Abstract
A novel centrosome protein, TCC52, was identified as a cancer‐testis (CT) antigen. The TCC52 gene was tissue‐restricted in normal tissues but highly expressed in lung cancer tissues and some cancer cell lines. Immunoglobulin G antibody specific to TCC52 was detected in serum samples from patients with prostate cancer (59.4%, 69/116), cholangiocarcinoma (17.6%, 6/34), laryngeal cancer (8%, 8/100) and lung cancer (5.6%, 4/71) in patients, rather than from healthy donors. Based on its restricted expression pattern and immunogenicity in some types of tumor, TCC52, as a novel CT antigen, would be a promising candidate for cancer immunotherapy. (Cancer Sci 2008; 99: 2274–2279)
Cancer‐testis (CT) antigen is a type of immunogenic human tumor antigen. Restricted expression in normal tissues and immunogenicity in cancer patients are two major characteristics of CT antigens.( 1 , 2 , 3 , 4 ) Therefore, CT antigens are the ideal targets for immunotherapy of human tumors.( 1 , 5 ) During the last two decades, more than 40 distinct CT antigens have been identified.( 6 , 7 ) However, a large number of identified CT antigens showed low humoral immune responses in cancer patients except NY‐ESO‐1 which holds the possibility to be one of the most immunogenic CT antigens.( 8 , 9 ) Thus, it is crucial to identify new CT antigens, particularly those that could elicit immune responses in humans.
Centrosomes are the microtubule‐organizing centers of animal cells, influencing both tissue architecture and the accuracy of chromosome segregation.( 10 ) Since the 19th century, the relationship between centrosome malfunction and tumorigenesis has been studied.( 11 ) Centrosome abnormalities could be observed in various types of cancer and always related to the cancer's occurrence development and poor prognosis.( 12 ) During our previous study on centrosome proteomes,( 13 ) one of the proteins that was caught in co‐immunoprecipitation, was identified as KIAA 1892 (WDR40A).( 14 ) Its expression was found to be tissue‐restricted in normal tissues and frequently in lung cancer tissues as well as certain types of cancer cell lines. Thus, we renamed it TCC52 (testis cancer centrosome‐related protein) and registered to GenBank (no. DQ518909). Furthermore, humoral responses to TCC52 were detected in four different types of cancer patients, indicating that TCC52 was the first identified CT antigen that was localized on the centrosome. It therefore might be a potential target for cancer immunotherapy. Our finding added more evidence supporting that centrosomes play a critical role in cancer progression.
Materials and Methods
Cell lines, tumor tissues and serum samples. All cancer cell lines (HeLa, BGC823, HepGII, HepII, MCF7, PC3M, K562, A375, A549, LTEP‐a‐2 and NCI‐H226) were maintained in Dulbecco's modified Eagle's medium (DMEM) medium (Invitrogen/Gibco, San Diego, CA, USA) containing 10% fetal calf serum. Twenty‐four lung cancer tissues and paired adjacent non‐cancerous tissues were obtained from General Hospital of Beijing Unit, PLA and Peking Union Medical College and Chinese Academy of Medical Sciences. Sera of cancer patients were collected from General Hospital of Beijing Unit, PLA, Second Hospital of Xi’an Jiaotong University, General Hospital of the Chinese People's Liberation Army and Third Hospital of Jilin University. Sera of healthy donors were obtained from a general health examination in staffs of the General Administration of Sport. All patient samples were obtained with informed consent with Institutional Review Board approval.
RNA extraction, cDNA synthesis, and reverse transcription polymerase chain reaction. RNA extraction, reverse transcription (RT) and polymerase chain reaction (PCR) were carried out according to our previous work.( 15 ) Normal tissue cDNA panels (multiple tissue cDNA panels [MTC] I and II) were purchased from Takara Biosciences Clontech (Palo Alta, CA, USA). Primers used in the expression analysis of TCC52 were: forward, 5′‐CCGAATTCTAT GGCCCGGAAAGTAGT‐3′; reversed, 5′‐TAGTCGACTTAACTCC AGAGCCCAGCA‐3′ (product, 1379 bp). Primers for β‐actin were used as an internal control.
TCC52 recombinant protein and preparation of antibody. TCC52 ORF (454 amino acids) was amplified from HeLa cell cDNA by using the following primers (forward, 5′‐ACGAATTCATGGCCCG GAAAGTAGT‐3′; reversed, 5′‐GGAAGCTTACTCCAGAGCCC AGCA‐3′). The PCR product was digested with EcoRI and HindIII and cloned into the expression vector pET30a (Stratagene, La Jolla, CA, USA). TCC52 recombinant protein fused with two 6His‐tags was purified by Ni+2 affinity chromatography by using the NiNTA agarose resin (Invitrogen). Following that, the polyclonal antibody against the full length TCC52 recombinant protein was prepared in the BALB/c mice.
Mass spectrometry. Protein digestion was performed essentially as reported,( 16 ) and peptides were desalted and concentrated using C18 extraction tips.( 17 ) The peptide mixture was analyzed with a Qstar Pulser I Quadrupole TOF‐MS mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA, USA).
Immunofluorescence. HeLa cells were grown on coverslips. Immunostaining was performed by following a previously published protocol with TCC52 antibody (home‐made), antimouse‐fluorescein isothiocyanate (FITC) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti‐γ‐tubulin antibody (Sigma Chemical, St Louis, MO, USA) and antimouse cy5 (Jackson Immunoresearch) and 4,6‐diamino‐2‐phenylindole (DAPI) (Calbiochem, San Diego, CA, USA).( 18 ) Immunofluorescence microscopy was performed with an FV3000 Olympus microscope (Olympus, Tokyo, Japan).
Immunohistochemistry. Paraffin‐embedded sections were heated at 60°C for 30 min, dewaxed and rehydrated with xylene and grades of alcohol. Immunohistochemistry was performed by following a previously published protocol with anti‐TCC52 anti‐sera (at 1:100 dilution) and horseradish peroxidase‐conjugated horse antimouse immunoglobulin (Ig)G (Vector Laboratories, Burlingame, CA, USA). Sections were incubated with 3,3′‐diaminobenzidine substrate until suitable staining developed and counterstained with hematoxylin solution. Pre‐immune sera were used as a negative control.
Enzyme‐linked immunosorbent assay. Detection of humoral response to TCC52 in cancer patients was done by an indirect enzyme‐linked immunosorbent assay (ELISA) screening system,( 8 ) according to our previous work.( 15 ) Data was analyzed statistically using the software SPSS ver. 13.0 (SPSS Software, Chicago, IL, USA).
Western blot. Total cell lysis or tissue lysis or purified recombinant protein TCC52 were subjected to immunoblotting with home‐made TCC52 polyclonal antibody (at 1:500 dilution) or human sera (at 1:100 dilution) or β‐actin antibody (Santa Cruz Biotechnology) as previously described.( 15 )
Results
TCC52, a new centrosome protein, detected in cancer cell lines. TCC52 was previously cloned and deposited in the GenBank database as KIAA1892. As deduced from its primary sequence, the human TCC52 is a polypeptide of 454 amino acids residues and its gene is located at chromosome 9p13.3 and exists as a single copy.
To understand the biological function of TCC52, in the present study, we produced the mouse polyclonal antibody against the full‐length TCC52 protein. The TCC52 recombinant protein was expressed in Escherichia coli cells and purified. The purified protein was separated by sodium dodecylsulfate polyacrylamide gel electrophoresis and stained by Coomassie blue (Fig. 1A). The purified single band was identified as TCC52 by mass spectrometry analysis (Fig. 1B). The polyclonal antibody against the full‐length TCC52 recombinant protein was then prepared in the BALB/c mice. To examine the quality of the antibody, home‐made TCC52 antibody was evaluated in HeLa and BGL823 by western blot. As shown in Fig. 1(C), a single band was observed with a higher molecular weight (~55 kDa) than predicted (52.5 kDa). The slighter migration may be due to its posttranslational modification in eukaryotic cells. To ensure the specificity of the result, both pre‐immune sera and sera against TCC52, but depleted of anti‐TCC52 Ig, were used as negative controls.
Figure 1.
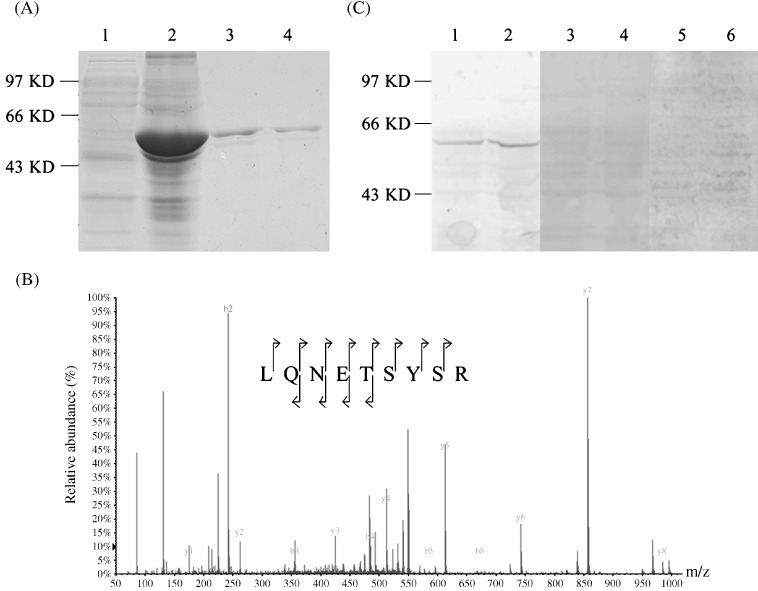
The construction of TCC52 recombinant protein and the evaluation of home‐made TCC52 polyclone antibody. (A) Escherichia coli cells (BL21 strain) transformed with His‐TCC52 were treated without (line 1) or with 0.4 mM isopropyithio‐β‐D‐galactoside (IPTG) for 6 h (line 2) and purified by using the NiNTA agarose resin (lines 3 and 4). (B) Identification of TCC52 by mass spectrometry (MS)/MS. MS/MS spectra of one doubly charged peptide of m/z of 549.26 corresponding to the sequences LQNETSYSR of TCC52. Predicted N‐ and C‐terminal fragments are marked b and y, respectively. Observed fragments are marked by arrows. (C) TCC52 was detected by western blot in proteins extract from HeLa cell (lanes 1, 3 and 5) and BGC823 cell (lanes 2, 4 and 6) by using home‐made TCC52 polyclonal antibody (lanes 1 and 2). Pre‐immune (lanes 3 and 4) and TCC52 antibody depleted of anti‐TCC52 immunoglobulins (lanes 5 and 6) were used as negative controls. Molecular weight markers are indicated.
To determine the subcellular localization of TCC52, an indirect immunofluorescence assay was performed with the home‐made TCC52 antibody on HeLa cells. γ‐Tubulin, a well‐known constitutive centrosome protein, was served as an indicator for the centrosome. As shown in Fig. 2, TCC52 was co‐localized with γ‐tubulin in both interphase and mitotic cells, which suggested that TCC52 might be a novel constitutive centrosome protein. Six random fields were observed for each phase. More than 95% cells in the same field showed a similar localization pattern as TCC52. The detailed localization pattern of TCC52 in mitotic cells, including metaphase, anaphase and telophase, further confirmed that TCC52 was a new centrosome protein. The same assays were carried out on several cancer cell lines from different tissues, which included BGL823 (the gastric cancer cell line), HepII (the laryngeal cancer cell line) and A549 (the lung adenocarcinoma cell line). Similar results were obtained (data not shown). Taken together, our data revealed that TCC52 existed in different cancer cell lines as a novel constitutive centrosome protein.
Figure 2.
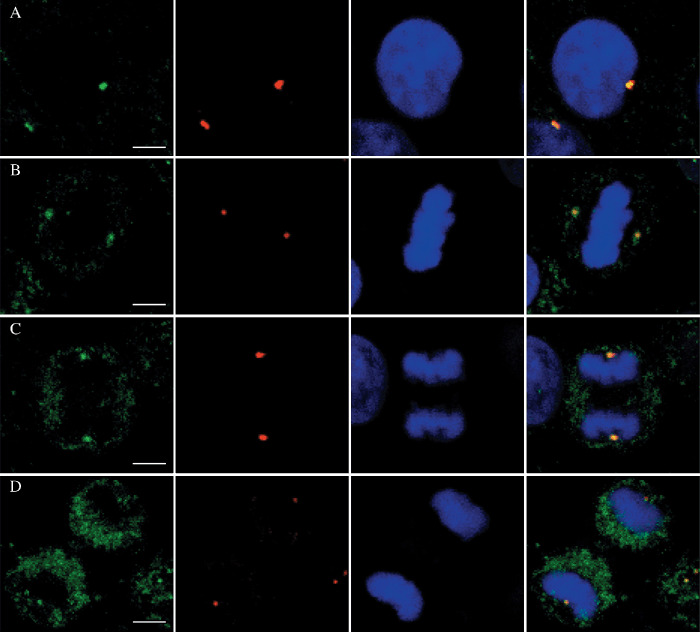
TCC52 is showed co‐localized with the centrosome during the cell cycle. Double staining with anti‐TCC52 (green) and anti‐γ‐tubulin (red) antibodies revealed a centrosomal localization for TCC52 during interphase (A), metaphase (B), anaphase (C) and telophase (D). 4,6‐diamino‐2‐phenylindole (DAPI) was used to visualize the DNA (blue). Merged pictures are shown at the right. Scale lines, 5 µm.
TCC52 mRNA was frequently expressed in lung cancer tissues and several cancer cell lines, but restrictedly expressed in testis among normal tissues. TCC52 mRNA expression was first examined in 16 normal human tissues (Takara Biosciences Clontech). The results demonstrated that the TCC52 gene was tissue‐restricted (mRNA detected in pancreas and spleen besides testis) (Fig. 3A). It was well known that CT antigens were restricted in normal gametogenic tissues, but aberrantly expressed in tumors. To test whether the expression pattern of TCC52 meet the criterion of CT antigens, we examined the mRNA expression of TCC52 in 38 paired lung cancer and adjacent non‐cancerous tissues. The results showed that the expression of TCC52 was detected in 89.47% (34/38) lung cancer tissues, whereas low expression or undetectable expression was found in the paired adjacent non‐cancerous tissues (Fig. 3B). Because the paired adjacent non‐cancerous lung tissues, even 5 cm distant from lung cancer tissues, may not be a physiological one, low expression of TCC52 may be due to some tumor cell infiltration. Additionally, TCC52 mRNA was also expressed in all of the examined tumor cell lines (Fig. 3C). However, no correlation between TCC52 expression and sex, age, clinical stages or nodular status was found in lung cancer patients (Table 1).
Figure 3.
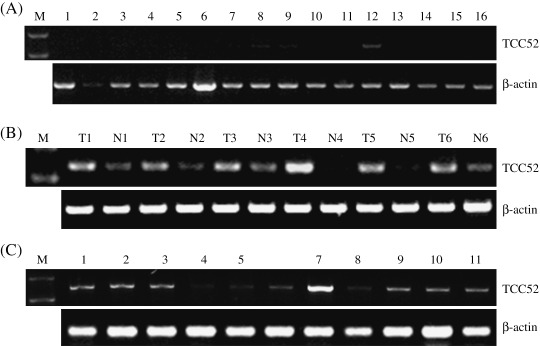
Expression of TCC52 mRNA. Expression of mRNA level was investigated by reverse transcription polymerase chain reaction (RT‐PCR). (A) TCC52 expression in panel of normal tissue cDNA specimens. Lanes: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal; 7, kidney; 8, pancreas; 9, spleen; 10, thymus; 11, prostate; 12, testis; 13, ovary; 14, small intestine; 15, colon; 16, leukocyte; M, molecular marker. (B) TCC52 expression in six pairs of lung cancer tissue specimens. T, lung cancer tissues; N, paired adjacent non‐cancerous lung tissues. (C) TCC52 mRNA expression in cancer cell lines: 1, HeLa; 2, MCF7; 3, K562; 4, LEPT‐A2; 5, HepGII; 6, HepII; 7, BGC823; 8, A375; 9, PC3; 10, A549; 11, H226; M, molecular weight marker.
Table 1.
Summary of TCC52 mRNA expression in lung cancer tissues
| mRNA expression | ||
|---|---|---|
| No. positive | No. negative | |
| No. of cases | 34 (89.47%) | 4 (10.52%) |
| Clinical stage | ||
| I and II | 8 | 1 |
| III and IV | 26 | 3 |
| Histological type | ||
| Adenocarcinoma | 7 | 3 |
| Squamous cell carcinoma | 15 | 15 |
| Adenosquamous carcinoma | 4 | 4 |
| Large cell lung carcinoma | 1 | 0 |
| Other types of lung cancer | 7 | 1 |
TCC52 protein was expressed in lung cancer tissues and several cancer cell lines. The existence of TCC52 protein was confirmed by western blot applied on the same sets of lung cancer tissues and tumor cell lines. The expression of TCC52 protein could be detected in all of the tested cell lines and lung cancer tissues except the T5 sample (Fig. 4A). In contrast, much lower expression of TCC52 could be detected in the paired adjacent non‐cancerous lung tissue samples. The data was consistent with that from mRNA expression of TCC52. The representative results are shown in Fig. 4.
Figure 4.
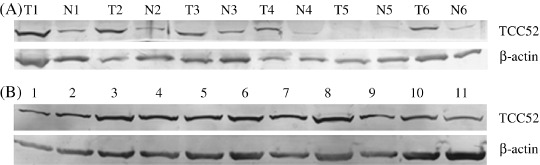
TCC52 protein expression in lung cancer tissues (A) and tumor cell lines (B). (A) T, lung cancer tissues; N, paired adjacent non‐cancerous lung tissues. (B) 1, HeLa; 2, BGC823; 3, HepGII; 4, HepII; 5, MCF7; 6, PC3; 7, K562; 8, A375; 9, A549; 10, LEPT‐A2; 11, H226.
The existence of TCC52 protein was also analyzed by immunohistochemistry in normal testis and lung cancer tissues. Immunohistochemistry carried out to analyze frozen sections of lung cancer tissues and paired adjacent non‐cancerous specimens, which were analyzed in RT‐PCR and western blot, could not show the expression of TCC52 clearly in our experiment. Immunohistochemistry was then performed by using paraffin‐embedded sections of another 15 lung cancer tissues, and normal testis as a positive control. Positive staining of TCC52 was observed in 14 of 15 lung cancer specimens. In cancer tissues, staining was predominantly in the cytoplasm of cancer cells. In the normal testis, TCC52 expression was generally found in the cytoplasm of spermatogonia and spermatocytes (Fig. 5).
Figure 5.
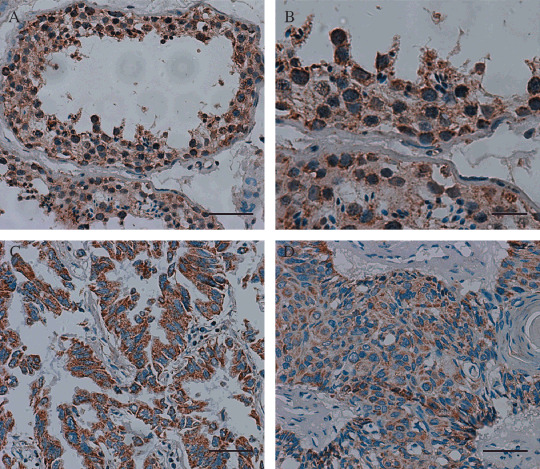
TCC52 protein expression in normal testis (A,B) and lung cancer tissues (C, adenocarcinoma; D, squamous). Cytoplasm staining was observed mainly in spermatogonia and spermatocytes and tumor cells. Scale lines, 50 µm (A,B,D) and 20 µm (B).
Humoral response against TCC52 in cancer patients. Although the expression pattern of TCC52 strongly supported that TCC52 might be a novel CT antigen, immunogenicity of TCC52 in vivo should also be taken into account. To evaluate the presence of antibodies against TCC52 in sera from cancer patients, we performed immunoblotting assay‐directed ELISA by using recombinant His‐tagged TCC52. His‐tagged protein served as an internal negative control. The mean absorbance value of healthy donors (0.32 for TCC52) + 3 standard deviations (SD; 0.12) was set as a cut‐off value (0.68). Among the 321 sera analyzed, 87 (27.1%) were reactive to TCC52 recombinant protein. The highest percentage of antibody response was detected in prostate cancer patients (59.4%, 69/116). The percentages of antibody response in the other three types of patients were 5.6% (4/71) in lung cancer patients, 17.6% (6/34) in cholangiocarcinoma patients and 8% (8/100) in laryngeal cancer patients, respectively. However, the response was not detected in healthy donors. The absorbance value of negative control in each experiment was below 0.01 and the absorbance value of nonspecific control for each positive serum was below the cut‐off value. Fig. 6(A) and Table 2 summarize the results, and Fig. 7 shows the typical titration curve of positive and negative sera. Western blot was performed to further confirm the presence of TCC52 antibodies in the aforementioned sera. Recombinant TCC52 protein served as antigen and the ELISA‐positive sera as primary antibody. TCC52 antibody‐positive sera were further confirmed to be positive by western blot. In contrast, healthy sera, the negative control and the nonspecific control remained negative, which were consistent with the results from the ELISA assay (Fig. 6B). To sum up, our results strongly support that TCC52 is a novel CT antigen and might be a target for cancer immunotherapy.
Figure 6.
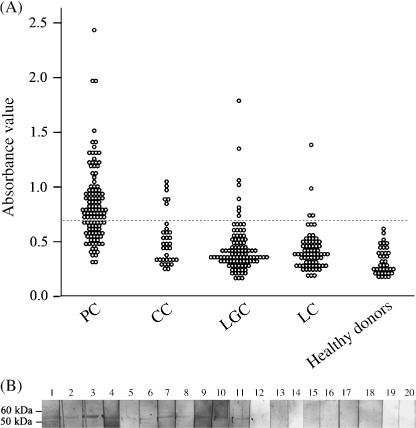
Humoral responses against TCC52 in cancer patients and healthy donors by enzyme‐linked immunosorbent assay (A) and western blot (B). (A) N, normal; CC, cholangiocarcinoma; LGC, laryngeal cancer; LC, lung cancer; PC, prostate cancer. (B) Lanes 1–11, positive reaction with sera from patients; lanes 13–20, negative reaction with sera from healthy donors; lane 12, negative control. Molecular weight markers are indicated.
Table 2.
Summary for humoral responses of TCC52 in serum samples from cancer patients and healthy donors
| Type † | OD value | P‐value* | Antibody response | ||
|---|---|---|---|---|---|
| Mean | SD | (%) | |||
| N (38) | 0.33 | 0.12 | – | – | – |
| CC (34) | 0.52 | 0.24 | <0.01 | 6/34 | 17.6 |
| LGC (100) | 0.44 | 0.24 | <0.01 | 8/100 | 8.0 |
| LC (71) | 0.41 | 0.18 | <0.05 | 4/71 | 5.6 |
| PC (116) | 0.83 | 0.34 | <0.01 | 69/116 | 59.4 |
| Total (321) | – | – | – | 87/321 | 27.1 |
A two‐tailed Student's t‐test was used to assess the statistical significance of the difference in absorbance values between patients and healthy donors.
† N, normal; CC, cholangiocarcinoma; LC, Lung cancer; LGC, laryngeal cancer; PC, prostate cancer; Total, total no. of patients.
Figure 7.
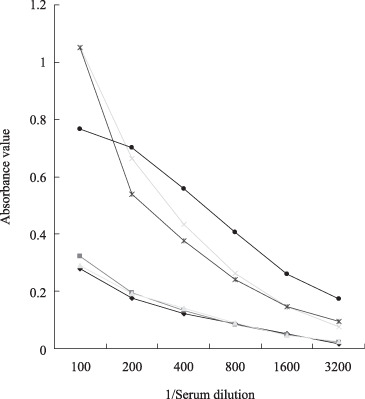
Antibody production against TCC52 in prostate patients. Serially diluted sera from three patients with prostate cancer ( ,
, ,
,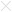 ) and sera from three healthy donors (
) and sera from three healthy donors ( ,
, ,
, ).
).
Discussion
Cancer‐testis antigen is a type of antigen whose expression is highly restricted, primarily in normal testis, but aberrantly in a wide range of different tumor types.( 1 , 2 , 3 ) Because of their restricted expression pattern, CT antigens have taken center stage in cancer antigen identification and vaccine development.( 1 , 2 ) In this study, we for the first time demonstrated that TCC52, a novel centrosome protein, shows tissue‐restricted expression pattern in normal tissues. Based on its expression profile and humoral immune response, it was reasonable to recognize TCC52 as a new member of CT antigens. To our knowledge, TCC52 was the first identified CT antigen that was localized on the centrosome. A growing body of experimental evidence provides the foundations for the essential roles of the centrosome in tumorigenesis. Although whether TCC52 represents a class of centrosome proteins which show a tissue‐restricted expression pattern in normal tissues remains to be elucidated, one thing that can be confirmed is that TCC52, as a potential target for cancer therapy, might play some roles in tumor treatment through modulation centrosome‐related functions. Moreover, the specific localization of TCC52 might provide valuable clue to further investigate the biological function of TCC52.
As opposed to the conventional view that CT antigens are expressed only in gametogenic tissues, low expression of TCC52 was observed in normal pancreas and spleen. It was not a surprising result because recent studies revealed that a couple of CT antigens were present in some other normal tissues.( 15 , 19 , 20 ) However, the expression of CT antigens in non‐gametogenic tissues was very low and unlikely to result in the presence of immunologically relevant levels of major histocompatibility complex/CT peptide complexes. Thus, this does not impair their immunotherapeutic potential in most cases.( 1 ) It has been suggested that CT antigen genes often form families on the X chromosome.( 3 ) Interestingly, unlike the other CT antigen genes, the TCC52 gene is localized to 9p13.3.
In addition to the analysis of mRNA expression, western blot and immunohistochemistry should be performed to confirm the expression of TCC52 protein in cancer. Western blot results clearly showed that TCC52 was expressed in lung cancer tissues (except T5) and all the examined cell lines. Because we know that the protein level sometimes does not necessarily reflect the mRNA level as a result of mRNA degradation, different translational regulation and protein degradation, it is not unexpected that TCC52 protein expression was not detected in T5 lung cancer tissue samples. Taken together, the results from western blotting provided further evidence to support TCC52 as a CT antigen in lung cancer. One thing necessary to note is that the low expression was detected in the adjacent non‐cancerous tissues. Although these adjacent tissues are not normal in that some of them express TCC52, we found that all of the tumor tissues overexpressed TCC52 compared with the adjacent tissues, which suggested that TCC52 might be involved in the progression of tumorigenesis.( 21 ) However, to elucidate this possibility, more tissue samples and the biological function of TCC52 should be investigated.
In addition to tissue restriction, inherent immunogenicity is another characteristic of some CT antigens. Approximately half of known CT antigens were reported to induce frequent humoral responses.( 1 , 8 , 22 ) Our results showed that high percentages of antibody response to TCC52 could be detected in patients with prostate cancer (59.4%), cholangiocarcinoma (17.6%), laryngeal cancer (8%) and lung cancer (5.6%), respectively. However, as for some CT antigens, humoral responses to CT antigens are relatively rare, occurring in less than 10% of cancer patients.( 8 , 9 , 23 , 24 ) The exceptional immunogenicity of TCC52 coupled with its broad distribution among many cancer types makes it a very good vaccine candidate, with the potential to be used in vaccines against many types of tumors.
In summary, we have identified a novel CT antigen, TCC52. Because it was the first identified CT antigen that localized on the centrosome, with the features of tissue‐restricted expression pattern in normal tissues and a high frequency of humoral responses in different types of cancer, the new centrosome protein might play a critical role in cancer progression and be a potential target for cancer immunotherapy.
Acknowledgments
We appreciate M. D. Xueyuan Xiao and Dr Fang He for critical reading of this manuscript. This work was supported in part by a Special Grant of the Major State Basic Research Plan of China (no. 2006CB910100), the Natural Science Foundation of Beijing (no. 05G294) and the Natural Science Foundation of Beijing and Science and Technology Commission of Beijing (Y020400 2040111).
References
- 1. Scanlan M, Simpson A, Old L. The cancer/testis genes: review, standardization, and commentary. Cancer Immun 2004; 4: 1–15. [PubMed] [Google Scholar]
- 2. Old L. Cancer vaccines 2003: opening address. Cancer Immun 2003, 3. [PubMed] [Google Scholar]
- 3. Scanlan M, Gure A, Jungbluth A, Old L, Chen Y. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev 2002; 188: 22–32. [DOI] [PubMed] [Google Scholar]
- 4. Old L. Cancer/testis (CT) antigens – a new link between gametogenesis and cancer. Cancer Immun 2001; 1: 1–7. [PubMed] [Google Scholar]
- 5. Fiszer D, Kurpisz M. Major histocompatibility complex expression on human, male germ cells: a review. Am J Reprod Immunol 1998; 40: 172–6. [DOI] [PubMed] [Google Scholar]
- 6. van der Bruggen P, Traversari C, Chomez P et al . A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991; 254: 1643–7. [DOI] [PubMed] [Google Scholar]
- 7. Traversari C, Bruggen P, Eynde B et al . Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics 1992; 35: 145–52. [DOI] [PubMed] [Google Scholar]
- 8. Stockert E, Jager E, Chen Y et al . A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J Exp Med 1998; 187: 1349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yakirevich E, Sabo E, Lavie O, Mazareb S, Spagnoli G, Resnick M. Expression of the MAGE‐A4 and NY‐ESO‐1 cancer‐testis antigens in serous ovarian neoplasms. Clin Cancer Res 2003; 9: 6453–60. [PubMed] [Google Scholar]
- 10. Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 2002; 14: 25–34. [DOI] [PubMed] [Google Scholar]
- 11. Boveri T. The Origin of Malignant Tumors . Baltimore, Maryland: Williams and Wilkins, 1929.
- 12. Wang X, Greenhalgh D, Jiang A et al . Analysis of centrosome abnormalities and angiogenesis in epidermal‐targeted p53172H mutant and p53‐knockout mice after chemical carcinogenesis: evidence for a gain of function. Mol Carcinog 1998; 23: 185–92. [DOI] [PubMed] [Google Scholar]
- 13. Xia L, Li Y, Yang D et al . Identification of new centrosome proteins by autoimmune patient sera. Sci China C Life Sci 2007; 50: 194–202. [DOI] [PubMed] [Google Scholar]
- 14. Nagase T, Kikuno R, Ohara O. Prediction of the coding sequences of unidentified human genes. XXI. The complete sequences of 60 new cDNA clones from brain which code for large proteins. DNA Res 2001; 8: 179–87. [DOI] [PubMed] [Google Scholar]
- 15. Luo C, Xiao X, Liu D et al . CABYR is a novel cancer‐testis antigen in lung cancer. Clin Cancer Res 2007; 13: 1288. [DOI] [PubMed] [Google Scholar]
- 16. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver‐stained polyacrylamide gels. Anal Chem 1996; 68: 850–8. [DOI] [PubMed] [Google Scholar]
- 17. Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 2003; 75: 663–70. [DOI] [PubMed] [Google Scholar]
- 18. Guo D, Hu K, Lei Y, Wang Y, Ma T, He D. Identification and characterization of a novel cytoplasm protein ICF45 that is involved in cell cycle regulation. J Biol Chem 2004; 279: 53498. [DOI] [PubMed] [Google Scholar]
- 19. Crew A, Clark J, Fisher C et al . Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel‐associated box in human synovial sarcoma. EMBO J 1995; 14: 2333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yousef G, Kyriakopoulou L, Scorilas A et al . Quantitative expression of the human kallikrein gene 9 (KLK9) in ovarian cancer: a new independent and favorable prognostic marker. Cancer Res 2001; 61: 7811–8. [PubMed] [Google Scholar]
- 21. Park S, Lim Y, Lee D et al . Identification and characterization of a novel cancer/testis antigen gene CAGE‐1. BBA-Gene Structure Expression 2003; 1625: 173–82. [DOI] [PubMed] [Google Scholar]
- 22. Parmigiani R, Bettoni F, Vibranovski M et al . Characterization of a cancer/testis (CT) antigen gene family capable of eliciting humoral response in cancer patients. Proc Natl Acad Sci USA 2006; 103: 18066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sugita Y, Wada H, Fujita S et al . NY‐ESO‐1 expression and immunogenicity in malignant and benign breast tumors. Cancer Res 2004; 64 (6): 2199–204. [DOI] [PubMed] [Google Scholar]
- 24. Jaeger E, Stockert E, Zidianakis Z et al . Humoral immune responses of cancer patients against ‘Cancer‐Testis’ antigen NY‐ESO‐1: correlation with clinical events. Int J Cancer 1999; 84: 506–10. [DOI] [PubMed] [Google Scholar]


