Abstract
The incidence of chronic lymphocytic leukemia is low in the Japanese population compared with populations in western countries, suggesting a role for genetic factors in the occurrence of this disease. We have previously shown that chronic lymphocytic leukemia in Japan rarely expresses the immunoglobulin heavy chain variable region (IGHV) 1‐69 gene (1 out of 43 patients, 2.3%), which is a gene most commonly expressed in chronic lymphocytic leukemia cases from western countries. In the current study, we extended the previous study by examining immunoglobulin heavy chain and light chain gene expression in 80 Japanese patients with chronic lymphocytic leukemia and in 52 Japanese patients with other leukemic chronic lymphoproliferative disorders. IGHV1‐69 gene expression was again quite low in our cohort, found in only two patients: one with chronic lymphocytic leukemia and the other with splenic marginal zone lymphoma. The IGHV4‐34 gene was most frequently expressed in chronic lymphocytic leukemia (27.5%), whereas it was rarely found in leukemic chronic lymphoproliferative disorders (7.7%, P = 0.005). There was also a significant difference in the expression of IGLV3‐21 between chronic lymphocytic leukemia and leukemic chronic lymphoproliferative disorders (29.4 vs 4.8%, P = 0.018). The IGLV3‐21 gene in the majority of chronic lymphocytic leukemia cases was associated with homologous complementarity determining region 3 sequences. Recent studies identified subsets of cases expressing almost identical B‐cell receptors. We found that two patients with chronic lymphocytic leukemia and the patient with splenic marginal zone lymphoma expressed IGHV4‐39/IGKV1‐39 and IGHV1‐69/IGKV3‐20, respectively, which belong to these subsets. (Cancer Sci 2009)
Abbreviations:
- IGHD
Immunoglobulin heavy chain diversity region
- IGHJ
Immunoglobulin heavy chain joining region
- IGHV
Immunoglobulin heavy chain variable region
- IGKJ
Immunoglobulin kappa chain joining region
- IGKV
Immunoglobulin kappa chain variable region
- IGLJ
Immunoglobulin lambda chain joining region
- IGLV
Immunoglobulin lambda chain variable region
Chronic lymphocytic leukemia (CLL) is a neoplasm of mature small B lymphocytes in the peripheral blood, bone marrow, and lymph nodes, usually expressing CD5 and CD23.( 1 ) The incidence of CLL is low in Asian countries, including Japan,( 2 , 3 , 4 , 5 ) whereas it is the most common type of leukemia in western countries. It has been shown that the mutational status of the immunoglobulin heavy chain variable region (IGHV) gene can predict the prognosis of CLL; unmutated IGHV genes correlate with a worse prognosis, whereas mutated IGHV genes correlate with a good prognosis.( 6 , 7 ) Overrepresentation of selected IGHV genes (highly restricted immunoglobulin gene expression) such as IGHV1‐69 and IGHV4‐34 has been noted in CLL.( 7 , 8 , 9 , 10 , 11 ) Moreover, several studies have shown that subgroups of CLL have very similar antigen‐binding sequences (stereotyped B‐cell antigen receptors, BCR), including complementarity determining region (CDR) 3, suggesting antigen selection during the maintenance and development of the disease.( 12 , 13 , 14 , 15 , 16 , 17 ) These findings have emphasized the hypothesis that antigens could play a role in the pathogenesis of CLL.
We previously analyzed IGHV expression in 43 Japanese patients with CLL and identified only one patient who expressed IGHV1‐69,( 18 ) whereas IGHV1‐69 is a gene most frequently expressed in CLL cases in western countries (approximately 10–20%).( 7 , 11 , 19 ) Although IGHV genes have been investigated in detail, few studies have analyzed immunoglobulin light chain genes,( 20 , 21 , 22 ) other than in selected cases of CLL, such as IGHV1‐69+ or IGHV3‐21+ cases.( 12 , 23 ) To further characterize CLL in the Japanese population, immunoglobulin light chain (kappa chain, IGK; lambda chain, IGL) genes in addition to IGHV genes were analyzed in 82 CLL cases and compared with those of 53 leukemic chronic lymphoproliferative disorders (CLPD), including hairy cell leukemia, prolymphocytic leukemia (PLL), and indolent lymphomas in the leukemic phase, such as mantle cell lymphoma (MCL), follicular lymphoma, splenic marginal zone lymphoma (SMZL), and lymphoplasmacytic lymphoma.
Materials and Methods
Patients. This study was approved by the local institutional review board. After obtaining written informed consent, we enrolled 82 patients with CLL and 53 patients with leukemic CLPD who were referred to our institution between March 1999 and May 2008. The median age at diagnosis was 70 years (range 37–92 years) in patients with CLL and 68 years (range 40–95 years) in patients with leukemic CLPD. At least five hematologists examined the blood smears. The cytology of biopsied samples was reevaluated at a monthly conference consisting of hematologists, radiologists, and pathologists, including a hematopathologist from Gunma Cancer Center Hospital.
Cell‐surface antigen analysis. Cell‐surface antigens were analyzed by flow cytometry using a FACSCalibur or a FACSCanto II flow cytometer (Becton Dickinson, San Jose, CA, USA) and the data were analyzed using CELLQuest or FACSDiva software (Becton Dickinson) as described previously.( 24 ) Fluorescein isothionate‐conjugated antimouse IgG1 (control), anti‐CD5, anti‐CD19, and antikappa antibodies were purchased from BD Pharmingen (San Diego, CA, USA). Phycoerythrin‐conjugated antimouse IgG1, anti‐CD10, anti‐CD11c, anti‐CD19, anti‐CD23, anti‐CD25, anti‐CD27, anti‐CD79b, anti‐CD103, anti‐IgM, anti‐IgD, anti‐IgG, and antilambda antibodies were purchased from BD Pharmingen. Peridinin chlorophyll protein‐conjugated anti‐CD45 antibody (for gating) was purchased from Becton Dickinson.
Analysis of immunoglobulin heavy chain, IGK, and IGL gene expression. DNA isolation, RNA isolation, and first‐strand cDNA synthesis were carried out according to reported methods.( 18 ) In a previous report of 44 patients with CLL,( 18 ) immunoglobulin heavy chain (IGH) genes were amplified from cDNA according to the method which written in the report of Fais et al.( 8 ) or from genomic DNA (gDNA) according to the method which written in the report of Küppers et al.( 25 ) In newly diagnosed additional CLL and leukemic CLPD cases, IGH genes were detected from gDNA using BIOMED‐2 VH FR1 and JH primers, IGK genes were detected using BIOMED‐2 Vκ and Jκ primers, and IGL genes were detected using BIOMED‐2 Vλ and Jλ primers.( 26 ) When two bands were amplified from gDNA, amplification was repeated from cDNA. Cell‐surface antigen analysis revealed that kappa light chain was expressed by 45% of CLL cells, lambda light chain was expressed by 45% of CLL cells, and no or very faint light chain was expressed by 10% of CLL cells. IGK genes were amplified from kappa+ cases, IGL genes from lambda+ cases, and both genes from no or very faint cases. Most of the amplified samples were directly sequenced using the forward primers as above, with the BigDye Terminator Cycle Sequencing FS Ready Reaction kit (Applied Biosystems, Foster City, CA, USA) on an ABI Prism 310 Genetic Analyzer (Applied Biosystems) and Sequencing Analysis Software (Applied Biosystems). Some samples were sequenced by TA cloning. Nucleotide sequences were compared with those in the IgBLAST (http://www.ncbi.nlm.nih.gov/igblast) and IMGT (http://imgt.cines.fr) databases. Unmutated immunoglobulin sequences were defined as those with <2% sequence deviation from the most similar germline immunoglobulin genes and mutated sequences were defined as those with 2% sequence differences.( 6 , 7 , 11 )
Statistical analysis. Comparisons of the frequency between groups were analyzed (Pearson's coefficient) by χ2‐test and Fisher's exact test using StatMateIII software (ATMS, Tokyo, Japan). A probability value of P < 0.05 was considered to indicate statistical significance.
Results
Patients and diagnosis. Table 1 shows the diagnoses of patients enrolled in this study. All cases of CLL had more than 5 × 109 B lymphocytes/L of blood,( 27 ) and all patients with leukemic CLPD showed at least 10% leukemic cells in their peripheral blood.( 1 ) The diagnosis was based on immunophenotypic analysis and cell morphology determined from Wright‐stained peripheral blood smears. All cases of CLL were positive for CD5, and CD23‐positive CLL cases were 90.1% (73/81) (Table 2). To distinguish MCL from CLL, both of which are CD5‐positive, cyclin D1 (CCND1) expression was analyzed using real‐time quantitative polymerase chain reaction (RQ‐PCR).( 28 ) This analysis confirmed low CCND1 expression in all patients with CLL and high expression in all patients with MCL. Hairy cell leukemic cells frequently expressed CD11c, CD25, and/or CD103. Prolymphocytes of PLL exceeded 55% of the lymphoid cells in the peripheral blood. Follicular lymphoma was diagnosed based on CD10 positivity, typical cell morphology, and the presence of t(14;18) in peripheral tumor cells or typical histological findings of lymph nodes or spleen. Splenic marginal zone lymphoma was diagnosed from histological samples of an enlarged spleen. Lymphoplasmacytic lymphoma was diagnosed from peripheral blood and bone marrow features. Unless typical phenotypic or genotypic markers were available, it was often difficult to diagnose indolent lymphomas in the leukemic phase without appropriate tissue biopsy samples, so 14 cases were diagnosed as unclassified CLPD, in which cases of CD5‐negative CLL might be included.
Table 1.
Numbers of patients with chronic lymphocytic leukemia (CLL) and leukemic chronic lymphoproliferative disorders (CLPD)
| Diagnosis | No. patients |
|---|---|
| CLL | 82 |
| Leukemic CLPD | 53 |
| Lymphoma in leukemic phase | |
| Mantle cell lymphoma | 15 |
| Follicular lymphoma | 7 |
| SMZL | 5 |
| Lymphoplasmacytic lymphoma | 5 |
| Hairy cell leukemia | 6 |
| Prolymphocytic leukemia | 1 |
| Unclassified CLPD | 14 |
CLPD, chronic lymphoproliferative disorder; SMZL, splenic marginal zone lymphoma.
Table 2.
Percentage of CD5‐positive or CD23‐positive cells in CD19‐positive cells immunoglobulin heavy chain variable region (IGHV), IGKV, and IGLV status in chronic lymphocytic leukemia (CLL) patients
| CLL patient | CD5 (%) | CD23 (%) | IGHV | Homology (%) | IGHD | IGHJ | K/L | IGKV/IGLV | IGKJ/LJ | Homology (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| CLL 1 | 98.0 | 85.1 | 1–2 | 94.2 | 2–15 | 4 | K | 3–20(A27) | 2 | 98.8 |
| CLL 2 | 86.2 | 97.4 | 1–3 | 90.0 | 3–9 | 5 | K | 3D‐20(A11) | 2 | 96.2 |
| CLL 3 | 89.7 | 87.5 | 1–8 | 94.2 | 1–1 | 5 | K | 3D‐20(A11) | 2 | 89.1 |
| CLL 4 | 87.1 | 98.0 | 1–8 | 94.9 | 6–13 | 4 | K | 3–11(L6) | 4 | 98.0 |
| CLL 5 | 98.0 | 54.7 | 1–18 | 98.5 | 3–10 | 6 | L | 2–11(V1‐3) | 3 | 92.9 |
| CLL 6 | 73.3 | 97.7 | 1–18 | 99.0 | 2–15 | 6 | L | 1–47(V1‐17) | 3 | 100 |
| CLL 7 | 98.4 | 97.4 | 1–46 | 99.1 | 3–9 | 6 | K | 3D‐20(A11) | 2 | 97.7 |
| CLL 8 | 100 | 69.7 | 1–69 | 98.0 | 5–12 | 6 | L | 3–1(V2‐1) | 4 | 99.5 |
| CLL 9 | 85.9 | 98.0 | 2–5 | 99.4 | 2–2 | 4 | K | 2D‐29(A2) | 1 | 99.3 |
| CLL 10 | 58.0 | 89.3 | 2–70 | 91.6 | 3–10 | 4 | L | 3–21(V2‐14) | 3 | 90.9 |
| CLL 11 | 89.4 | 38.5 | 3–7 | 93.7 | 2–2 | 3 | L | 1–44(V1‐16) | 1 | 96.0 |
| CLL 12 | 100 | 18.2 | 3–7 | 99.3 | 3–9 | 4 | L | 3–21(V2‐14) | 3 | 100 |
| CLL 13 | 51.9 | 40.9 | 3–7 | 87.0 | 4–17 | 3 | K | 1–33(O18) | 4 | 100 |
| CLL 14 | 99.4 | 52.5 | 3–7 | 94.8 | 2–15 | 5 | K | NA | NA | NA |
| CLL 15 | 96.0 | 96.0 | 3–11 | 99.6 | 3–3 | 4 | L | 1–44(V1‐16) | 2 | 98.0 |
| CLL 16 | 91.5 | 63.2 | 3–15 | 94.8 | 2–2 | 2 | L | 3–25(2–17) | 2 | 96.0 |
| CLL 17 | 94.2 | 90.1 | 3–15 | 96.7 | 2–2 | 4 | L | 10–54(V1‐20) | 3 | 83.9 |
| CLL 18 | 43.7 | 97.6 | 3–15 | 93.0 | 6–19 | 4 | L | 3–21(V2‐14) | 3 | 98.0 |
| CLL 19 | 98.2 | 97.3 | 3–21 | 96.3 | 3–10 | 6 | L | 3–19(V2‐13) | 2 | 96.0 |
| CLL 20 | 97.9 | 9.2 | 3–21 | 99.4 | 3–9 | 4 | L | 2–14(V1‐4) | 2 | 94.7 |
| CLL 21 | 32.1 | 35.9 | 3–21 | 99.6 | 2–15 | 5 | L | 2–14(V1‐4) | 2 | 100 |
| CLL 22 | 85.2 | 92.7 | 3–21 | 91.0 | 3–16 | 5 | L | 3–21(V2‐14) | 3 | 97.0 |
| CLL 23 | 100 | 96.6 | 3–21 | 96.0 | 5–21 | 4 | L | 3–21(V2‐14) | 3 | 98.0 |
| CLL 24 | 69.9 | 51.8 | 3–21 | 94.4 | 4–23 | 4 | K | 3–20(A27) | 1 | 95.8 |
| CLL 25 | 47.1 | 14.3 | 3–23 | 88.1 | 1–26 | 4 | L | 3–1(V2‐1) | 2 | 97.0 |
| CLL 26 | 85.0 | 96.0 | 3–23 | 96.6 | 1–26 | 4 | L | 3–21(V2‐14) | 3 | 97.0 |
| CLL 27 | 83.0 | 98.1 | 3–23 | 95.7 | 2–21 | 4 | K | 3–20(A27) | 3 | 95.3 |
| CLL 28 | 100 | 98.2 | 3–23 | 100 | 3–9 | 4 | K | 1–33(O18) | 4 | 100 |
| CLL 29 | 74.0 | 63.5 | 3–23 | 99.1 | 6–19 | 2 | K | 4–1(B3) | 1 | 92.0 |
| CLL 30 | 98.0 | 85.3 | 3–23 | 91.1 | 7–27 | 4 | K | 2–30(A17) | 2 | 91.0 |
| CLL 31 | 95.2 | 99.3 | 3–23 | 92.9 | 2–21 | 6 | L | 1–51(V1‐19) | 5 | 98.8 |
| CLL 32 | 95.2 | 24.7 | 3–30 | 94.7 | 3–22 | 3 | K | 1–37(O14) | 4 | 98.7 |
| CLL 33 | 32.5 | 90.2 | 3–30 | 94.6 | 6–6 | 4 | L | 1–44(V1‐16) | 3 | 92.9 |
| CLL 34 | 42.4 | 93.5 | 3–30 | 85.3 | 2–2 | 5 | L | 1–51(V1‐19) | 3 | 96.7 |
| CLL 35 | 93.2 | 98.2 | 3–30 | 88.0 | 3–3 | 5 | K | 3–11(L6) | 4 | 94.0 |
| CLL 36 | 30.7 | 28.6 | 3–30 | 95.6 | 2–15 | 6 | K | 3–11(L6) | 5 | 100 |
| CLL 37 | 98.0 | 100 | 3–33 | 92.5 | 3–16 | 6 | K | 1D‐17(L14) | 1 | 95.5 |
| CLL 38 | 52.9 | 89.3 | 3–43 | 89.2 | 4–23 | 4 | K | 1D‐17(L14) | 2 | 98.4 |
| CLL 39 | 100 | 98.0 | 3–48 | 98.3 | 3–22 | 4 | L | 3–21(V2‐14) | 1 | 98.0 |
| CLL 40 | 100 | 77.5 | 3–48 | 90.0 | 3–16 | 4 | L | 3–21(V2‐14) | 3 | 96.0 |
| CLL 41 | 44.4 | 41.5 | 3–48 | 91.4 | 7–27 | 4 | K | 2–30(A17) | 6 | 88.2 |
| CLL 42 | 100 | 2.9 | 3–49 | 91.9 | 3–10 | 4 | K | 1–39(O12) | 1 | 95.0 |
| CLL 43 | 100 | 98.0 | 3–53 | 99.0 | 3–10 | 6 | K | 3–11(L6) | 2 | 99.0 |
| CLL 44 | 100 | 91.1 | 3–53 | 95.9 | 1–26 | 2 | K | 3D‐20(A11) | 4 | 92.3 |
| CLL 45 | 89.7 | 100 | 3–64 | 95.7 | 4–17 | 4 | L | 2–14(V1‐4) | 1 | 98.7 |
| CLL 46 | 61.7 | 98.0 | 3–64 | 91.0 | 1–26 | 3 | L | 3–21(V2‐14) | 3 | 92.0 |
| CLL 47 | 36.5 | 78.0 | 3–73 | 91.6 | 3–9 | 4 | K | 2–28(A19) | 4 | 96.0 |
| CLL 48 | 97.5 | 52.5 | 3–74 | 93.0 | 1–1 | 6 | L | 2–23(V1‐7) | 7 | 95.0 |
| CLL 49 | 85.5 | 11.3 | 3–74 | 89.4 | 5–24 | 6 | L | 3–25(V2‐17) | 3 | 94.7 |
| CLL 50 | 36.9 | 64.9 | 4–34 | 96.9 | 6–19 | 4 | L | 2–23(V1‐7) | 2 | 94.4 |
| CLL 51 | 35.9 | 32.8 | 4–34 | 92.2 | NA | NA | L | 3–10(V2‐7) | NA | 91.9 |
| CLL 52 | 98.0 | 98.0 | 4–34 | 94.0 | 6–19 | 5 | L | 1–40(V1‐13) | 2 | 97.2 |
| CLL 53 | 100 | 100 | 4–34 | 94.0 | 2–21 | 5 | L | 3–21(V2‐14) | 1 | 100 |
| CLL 54 | 36.2 | 93.3 | 4–34 | 90.0 | 4–17 | 4 | L | 2–14(V1‐4) | 3 | 90.0 |
| CLL 55 | 52.0 | 29.3 | 4–34 | 92.5 | 3–22 | 1 | K | 2–30(A17) | NA | 97.4 |
| CLL 56 | 78.7 | 82.5 | 4–34 | 86.0 | 3–10 | 4 | K | 3–11(L6) | 4 | 91.5 |
| CLL 57 | 33.0 | 90.0 | 4–34 | 92.3 | 2–15 | 6 | K | 2–28(A19) | 1 | 96.0 |
| CLL 58 | 84.2 | 62.1 | 4–34 | 90.1 | 1–26 | 3 | K | NA | NA | NA |
| CLL 59 | 100 | 94.4 | 4–34 | 98.3 | 5–12 | 6 | K | 1–9(L8) | 3 | 94.0 |
| CLL 60 | 98.0 | 51.0 | 4–34 | 94.6 | 3–22 | 3 | K | 3–11(L6) | 2 | 87.6 |
| CLL 61 | 75.8 | 100 | 4–34 | 88.4 | 1–26 | 6 | K | 2–30(A17) | 2 | 95.7 |
| CLL 62 | 44.3 | 93.8 | 4–34 | 93.0 | 2–21 | 6 | K | 2–28(A19) | 1 | 89.0 |
| CLL 63 | 40.3 | 95.0 | 4–34 | 87.0 | 3–3 | 5 | K | 2–28(A19) | 1 | 93.0 |
| CLL 64 | 80.9 | 98.0 | 4–34 | 99.5 | 7–27 | 4 | K | 3–15(L‐2) | 1 | 99.0 |
| CLL 65 | 86.3 | 95.3 | 4–34 | 97.0 | 7–27 | 4 | K | 1–9(L8) | 4 | 96.0 |
| CLL 66 | 71.8 | 90.9 | 4–34 | 92.0 | 5–18 | 6 | K | 3–11(L6) | 2 | 92.0 |
| CLL 67 | 86.1 | 92.0 | 4–34 | 89.0 | 3–10 | 5 | K | 3D‐15(L16) | 4 | 95.0 |
| CLL 68 | 55.7 | 72.1 | 4–34 | 93.2 | 3–10 | 5 | K | 1–5(L12) | 1 | 98.2 |
| CLL 69 | 58.1 | 98.0 | 4–34 | 90.6 | 2–15 | 6 | K | 1–5(L12) | 2 | 94.1 |
| CLL 70 | 37.9 | 99.4 | 4–34 | 91.0 | 1–26 | 4 | K | 1–5(L12) | 2 | 86.8 |
| CLL 71 | 81.6 | 100 | 4–39 | 94.0 | 1–26 | 4 | L | 3–25(V2‐17) | 1 | 93.9 |
| CLL 72 | 91.2 | 81.5 | 4–39 | 99.7 | 6–13 | 5 | K | 1–39(O12) | 1 | 100 |
| CLL 73 | 82.9 | 48.8 | 4–39 | 98.0 | 6–13 | 5 | K | 1–39(O12) | 2 | 98.0 |
| CLL 74 | 61.5 | 83.1 | 4–4 | 84.7 | 2–8 | 5 | K | 3–20(A27) | 1 | 98.0 |
| CLL 75 | 68.8 | 71.4 | 4–4 | 95.5 | 2–21 | 3 | L | 2–23(V1‐7) | 3 | 96.0 |
| CLL 76 | 43.8 | 63.4 | 4–34 | 92.8 | 6–19 | 2 | L | 1–40(V1‐13) | 3 | 95.1 |
| CLL 77 | 69.1 | 40.9 | 4–61 | 85.4 | 7–27 | 5 | K | 2–30(A17) | 2 | 94.6 |
| CLL 78 | 88.9 | 95.6 | 5–51 | 89.0 | 7–27 | 4 | L | NA | NA | NA |
| CLL 79 | 98.0 | 74.1 | 5–51 | 96.4 | 3–10 | 4 | K | 2–28(A19) | 2 | 100 |
| CLL 80 | 98.2 | 100 | 6–1 | 92.4 | 3–10 | 3 | K | 2–28(A19) | 2 | 94.0 |
All CLL cases were positive for CD5. CD23‐positive cases were 90.0% (72/80). The cut‐off point for CD23‐positivity in CLL cells was ≥30%. Shaded cells indicate CD23 expression <30%. For all IGKV genes, the gene name in parentheses indicates the Zachau nomenclature. For all IGLV genes, the gene name in parentheses indicates the Kawasaki nomenclature. K, kappa chain; L, lambda chain; NA, not available.
IGHV gene expression. The IGHV gene status was determined in 80 of 82 patients with CLL and 52 of 53 patients with leukemic CLPD, because no bands were amplified in samples from two CLL cases and one leukemic CLPD case. Figure 1 shows the proportions of IGHV expression in patients with CLL and leukemic CLPD. IGHV can be divided into seven families (family 1 to family 7) based on sequence homology. The patient numbers of each of the seven IGHV families in the 80 patients with CLL and the 52 patients with leukemic CLPD were as follows. In patients with CLL, family 1 was found in eight (10.0%), family 2 in two (2.5%); family 3 in 39 (48.8%); family 4 in 28 (35.0%); family 5 in two (2.5%); family 6 in one (1.3%); and family 7 in 0 (0%). In patients with leukemic CLPD, family 1 was found in six (11.5%); family 2 in two (3.8%); family 3 in 31 (59.6%); family 4 in 11 (21.2%); family 5 in one (1.9%); family 6 in 0 (0%); and family 7 in one (1.9%). IGHV family 34 was most prevalent in CLL and CLPD cases. One remarkable finding was that the expression of IGHV family 1 by CLL in our study was low compared with those in western countries and the difference was statistically significant compared with large CLL cohorts in North America( 13 ) (n = 1188, P < 0.001) and Europe( 14 ) (n = 1967, P = 0.003). Subfamily analysis in CLL revealed that IGHV4‐34 was the most frequent (22, 27.5%) followed by IGHV3‐23 (7, 8.8%), IGHV3‐21 (6, 7.5%), and IGHV3‐30 (5, 6.3%). These four IGHV genes accounted for 50% of all CLL cases. In leukemic CLPD, IGHV3‐23 was the most frequent (8, 15.4%) followed by IGHV3‐21 (7, 13.5%), IGHV3‐30 (7, 13.5%), and IGHV4‐34 (4, 7.7%) and these genes accounted for 50%. Only one patient showed IGHV1‐69 expression in our CLL cohort. There was a statistically significant difference in IGHV1‐69 expression between our result and the results from North America( 13 ) (n = 1846, P = 0.001) or Europe( 14 ) (P = 0.002). In leukemic CLPD, IGHV1‐69 was also expressed in only one patient with SMZL. IGHV3‐21, which is preferentially expressed in Scandinavian patients with CLL, was associated with biased expression of lambda light chain in both CLL (5/6) and in leukemic CLPD (6/7) in this cohort, as previously described.( 12 ) Comparison of IGHV status between the CLL and leukemic CLPD groups revealed a difference in IGVH4‐34 expression (Fig. 1, P = 0.005): 22 of 80 cases of CLL (27.5%) and 4 of 52 cases of leukemic CLPD (7.7%, three MCL and one unclassified CLPD).
Figure 1.
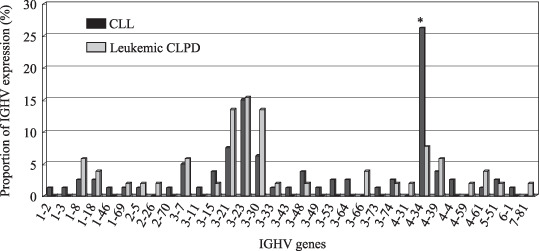
Proportions of immunoglobulin heavy chain variable region (IGHV) expression in patients with chronic lymphocytic leukemia (CLL) and leukemic chronic lymphoproliferative disorders (CLPD). IGHV gene status was determined in 80 patients with CLL and 52 patients with leukemic CLPD. Between the CLL and leukemic CLPD groups, a difference in IGVH4‐34 expression was noted: 22/80 (27.5%) in CLL and 4/52 (7.7%, 3 MCL and 1 unclassified CLPD) in leukemic CLPD (*P = 0.00517).
IGK gene expression. A total of 14 kinds of functional IGKV genes were expressed in 43 of 45 CLL cases. In these, frequently expressed IGKV genes were IGKV3‐20(A27)/3D‐20(A11) (8, 18.6%), IGKV3‐11(L6) (7, 16.3%), and IGKV2‐28(A19) (6, 14.0%). Previous reports described that IGKV3‐20(A27) and IGKV1‐39(O12)/1D‐39(O2) were the two most frequently expressed IGKV genes in western countries.( 20 , 21 ) In addition, 10 kinds of functional IGKV genes were expressed in 26 leukemic CLPD cases. Preferentially expressed IGKV genes were IGKV1‐39(O12)/1D‐39(O2) (7, 26.9%), IGKV3‐20(A27)/3D‐20(A11) (5, 19.2%), and IGKV2‐28(A19) (4, 15.4%). In IGKV expression between patients with CLL and those with leukemic CLPD, the prevalence of IGKV3‐11 expression was significantly different (P = 0.030): 7 of 43 patients with CLL versus none of 26 patients with leukemic CLPD (Fig. 2).
Figure 2.
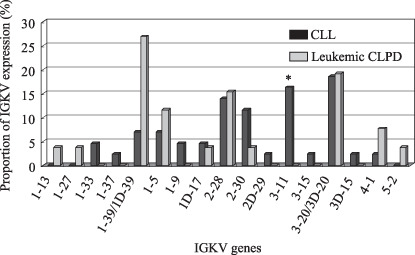
Proportions of IGKV expression in patients with chronic lymphocytic leukemia (CLL) and leukemic chronic lymphoproliferative disorders (CLPD). Fourteen kinds of functional IGKV genes were expressed in 43 CLL cases and 10 kinds were expressed in 26 leukemic CLPD cases. The prevalence of IGKV3‐11 expression was significantly different (*P = 0.0299): 7/43 in CLL and 0/26 in leukemic CLPD.
IGL gene expression. In 34 of 35 CLL cases, 13 kinds of functional IGLV genes were expressed. In these, the most frequently expressed IGLV genes were IGLV3‐21(V2‐14) (10, 29.4%) and IGLV2‐14(V1‐4) (4, 11.8%). Similar to the previous reports investigating IGLV expression in patients with unselected CLL,( 20 , 21 , 22 ) IGLV3‐21 was the most prevalent gene in this study. Of the IGLV3‐21(V2‐14) genes, only 2 of 10 were associated with IGHV3‐21. This is markedly different from other countries in which IGLV3‐21+ cases of CLL had highly biased IGLV3‐21 expression.( 12 ) In 21 of 26 leukemic CLPD cases, 11 kinds of functional IGLV genes were expressed. Preferentially expressed IGLV genes were IGLV2‐14(V1‐4) (4, 15.4%) and IGLV3‐19(V2‐13) (3, 11.5%). In IGLV expression between patients with CLL and those with leukemic CLPD, the prevalence of IGLV3‐21 expression was significantly different (P = 0.026): 10 of 34 in CLL versus 1 of 21 (a case of MCL) in leukemic CLPD (Fig. 3). By analysis of IGLV and IGLJ pairing, preferential pairing of IGLV3‐21 and IGLJ3 was noted (8/10) and some of the light chain CDR3 samples showed highly homologous sequences (Table 3). In leukemic CLPD, one MCL case with t(11;14) determined by fluorescence in situ hybridization and very high CCND1 expression determined by RQ‐PCR had the IGLV3‐21 gene, which was rearranged to IGLJ3 and had the consensus CDR3 sequences.
Figure 3.
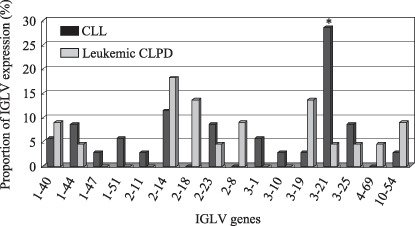
Proportions of IGLV expression in patients with chronic lymphocytic leukemia (CLL) and leukemic chronic lymphoproliferative disorders (CLPD). Thirteen kinds of functional IGLV genes were expressed in 34 CLL cases and 11 kinds were expressed in 21 leukemic CLPD cases. The prevalence of IGLV3‐21 expression was significantly different (P = 0.0264): 10/34 in CLL and 1/21 (a MCL case) in leukemic CLPD.
Table 3.
IGLJ expression and amino acid sequences of IGLV3‐21
| IGLJ | CDR3 | |
|---|---|---|
| Consensus | 3 | QVWDS(G/S)SDHPWV |
| CLL | 3 | ‐ ‐ ‐ ‐ ‐S ‐ ‐ ‐ ‐ ‐ ‐ |
| CLL | 3 | ‐ ‐ ‐ ‐ ‐G ‐ ‐ ‐ ‐ ‐ ‐ |
| CLL | 3 | ‐ ‐ ‐ ‐ ‐G ‐ ‐ ‐ ‐ ‐ ‐ |
| CLL | 3 | ‐ ‐ ‐ ‐ ‐G ‐ ‐ ‐ ‐ ‐ ‐ |
| CLL | 3 | ‐ ‐ ‐ ‐ ‐S ‐ RSS ‐ ‐ |
| CLL | 3 | ‐ ‐ ‐ ‐ ‐S ‐ RSIRRV |
| CLL | 3 | ‐ ‐ ‐ ‐ ‐S ‐ RSIRRV |
| CLL | 3 | ‐ ‐ ‐ ‐ TS R ‐ QGV |
| CLL | 3 | ‐ ‐ ‐ ‐ YS R ‐ QV |
| CLL | 1 | ‐ ‐ ‐ ‐ GS ‐ ‐ ‐ ‐ Y ‐ |
| CLL | 1 | ‐ ‐ ‐ ‐ ‐S ‐ ‐ Q ‐ Y ‐ |
| MCL | 3 | ‐ ‐ ‐ ‐ ‐S ‐ ‐ ‐ ‐ ‐ ‐ |
Homology to the consensus sequence is indicated by dots. CDR, complementarity determining region; CLL, chronic lymphocytic leukemia; MCL, mantle cell lymphoma.
IGHV, IGKV, and IGLV mutation status. In patients with CLL, the mutation status of 80 of 82 IGHV genes, 43 of 45 IGKV genes, and 34 of 35 IGLV genes was successfully analyzed. Because the number of nucleotide differences was estimated from framework region 1 to CDR3 according to recommendations by Ghia et al.,( 29 ) the proportion of patients with unmutated IGHV genes was lower in this report than in our previous report, in which CDR1, CDR2, and CDR3 were not evaluated.( 18 )
In the present study, 21.3% of IGHV genes, 34.9% of IGKV genes, and 32.4% of IGLV genes were unmutated. Analysis of individual IGHV genes identified that 20 of 22 IGHV4‐34 genes in CLL were in the mutated group and the prevalence was significantly higher than that of the other IGHV genes in CLL (P = 0.002). In patients with leukemic CLPD, regarding the mutation status of 52 of 53 IGHV genes, all 26 IGKV genes and 21 of 26 IGLV genes were successfully analyzed. Of these, 21.2% of IGHV genes, 30.8% of IGKV genes, and 52.4% of IGLV genes were unmutated. Among the types of leukemic CLPD, MCL had the highest proportion of unmutated cases: 60.0% in IGHV genes, 100% in IGKV genes, and 87.5% in IGLV genes.
Immunoglobulin heavy and light chain gene pairings. Recently, subgroups of CLL cases with remarkable similarity in the sequences of the immunoglobulin heavy and light chains, including CDR3, were reported (stereotyped BCR). In the present study, cases of CLL belonging to such subgroups were identified as well (Table 4). We identified two patients with CLL who had the same IGHV4‐39, IGH diversity region (IGHD) 6–13, IGH joining region (IGHJ) 5, and IGKV1‐39(O12), IGKJ1, or IGKJ2 segments with remarkably similar heavy and light chain CDR3 sequences, as described previously.( 15 , 16 ) The IGHV1‐69 gene expressed in a patient with CLL consisted of IGHD5‐12, IGHJ6, IGLV3‐1(V2‐1), and IGLJ4 and did not belong to any subset of stereotyped CLL BCR reported so far. In patients with leukemic CLPD, only one patient with SMZL expressed the IGHV1‐69 gene; in this case, the disease was diagnosed by typical histology from splenectomized specimens. Interestingly, the IGHV1‐69 gene was associated with IGHD5‐24, IGHJ3, IGKV3‐20(A27), and IGKJ1, which have been previously reported as stereotypical BCR gene subsets in patients with CLL.( 16 ) There were no significant differences in clinical parameters such as lymphocyte count, hepatosplenomegaly, or lymphadenopathy in association with IGK or IGL expression, IGK or IGL mutated status, or the existence of stereotyped BCR.
Table 4.
Immunoglobulin expression and amino acid sequences of heavy chain and light chain complementarity determining region (CDR) 3
| IGHV | IGHD | IGHJ | Heavy chain CDR3 | IGKV | IGKJ | Light chain CDR3 | |
|---|---|---|---|---|---|---|---|
| Consensus | 4‐39 | 6‐13 | 5 | AR GYSSSWY NWFDP | 1‐39 | 1 or 2 | QQSYSTPRT |
| CLL | 4‐39 | 6‐13 | 5 | ‐ ‐ LT A ‐ ‐ ‐ ‐ ‐ ‐ SGI ‐ ‐ ‐ ‐ ‐ | 1‐39 | 1 | ‐ ‐ ‐ ‐ ‐ ‐ ‐ ‐ ‐ |
| CLL | 4‐39 | 6‐13 | 5 | ‐ ‐ RL ‐ ‐ ‐ ‐ ‐ ‐ ‐ GV ‐ ‐ ‐ ‐ ‐ | 1‐39 | 2 | ‐ ‐ ‐ ‐ ‐ ‐ ‐ ‐ ‐ |
| Consensus | 1‐69 | 5‐24 | 3 | AR VEMATI AFI | 3‐20 | 1 | QQYVGSPRT |
| SMZL | 1‐69 | 5‐24 | 3 | ‐ ‐ EG ‐ ‐ ‐ ‐ ‐ ‐ RGFG ‐ ‐ ‐ | 3‐20 | 1 | ‐ ‐ ‐ GT ‐ R ‐ ‐ |
Homology to the consensus sequence is indicated by dots. CLL, chronic lymphocytic leukemia; SMZL, splenic marginal zone lymphoma.
Discussion
Three previous reports, including ours, suggested that the proportion of CLL expressing IGHV family 1 in Japan was not as prominent as in western countries, although the patient number analyzed was limited.( 18 , 30 , 31 ) With a larger number of patients with CLL in the present study, we added further evidence that IGHV family 1 expression in Japan is significantly lower than in other countries. Recently, the frequency of IGHV family expression was investigated in Iranian and Chinese patients with CLL and the studies revealed that IGHV family 1 expression in both populations was lower than in western countries.( 32 , 33 ) Regarding the individual expression of IGHV genes, we reported for the first time that IGHV1‐69 CLL is very rare in Japan (1/44),( 18 ) which was confirmed by the present study. Moreover, in a recent report from China,( 33 ) only one IGHV1‐69+ case was identified in 65 patients with CLL, suggesting that IGHV family 1 and IGHV1‐69 expression is rare in Asian patients with CLL. Similar to reports from Scandinavian countries, IGHV3‐21 cases showed biased lambda light chain expression in leukemic CLPD (6/7) as well as in CLL (5/6), but were associated with neither overexpression of IGLV3‐21 (V2‐14) nor specific CDR3 features in our cohort in contrast to previous observations.( 12 ) The expression of IGHV was significantly different between CLL and leukemic CLPD cases. IGHV4‐34, which was the most preferentially expressed in patients with CLL (22/80), was rarely expressed in patients with leukemic CLPD (4/52, P = 0.005). Because three of the four patients with IGHV4‐34+ leukemic CLPD were MCL‐ and CD5‐positive, only one CD5‐negative patient (with unclassified CLPD) expressed IGHV4‐34. In normal B‐cell development, naive IGHV4‐34 B cells are positively selected and mostly restricted to the follicular mantle zone, but these cells are largely excluded from the germinal centers.( 34 ) This mechanism may be relevant to IGHV4‐34 expression being underrepresented in leukemic CLPD other than MCL, which mainly consists of germinal center‐derived or postgerminal center‐derived lymphomas and leukemias. Similarly, complete lack of IGHV4‐34 gene rearrangement was reported in multiple myeloma.( 35 )
In patients with CLL in the present study, frequently expressed IGKV genes were IGKV3‐20(A27) (18.6%), IGKV3‐11(L6) (16.3%), and IGKV2‐28(A19) (14.0%). IGKV3‐20(A27) (13–37%) was one of the most preferentially expressed IGKV genes in previous reports, but IGKV3‐11(L6) and IGKV2‐28(A19) were not as frequently expressed (5–7% and 4–5%, respectively).( 20 , 21 , 22 ) However, IGKV3‐20(A27) was also a frequently expressed IGKV gene in normal peripheral blood IgM+ B cells.( 36 ) Meanwhile, in an analysis of IGLV expression, the most frequently expressed gene was IGLV3‐21(V2‐14) (29.4%), which was uniformly observed in IgM‐expressing CLL.( 20 , 21 , 22 ) IGKV3‐20(A27) expression in CLL cells was significantly more frequent than in normal B cells.( 37 , 38 ) Conversely, in leukemic CLPD, only one patient with MCL expressed IGLV3‐21(V2‐14) (4.8%, P = 0.002). Ghiotto et al. also revealed that IGLV3‐21(V2‐14) expression was not found in class‐switched IgG‐ or IgA‐expressing cells, which had more mutations in the IGLV genes than IgM‐expressing cells.( 21 ) These findings suggest that frequent IGLV3‐21(V2‐14) expression may be a unique characteristic of CLL with IgM‐expressing cells. In an analysis of IGLV and IGLJ pairing, the predominant IGLV3‐21(V2‐14) and IGLJ3 combination (8/10) associated with the homologous CDR3 sequences was observed (Table 2), which was a characteristic feature of IGLV3‐21+ CLL.( 22 , 39 ) Interestingly, the IGLV3‐21(V2‐14) gene, which was expressed by the patient with MCL mentioned above, was also associated with IGLJ3 and the consensus homologous CDR3 sequences seen in CLL (Table 2).
Of the 80 Japanese patients with CLL, 17 (21.3%) had unmutated IGHV and 63 (78.7%) had mutated IGHV. The proportion of CLL with mutated IGHV was higher compared to previous reports from western countries. It may be partly explained by the fact that the commonly unmutated IGHV1‐69 type was rare (1.3%), but the commonly mutated IGHV4‐34 type was frequent (27.5%) in the Japanese patients with CLL.
Recently, studies of BCR in patients with CLL identified that subsets of cases expressed almost identical BCR.( 12 , 13 , 14 , 15 , 16 , 17 , 40 , 41 ) In our cohort, we found two patients with CLL with IGHV4‐39 and IGKV1‐39(O12) expression, which belonged to one such subgroup( 15 , 16 ) (Table 3). Only one patient with SMZL with IGHV1‐69 expression was identified among patients with leukemic CLPD and the IGHV1‐69 was associated with IGHD5‐24, IGHJ3, IGKV3‐20(A27), and IGLKJ1. Surprisingly, this combination is identical to one of the stereotypical BCR gene subsets, which is thought to be CLL specific( 16 ) (Table 3). This may suggest that this stereotyped BCR is not CLL specific or that SMZL shares a common mechanism with CLL for the acquisition of this particular immunoglobulin heavy and light chain pairing.
Our data identified the differences and similarities of immunoglobulin gene status between Japan and western countries, and elucidated the characteristics of CLL further by a comparison with leukemic CLPD. Analysis of the presence of IGHV confirmed that IGHV1‐69 gene presence in Japanese patients with CLL was quite low, and newly identified that this gene was also rarely found in Japanese patients with leukemic CLPD. IGHV4‐34 was most frequently expressed by CLL but rarely found in leukemic CLPD. There was also characteristic light chain usage in CLL and leukemic CLPD. By analysis of IGHV and IGLV genes, some patients showed highly homologous sequences, including CDR3.
Acknowledgments
We acknowledge Dr Masaru Kojima for carrying out the pathological diagnoses. We are also indebted to Drs Arito Yamane, Takeki Mitsui, Morio Sawamura, Takafumi Matsushima, and Shuichi Miyawaki for providing patient samples and valuable advice. This research was supported in part by Grants‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and Initiatives for Attractive Education in Graduate Schools from MEXT.
References
- 1. Jaffe ES, Harris NL, Stein H, Vardiman JW, eds. World Health Organization Classification of Tumors. Tumors of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press, 2001. [Google Scholar]
- 2. Nishiyama H, Mokuno J, Inoue T. Relative frequency and mortality rate of various types of leukemia in Japan. Gann 1969; 60: 71–81. [PubMed] [Google Scholar]
- 3. Weiss NS. Geographical variation in the incidence of the leukemias and lymphomas. Natl Cancer Inst Monogr 1979; 53: 139–42. [PubMed] [Google Scholar]
- 4. Tamura K, Sawada H, Izumi Y et al . Chronic lymphocytic leukemia (CLL) is rare, but the proportion of T‐CLL is high in Japan. Eur J Haematol 2001; 67: 152–7. [DOI] [PubMed] [Google Scholar]
- 5. Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood 2006; 107: 265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Damle RN, Wasil T, Fais F et al . IgV gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94: 1840–7. [PubMed] [Google Scholar]
- 7. Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated IgV(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94: 1848–54. [PubMed] [Google Scholar]
- 8. Fais F, Ghiotto F, Hashimoto S et al . Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 1998; 102: 1515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol 2003; 21: 841–94. [DOI] [PubMed] [Google Scholar]
- 10. Johnson TA, Rassenti LZ, Kipps TJ. Ig VH1 genes expressed in B cell chronic lymphocytic leukemia exhibit distinctive molecular features. J Immunol 1997; 158: 235–46. [PubMed] [Google Scholar]
- 11. Schroeder HW Jr, Dighiero G. The pathogenesis of chronic lymphocytic leukemia: analysis of the antibody repertoire. Immunol Today 1994; 15: 288–94. [DOI] [PubMed] [Google Scholar]
- 12. Tobin G, Thunberg U, Johnson A et al . Chronic lymphocytic leukemias utilizing the VH3‐21 gene display highly restricted Vlambda2‐14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood 2003; 101: 4952–7. [DOI] [PubMed] [Google Scholar]
- 13. Widhopf GF, Rassenti LZ 2nd, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood 2004; 104: 2499–504. [DOI] [PubMed] [Google Scholar]
- 14. Murray F, Darzentas N, Hadzidimitriou A et al . Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood 2008; 111: 1524–33. [DOI] [PubMed] [Google Scholar]
- 15. Ghiotto F. Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia. J Clin Invest 2004; 113: 1008–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stamatopoulos K, Belessi C, Moreno C et al . Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood 2007; 109: 259–70. [DOI] [PubMed] [Google Scholar]
- 17. Messmer BT, Albesiano E, Efremov DG et al . Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med 2004; 200: 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koiso H, Yamane A, Mitsui T et al . Distinctive immunoglobulin VH gene usage in Japanese patients with chronic lymphocytic leukemia. Leuk Res 2006; 30: 272–6. [DOI] [PubMed] [Google Scholar]
- 19. Kipps TJ, Tomhave E, Pratt LF, Duffy S, Chen PP, Carson DA. Developmentally restricted immunoglobulin heavy chain variable region gene expressed at high frequency in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 1989; 86: 5913–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stamatopoulos K, Belessi C, Hadzidimitriou A et al . Immunoglobulin light chain repertoire in chronic lymphocytic leukemia. Blood 2005; 106: 3575–83. [DOI] [PubMed] [Google Scholar]
- 21. Ghiotto F, Fais F, Albesiano E et al . Similarities and differences between the light and heavy chain Ig variable region gene repertoires in chronic lymphocytic leukemia. Mol Med 2006; 12: 300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abusedra A, Joshi R, Bybee A, Apperley JF, Wagner SD. V lambda genes in chronic lymphocytic leukemia: highly skewed V gene segment usage with similar CDR3 sequences. Leukemia 2008; 22: 1073–5. [DOI] [PubMed] [Google Scholar]
- 23. Widhopf GF, Goldberg CJ 2nd, Toy TL et al . Nonstochastic pairing of immunoglobulin heavy and light chains expressed by chronic lymphocytic leukemia B cells is predicated on the heavy chain CDR3. Blood 2008; 111: 3137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Isoda A, Yokohama A, Matsushima T, Tsukamoto N, Nojima Y, Karasawa M. The naive T‐lymphocyte compartment is well preserved in patients with chronic myelogenous leukaemia in chronic phase. Br J Haematol 2002; 119: 949–55. [DOI] [PubMed] [Google Scholar]
- 25. Küppers R, Willenbrock K, Rajewsky K, Hansmann ML. Detection of clonal B‐cell populations in paraffin‐embedded tissues by polymerase chain reaction. Am J Pathol 1993; 44: 230–9. [PMC free article] [PubMed] [Google Scholar]
- 26. Van Dongen JJ, Langerak AW, Brüggemann M et al . Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T‐cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED‐2 concerted action BMH4‐CT98‐3936. Leukemia 2003; 17: 2257–317. [DOI] [PubMed] [Google Scholar]
- 27. Marti GE, Rawstron AC, Ghia P et al . The international familial CLL consortium. Diagnostic criteria for monoclonal B‐cell lymphocytosis. Br J Haematol 2005; 130: 325–32. [DOI] [PubMed] [Google Scholar]
- 28. Koiso H, Tsukamoto N, Miyawaki S, Shinonome S, Nojima Y, Karasawa M. Quantitative analysis of Cyclin D1 and CD23 expression in mantle cell lymphoma and B‐chronic lymphocytic leukemia. Leuk Res 2002; 26: 809–15. [DOI] [PubMed] [Google Scholar]
- 29. Ghia P, Stamatopoulos K, Belessi C et al . ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia 2007; 21: 1–3. [DOI] [PubMed] [Google Scholar]
- 30. Ikematsu W, Ikematsu H, Okamura S, Otsuka T, Harada M, Niho Y. Surface phenotype and Ig heavy‐chain gene usage in chronic B‐cell leukemias: expression of myelomonocytic surface markers in CD5‐chronic B‐cell leukemia. Blood 1994; 83: 2602–10. [PubMed] [Google Scholar]
- 31. Nakamura N, Kuze T, Hashimoto Y et al . Analysis of the immunoglobulin heavy chain gene variable region of 101 cases with peripheral B cell neoplasms and B cell chronic lymphocytic leukemia in the Japanese population. Pathol Int 1999; 49: 595–600. [DOI] [PubMed] [Google Scholar]
- 32. Farsangi MH, Jeddi‐Tehrani M, Sharifian RA et al . Analysis of the immunoglobulin heavy chain variable region gene expression in Iranian patients with chronic lymphocytic leukemia. Leuk Lymphoma 2007; 48: 109–16. [DOI] [PubMed] [Google Scholar]
- 33. Chen L, Zhang Y, Zheng W et al . Distinctive IgVH gene segments usage and mutation status in Chinese patients with chronic lymphocytic leukemia. Leuk Res 2008; 32: 1491–1498. [DOI] [PubMed] [Google Scholar]
- 34. Pugh‐Bernard AE, Silverman GJ, Cappione AJ et al . Regulation of inherently autoreactive VH4‐34 B cells in the maintenance of human B cell tolerance. J Clin Invest 2001; 108: 1061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rettig MB, Vescio RA, Cao J et al . VH gene usage is multiple myeloma: complete absence of the VH4.21 (VH4‐34) gene. Blood 1996; 87: 2846–52. [PubMed] [Google Scholar]
- 36. Foster SJ, Brezinschek HP, Brezinschek RI, Lipsky PE. Molecular mechanisms and selective influences that shape the kappa gene repertoire of IgM+ B cells. J Clin Invest 1997; 99: 1614–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ignatovich O, Tomlinson IM, Jones PT, Winter G. The creation of diversity in the human immunoglobulin V (lambda) repertoire. J Mol Biol 1997; 268: 69–77. [DOI] [PubMed] [Google Scholar]
- 38. De Wildt RM, Hoet RM, Van Venrooij WJ, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol 1999; 285: 895–901. [DOI] [PubMed] [Google Scholar]
- 39. Matthews C, Catherwood MA, Morris TC, Alexander HD. V(H)348 and V(H)353, as well as V(H)321, gene rearrangements define unique subgroups in CLL and are associated with biased lambda light chain restriction, homologous LCDR3 sequences and poor prognosis. Leuk Res 2007; 31: 231–4. [DOI] [PubMed] [Google Scholar]
- 40. Hervé M, Xu K, Ng YS et al . Unmutated and mutated chronic lymphocytic leukemias derive from self‐reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest 2005; 115: 1636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ghia EM, Jain S, Widhopf GF 2nd et al . Use of IGHV3‐21 in chronic lymphocytic leukemia is associated with high‐risk disease and reflects antigen‐driven, post‐germinal center leukemogenic selection. Blood 2008; 111: 5101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


