified as prominent events promoting tumor progression of hemopoietic and solid malignancies.( 46 , 47 , 48 ) Overexpression of a caspase 10 subclass dramatically enhances NF‐κB activity in a dose‐ and time‐dependent manner.( 49 ) NAK phosphorylates a NF‐κB subunit and increases its activity.( 50 ) In this study, the correlation between underexpression of NAK and the presence of lymph node metastases was revealed (Table 4); although NAK has been reported as a mediator of tumor angiogenesis,( 51 ) this correlation had not been reported to date and, again, additional investigation is necessary. To the best of our knowledge, aberrant expression of these proteins has not been previously reported in ESCC. Our results also suggest that these members of the NF‐κB signaling pathway may play an important role in the carcinogenesis of ESCC.
In this study, the reproducibility of the antibody microarray results was poor (Fig. 2) and varied between the experiments, with the correlation coefficient ranging from 0.7172 to 0.9332. It remains a challenge to improve the reproducibility of the results obtained using antibody microarrays such as the ones in our study. To achieve this, Ellmark et al. used eight replicates in a microarray and excluded the two highest and two lowest replicates, and each data point represented the mean value of the remaining four replicates.( 30 ) When space allows, the antibody microarrays should therefore be spotted with more replicates in the same microarray to improve reproducibility. Furthermore, as the degree of reproducibility was different based on the signal intensity, the threshold may also need to be changed as a function of signal intensity.
As spots with lower signal intensity had poor reproducibility (Fig. 2), it may be better to pre‐exclude them from data analysis. The amount and affinity of the antibodies in the microarray should be best optimized so that the dynamic range of all spots is within a linearly measurable range. However, the affinity to antigens is different between antibodies. In addition, the amount of individual antigens is also variable between the samples. It seems that these problems may be inherent to antibody microarray experiments. Therefore, extensive validation studies by other methods should be carried out for results obtained by antibody microarrays.
As tumor tissues contain many types of tumor and non‐tumor cells, recovery of specific cell populations by laser microdissection prior to protein extraction is generally required for accurate expression analysis. However, as 100 µg of each protein is required for every antibody microarray experiment, and laser microdissection can collect a small amount of protein, we cannot avoid using homogenized tissue samples. The advantages and limitations of antibody microarrays, western blotting, and immunohistochemistry in the analysis of protein expression are discussed in the Supplemental text. In our study, two proteins identified as being overexpressed in tumor tissues were shown to derive from infiltrating inflammatory cells in the tumor stroma rather than the tumor itself, a finding that stresses the need for immunohistochemical studies in order to localize the identified proteins in the tissues.
In conclusion, through the use of antibody microarrays we identified a novel association of mitotic checkpoint gene products and the NF‐κB pathway with esophageal cancer. These proteins will not only be useful in better understanding the molecular background of ESCC but will also be candidates for biomarkers or therapeutic targets. Although antibody microarrays generate expression data in a high‐throughput way, validation of the results by other methods is required due to its limited reproducibility.
Disclosure Statement
The authors declare that they have no competing interests.
Abbreviations
| AP | activator protein |
| BubR | budding uninhibited by benzidazoles 1 homolog beta |
| Cdk | cyclin‐dependent kinase |
| ESCC | esophageal squamous cell carcinoma |
| HABH | alkylation repair homolog |
| Mad | mitotic arrest deficient‐like 1 |
| NAK | nuclear factor‐κB‐activating kinase |
| NF | nuclear factor |
| NG | neurite growth |
Supporting information
Supplemental text
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgments
This work was supported by a grant from the Ministry of Health, Labor, and Welfare and by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation of Japan.
References
- 1. Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer 1999; 80: 827–41. [DOI] [PubMed] [Google Scholar]
- 2. Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer 1999; 83: 18–29. [DOI] [PubMed] [Google Scholar]
- 3. Ilson DH. Oesophageal cancer: new developments in systemic therapy. Cancer Treat Rev 2003; 29: 525–32. [DOI] [PubMed] [Google Scholar]
- 4. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003; 349: 2241–52. [DOI] [PubMed] [Google Scholar]
- 5. Mariette C, Balon JM, Piessen G, Fabre S, Van Seuningen I, Triboulet JP. Pattern of recurrence following complete resection of esophageal carcinoma and factors predictive of recurrent disease. Cancer 2003; 97: 1616–23. [DOI] [PubMed] [Google Scholar]
- 6. Metzger R, Schneider PM, Warnecke‐Eberz U, Brabender J, Holscher AH. Molecular biology of esophageal cancer. Onkologie 2004; 27: 200–6. [DOI] [PubMed] [Google Scholar]
- 7. Hatakeyama H, Kondo T, Fujii K et al . Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics 2006; 6: 6300–16. [DOI] [PubMed] [Google Scholar]
- 8. Liu WL, Zhang G, Wang JY et al . Proteomics‐based identification of autoantibody against CDC25B as a novel serum marker in esophageal squamous cell carcinoma. Biochem Biophys Res Commun 2008; 375: 440–5. [DOI] [PubMed] [Google Scholar]
- 9. Homs MY, Kuipers EJ, Siersema PD. Palliative therapy. J Surg Oncol 2005; 92: 246–56. [DOI] [PubMed] [Google Scholar]
- 10. Katlic MR, Wilkins EW Jr, Grillo HC. Three decades of treatment of esophageal squamous carcinoma at the Massachusetts General Hospital. J Thorac Cardiovasc Surg 1990; 99: 929–38. [PubMed] [Google Scholar]
- 11. Brockmann JG, St Nottberg H, Glodny B, Heinecke A, Senninger NJ. CYFRA 21‐1 serum analysis in patients with esophageal cancer. Clin Cancer Res 2000; 6: 4249–52. [PubMed] [Google Scholar]
- 12. Crnogorac‐Jurcevic T, Gangeswaran R, Bhakta V et al . Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology 2005; 129: 1454–63. [DOI] [PubMed] [Google Scholar]
- 13. Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas – a tool for pathology. J Pathol 2008; 216: 387–93. [DOI] [PubMed] [Google Scholar]
- 14. Sobin L, Wittekind C. UICC TNM Classification of Malignant Tumors, 6th edn. New York: Wiley‐Liss, 2002. [Google Scholar]
- 15. Chen G, Gharib TG, Huang CC et al . Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 2002; 1: 304–13. [DOI] [PubMed] [Google Scholar]
- 16. Varambally S, Yu J, Laxman B et al . Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005; 8: 393–406. [DOI] [PubMed] [Google Scholar]
- 17. Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 1999; 19: 1720–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uemura N, Nakanishi Y, Kato H et al . Transglutaminase 3 as a prognostic biomarker in esophageal cancer revealed by proteomics. Int J Cancer 2009; 124: 2106–15. [DOI] [PubMed] [Google Scholar]
- 19. Nishimori T, Tomonaga T, Matsushita K et al . Proteomic analysis of primary esophageal squamous cell carcinoma reveals downregulation of a cell adhesion protein, periplakin. Proteomics 2006; 6: 1011–18. [DOI] [PubMed] [Google Scholar]
- 20. Madoz‐Gurpide J, Canamero M, Sanchez L, Solano J, Alfonso P, Casal JI. A proteomics analysis of cell signaling alterations in colorectal cancer. Mol Cell Proteomics 2007; 6: 2150–64. [DOI] [PubMed] [Google Scholar]
- 21. Smith L, Watson MB, O’Kane SL, Drew PJ, Lind MJ, Cawkwell L. The analysis of doxorubicin resistance in human breast cancer cells using antibody microarrays. Mol Cancer Ther 2006; 5: 2115–20. [DOI] [PubMed] [Google Scholar]
- 22. Celis JE, Moreira JM, Cabezon T et al . Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients: toward dissecting the molecular circuitry of epithelial–adipocyte stromal cell interactions. Mol Cell Proteomics 2005; 4: 492–522. [DOI] [PubMed] [Google Scholar]
- 23. Ingvarsson J, Wingren C, Carlsson A et al . Detection of pancreatic cancer using antibody microarray‐based serum protein profiling. Proteomics 2008; 8: 2211–19. [DOI] [PubMed] [Google Scholar]
- 24. Orchekowski R, Hamelinck D, Li L et al . Antibody microarray profiling reveals individual and combined serum proteins associated with pancreatic cancer. Cancer Res 2005; 65: 11 193–202. [DOI] [PubMed] [Google Scholar]
- 25. Sanchez‐Carbayo M, Socci ND, Lozano JJ, Haab BB, Cordon‐Cardo C. Profiling bladder cancer using targeted antibody arrays. Am J Pathol 2006; 168: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller JC, Zhou H, Kwekel J et al . Antibody microarray profiling of human prostate cancer sera: antibody screening and identification of potential biomarkers. Proteomics 2003; 3: 56–63. [DOI] [PubMed] [Google Scholar]
- 27. Knezevic V, Leethanakul C, Bichsel VE et al . Proteomic profiling of the cancer microenvironment by antibody arrays. Proteomics 2001; 1: 1271–8. [DOI] [PubMed] [Google Scholar]
- 28. Weber A, Hengge UR, Stricker I et al . Protein microarrays for the detection of biomarkers in head and neck squamous cell carcinomas. Hum Pathol 2007; 38: 228–38. [DOI] [PubMed] [Google Scholar]
- 29. Tannapfel A, Anhalt K, Hausermann P et al . Identification of novel proteins associated with hepatocellular carcinomas using protein microarrays. J Pathol 2003; 201: 238–49. [DOI] [PubMed] [Google Scholar]
- 30. Ellmark P, Ingvarsson J, Carlsson A, Lundin BS, Wingren C, Borrebaeck CA. Identification of protein expression signatures associated with Helicobacter pylori infection and gastric adenocarcinoma using recombinant antibody microarrays. Mol Cell Proteomics 2006; 5: 1638–46. [DOI] [PubMed] [Google Scholar]
- 31. Hoyt MA. A new view of the spindle checkpoint. J Cell Biol 2001; 154: 909–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Jin DY, Wong YC et al . Correlation of defective mitotic checkpoint with aberrantly reduced expression of MAD2 protein in nasopharyngeal carcinoma cells. Carcinogenesis 2000; 21: 2293–7. [DOI] [PubMed] [Google Scholar]
- 33. Tanaka K, Nishioka J, Kato K et al . Mitotic checkpoint protein hsMAD2 as a marker predicting liver metastasis of human gastric cancers. Jpn J Cancer Res 2001; 92: 952–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X, Jin DY, Ng RW et al . Significance of MAD2 expression to mitotic checkpoint control in ovarian cancer cells. Cancer Res 2002; 62: 1662–8. [PubMed] [Google Scholar]
- 35. Li GQ, Li H, Zhang HF. Mad2 and p53 expression profiles in colorectal cancer and its clinical significance. World J Gastroenterol 2003; 9: 1972–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeong SJ, Shin HJ, Kim SJ et al . Transcriptional abnormality of the hsMAD2 mitotic checkpoint gene is a potential link to hepatocellular carcinogenesis. Cancer Res 2004; 64: 8666–73. [DOI] [PubMed] [Google Scholar]
- 37. Tanaka K, Mohri Y, Ohi M et al . Mitotic checkpoint genes, hsMAD2 and BubR1, in oesophageal squamous cancer cells and their association with 5‐fluorouracil and cisplatin‐based radiochemotherapy. Clin Oncol (R Coll Radiol) 2008; 20: 639–46. [DOI] [PubMed] [Google Scholar]
- 38. Shichiri M, Yoshinaga K, Hisatomi H, Sugihara K, Hirata Y. Genetic and epigenetic inactivation of mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to survival. Cancer Res 2002; 62: 13–17. [PubMed] [Google Scholar]
- 39. Grabsch H, Takeno S, Parsons WJ et al . Overexpression of the mitotic checkpoint genes BUB1, BUBR1, and BUB3 in gastric cancer – association with tumour cell proliferation. J Pathol 2003; 200: 16–22. [DOI] [PubMed] [Google Scholar]
- 40. Lee EA, Keutmann MK, Dowling ML, Harris E, Chan G, Kao GD. Inactivation of the mitotic checkpoint as a determinant of the efficacy of microtubule‐targeted drugs in killing human cancer cells. Mol Cancer Ther 2004; 3: 661–9. [PubMed] [Google Scholar]
- 41. Doree M, Hunt T. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner? J Cell Sci 2002; 115: 2461–4. [DOI] [PubMed] [Google Scholar]
- 42. Murray AW. Recycling the cell cycle: cyclins revisited. Cell 2004; 116: 221–34. [DOI] [PubMed] [Google Scholar]
- 43. Hansel DE, Dhara S, Huang RC et al . CDC2/CDK1 expression in esophageal adenocarcinoma and precursor lesions serves as a diagnostic and cancer progression marker and potential novel drug target. Am J Surg Pathol 2005; 29: 390–9. [DOI] [PubMed] [Google Scholar]
- 44. Van Waes C. Nuclear factor‐kappaB in development, prevention, and therapy of cancer. Clin Cancer Res 2007; 13: 1076–82. [DOI] [PubMed] [Google Scholar]
- 45. Abdel‐Latif MM, O'Riordan J, Windle HJ et al . NF‐kappaB activation in esophageal adenocarcinoma: relationship to Barrett's metaplasia, survival, and response to neoadjuvant chemoradiotherapy. Ann Surg 2004; 239: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aggarwal BB. Nuclear factor‐kappaB: the enemy within. Cancer Cell 2004; 6: 203–8. [DOI] [PubMed] [Google Scholar]
- 47. Shaulian E, Karin M. AP‐1 as a regulator of cell life and death. Nat Cell Biol 2002; 4: E131–6. [DOI] [PubMed] [Google Scholar]
- 48. Eferl R, Wagner EF. AP‐1: a double‐edged sword in tumorigenesis. Nat Rev Cancer 2003; 3: 859–68. [DOI] [PubMed] [Google Scholar]
- 49. Wang H, Wang P, Sun X et al . Cloning and characterization of a novel caspase‐10 isoform that activates NF‐kappa B activity. Biochim Biophys Acta 2007; 1770: 1528–37. [DOI] [PubMed] [Google Scholar]
- 50. Bouwmeester T, Bauch A, Ruffner H et al . A physical and functional map of the human TNF‐alpha/NF‐kappa B signal transduction pathway. Nat Cell Biol 2004; 6: 97–105. [DOI] [PubMed] [Google Scholar]
- 51. Czabanka M, Korherr C, Brinkmann U, Vajkoczy P. Influence of TBK‐1 on tumor angiogenesis and microvascular inflammation. Front Biosci 2008; 13: 7243–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


