Abstract
Human tetraspanin proteins are a group of 33 highly hydrophobic membrane proteins that can form complexes in cholesterol‐rich microdomains, distinct from lipid rafts, on the cell surface in a dynamic and reversible way. These complexes are composed of a core of several tetraspanin proteins that organize other membrane proteins such as integrins, human leukocyte antigen (HLA) antigens and some growth factor receptors. Although most tetraspanin proteins have been studied individually, tetraspanin proteins and their complexes can have effects on cellular adhesion and motility, interactions with stroma or affect signaling by growth factors, and for most of them no ligand has been identified. Functionally these proteins have been mostly studied in cells of lymphoid lineage, but they are present in all cell types. Data is also available for some tumors, where some tetraspanins have been identified as metastasis suppressors, but their significance is still not clear. Some of their implications in tumor biology and the areas that deserve further study are outlined. (Cancer Sci 2007; 98: 1666–1677)
Tetraspanin proteins
Structural characteristics defining tetraspanin proteins. Membrane proteins that play a role in cellular interactions with other cells or stroma, as well as in signaling pathways, can have a major impact on the biological characteristics of tumor cells, particularly in tumor adhesion and dissemination properties. There is a new group of membrane proteins, the tetraspanins, which are starting to acquire relevance in cell biology( 1 , 2 ) but up to now have received very little attention in the context of cancer biology.( 3 )
In the human proteome, the tetraspanin family is composed of 33 proteins that are defined by their unique structural features (Fig. 1). Briefly, these proteins have four transmembrane domains with short intracytosolic N‐ and C‐terminal regions, and two extracellular (EC) loops.( 4 ) The EC1 loop is short, between the first and second transmembrane domains, and the EC2 loop is large, between the third and fourth transmembranes region, with more than 100 amino acids and some distinctive features, like a conserved CCG motif of unknown function and several conserved cysteines present in all members of the family. The conserved residues permit the identification of a protein signature,( 5 ) and the EC2 structure identifies at least three tetraspanin subgroups based on their different folding characteristics.( 6 ) The hydrophobic transmembrane regions also contain conserved polar residues (Fig. 1). The short C‐terminal region is likely to provide a link to intracellular signaling molecules.( 7 )
Figure 1.
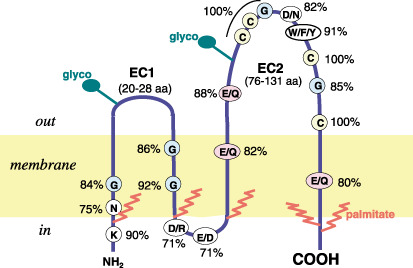
Structure of a tetraspanin protein. The conserved residues are indicated, as is the location of palmitoylation (red) and glycosylation (green). Not all tetraspanins undergo these modifications to the same extent. Glyco, glycosylation.
Tetraspanins can form complexes on the membrane and interact with many different molecules, thus the composition of the complex, known as the tetraspanin web, is very likely to condition biological effects.( 4 , 8 ) Biologically more important than individual components are the different combinations of tetraspanins and associated proteins that can be present on the cellular surface, thus permitting large variability. However, these proteins have been mostly studied from an individual protein point of view and most of the information has been obtained in cells of the hematopoietic system, although many of these proteins are expressed in most cell types.( 1 , 9 ) The tetraspanin proteins can associate with each other, although most of the information has been obtained from a subset of them, CD9, CD81, CD82 and CD151, which have been studied extensively.( 2 , 10 ) Other tetraspanin proteins less well characterized, but also detected in complexes, are CD53, CD63 and CD37. Information on the remaining tetraspanins is negligible.
In this context, some general principles have emerged, although there is still a long way to go regarding their physiological role and cell‐specific functions, particularly in epithelial cells.( 2 ) Therefore the apparent functional redundancy in their biological effects is in part complicated by the fact that they have been studied individually, rather than as a complex of multiple subunits. This requires that tetraspanin biology should be considered as a compartmentalized system. Furthermore, there are tetraspanins with highly specialized functions, such as uroplakins in the bladder where they form a structure that seals the epithelium,( 11 ) and peripherin–retinal degeneration, slow (RDS) and retinal outer segment membrane protein (ROM1) in the photoreceptors of the retina.( 12 )
Tetraspanin‐enriched microdomains and their components. The hydrophobic nature of these proteins clearly indicates that they are likely to be associated with themselves or with other membrane proteins. They therefore have the potential to form microdomains rich in cholesterol and gangliosides on the plasma membrane, known as tetraspanin webs or tetraspanin‐enriched microdomains, which are different from lipid rafts( 1 , 13 ) and can organize different types of membrane proteins,( 14 ) including growth factor receptors such as heparin‐binding epidermal growth factor (HB‐EGF). This is of particular relevance as HB‐EGF is a receptor related to tumor dissemination,( 15 ) proteins with immunoglobulin domains and members of the human leukocyte antigen (HLA) system. The combination of tetraspanin expression differs depending on cell type and differentiation. Lately, some web components have been confirmed by proteomic approaches.( 16 , 17 ) Table 1 summarizes the proteins identified as interacting with tetraspanins.
Table 1.
Proteins that interact with tetraspanins
| CD9 | CD53 | CD81 | CD82 | CD151 | CD63 | CD37 | |
|---|---|---|---|---|---|---|---|
| α3β1 | + | + | + | + | + | + | + |
| α4β1 | + | + | + | + | ND | + | ND |
| α6β1 | + | + | + | + | + | + | ND |
| α5β1 | + | ND | + | + | ND | + | ND |
| Precursor β1 | + | ND | – | – | ND | – | ND |
| αIIbβ3 | + | ND | ND | ND | ND | ND | ND |
| α6β4 | – | ND | – | ND | + | – | ND |
| CD11/CD18 | ND | ND | ND | ND | ND | + | ND |
| HLA‐DR | ND | + | + | + | ND | + | + |
| HLA‐DM | ND | ND | ND | + | ND | + | ND |
| HLA‐DQ | ND | + | + | + | ND | ND | ND |
| HLA‐DO | ND | ND | ND | + | ND | + | ND |
| EGF‐R | ND | ND | ND | + | ND | ND | ND |
| TGF‐α | + | ND | ND | ND | ND | ND | ND |
| FGFR | + | ND | + | ND | ND | ND | ND |
| c‐Met | + | ND | ND | + | ND | ND | ND |
| HB‐EGF | + | ND | –? | –? | ND | ND | ND |
| FRPP | + | ND | + | ND | ND | ND | ND |
| EWI‐2 | + | ND | + | + | ND | ND | ND |
| EWI‐F | ND | ND | + | ND | ND | ND | ND |
| CD36 | + | ND | ND | ND | ND | ND | ND |
| CD9P‐1 | + | ND | + | + | + | + | ND |
| CD2 | ND | + | ND | ND | ND | ND | ND |
| CD4 | ND | ND | + | + | ND | ND | ND |
| CD8 | ND | ND | + | + | ND | ND | ND |
| CD21‐CD19‐Leu13 | ND | ND | + | ND | ND | ND | ND |
| CD19 | + | ND | + | + | ND | ND | ND |
| CD20 | ND | + | + | + | ND | ND | ND |
| CD46 | + | ND | + | + | + | ND | ND |
| Protein kinase C | + | + | + | + | + | – | – |
| Phosphatidylinositol 4‐kinase | + | – | + | – | + | + | ND |
| Dectin‐1 | ND | ND | ND | ND | ND | ND | + |
| Syntenin 1 | ND | ND | ND | ND | ND | + | ND |
| γ‐Glutamyl transpeptidase | ND | + | + | + | ND | ND | ND |
Green (+), positive interaction; red (–), no interaction; EGF‐R, epidermal growth factor receptor; EWI, Ewing; FGFR, fibroblast growth factor receptor; FRPP, prostaglandin F2α receptor regulatory protein; HB‐EGF, heparin‐binding epidermal growth factor; HLA, human leukocyte antigen; ND, not determined; TGF, transforming growth factor.
Tetraspanin protein interactions
Tetraspanin interactions with integrins. Integrins represent the main group of proteins interacting with tetraspanins, and considering their relevance in processes such as adhesion to the extracellular matrix, cell motility, invasion and angiogenesis,( 18 , 19 , 20 , 21 ) it is surprising that the effects of tetraspanins on integrin function has only being studied in detail using mostly CD151; thus the functional consequences of interacting selectively with one tetraspanin or another has not yet been characterized. The main integrins found to be associated with tetraspanins are those containing the β1 chain, a major component for attachment to the extracellular matrix,( 22 ) with variation in the α subunit, such as the dimers α3β1, α4β1 and α6β1, which are detected in most cell types. The expression of other such dimers, such as α2β1, α5β1 and α6β4, is more restricted.( 23 ) Some interactions appear to be specific for a tetraspanin, as is the case for α1β1 with CD9, but others such as α4β1 are associated with CD9, CD53, CD81, CD82 and CD151.( 24 ) The association of α3 integrin with CD9 is promoted by the GM3 ganglioside.( 25 ) The direct interaction between a tetraspanin and an integrin has been specifically demonstrated in the case of the CD151–α3β1 interaction.( 24 ) The interaction takes place through its EC2 loop and requires the integrity of the CCG and PxxCC motifs of the tetraspanin and the α subunit of the integrin;( 23 ) glycosylation of this loop was also necessary for the interaction with CD82.( 26 ) In general it can be concluded that one α integrin can bind to more than one type of tetraspanin. However, the β chain also contributes to the interaction, as exemplified by CD151, which interacts with α6β1 and α6β4.( 27 )
These CD151–integrin interactions strengthen the attachment to the extracellular matrix.( 28 ) Integrin β1 is an important element in anchoring epithelial stem cells to basal membranes and therefore might affect expansion of tumor cells,( 21 ) but whether this role is modulated by tetraspanins is not known. The integrin β1 chain participates in signaling by the activation of the integrin‐linked kinase,( 29 , 30 ) but modulation of this kinase by tetraspanins has not been studied. Integrin activation or binding to ligands does not affect their interaction with tetraspanins,( 23 ) but tetraspanins appear to affect postligand effects such as modulating actin dynamics, which are functionally reflected in migration and cell adhesion properties. The tetraspanin complex with integrins is in a low‐affinity conformation, and changes in affinity do not alter integrin interaction with tetraspanins. The tetraspanin–integrin complexes are functional in adhesion assays in a cadherin‐independent manner, and might provide spatial cues for cellular polarization.( 31 )
Tetraspanin interaction with growth factor receptors and membrane proteins. Many growth factors and their receptors play an important role in tumor biology, and some were among the first oncogenes identified. Among the growth factor receptors found to be associated with tetraspanins there are several types (Table 1), including receptors with immunoglobulin domains, particularly those of the Ewing (EWI) family. EWI‐2 can be crosslinked to CD9 and CD81( 32 ) or CD82 in prostate cancer cells.( 33 ) EWI‐2 also modulates lymphocyte activation mediated by integrins( 34 ) and growth factor receptors with tyrosine‐kinase activity, such as c‐Met/hepatocyte growth factor receptor (HGF‐R), which interacts with CD82.( 35 ) The interaction of the ganglioside GM2 with CD82‐c‐Met inhibits its cross‐talk with integrin signaling and reduces c‐Met activation signals.( 36 ) It is interesting to note that c‐Met/HGF‐R is the receptor for scatter factor or hepatocyte growth factor, which is implicated in epithelial–mesenchimal transition, a fundamental process in the dissemination of tumor cells.( 37 ) These interactions have been studied mainly in the context of an individual tetraspanin, without taking into consideration their free or web organization, which can affect the magnitude or specificity of their effects. In epithelial cells CD9 interacts with epithelial cell adhesion molecule.( 17 ) However, a study of interactions between different tetraspanins and cadherins, and their functional consequences, has not yet been carried out.
The interaction of CD9 with transforming growth factor (TGF)‐α has been shown to regulate epidermal growth factor receptor (EGFR) activation and cell proliferation in epithelial cells.( 38 ) In macrophages, activation by interleukin (IL)‐6 requires the interaction between dectin‐1 and CD37.( 39 ) But in general it can be concluded that knowledge concerning tetraspanin effects on signaling by different types of growth factor receptors is limited and patchy at best.
Proteins of the HLA family, both class II and class I, constitute another major group of membrane proteins associated with tetraspanins.( 40 , 41 ) However, whether tetraspanin proteins modulate signals or functions mediated by these HLA antigens has not been characterized, but they are likely to affect antigen presentation.( 41 ) In dendritic cells the lateral interaction between CD9 and MHC class II antigen facilitates T‐cell activation.( 42 ) It will be important to determine their functional consequences in tumor cells were HLA antigen expression is frequently downregulated to facilitate immune evasion.
Tetraspanin interactions with intracellular signaling molecules. Tetraspanins are considered to be molecular facilitators because they participate in or modulate several signaling and biological processes.( 10 ) All of these effects have been reported in very heterogeneous cell types, but the implications is clear: tetraspanins can influence intracellular signaling, directly or indirectly, and thus can modulate signals initiated at other membrane receptors. However, as their characterization continues these effects are likely to be more specific than previously thought.
Most of the information on tetraspanin signaling has been obtained using specific monoclonal antibodies. In this regard CD9, CD82, CD81 and CD53 are the best‐characterized antigens.( 43 ) Among signals detected in response to tetraspanin antigen ligation by monoclonal antibodies were calcium mobilization by CD9, CD53, CD81 or CD82, protein kinase C (PKC) activation, increased levels of diacylglycerol, and activation of phosphatidylinositol 3‐kinase and phospholipase Cγ.( 43 )
Another component of tetraspanin signaling is determined by a direct physical interaction between some tetraspanins, such as CD151, and type II phosphatidylinositol 4‐kinase (PI4K); in this case the tetraspanin functions as the connector between the associated integrin and the PI4K molecule.( 44 , 45 ) Only some tetraspanins, CD9, CD63, CD81, A15 and CD151, are associated with PI4K, and not others such as CD82, CD53 or CD37; this interaction does not seem to require previous binding to an integrin, therefore it may be mediated by an as yet unidentified protein, and will have functional consequences depending on the pattern of expression in different cell types.
CD53 ligation activates PKC;( 46 , 47 , 48 ) it was later demonstrated that following cell stimulation, PKC binds to the intracellular side of CD9, CD53, CD81, CD82 and CD151.( 49 ) This bound PKC is able to phosphorylate the integrin α3 subunit interacting with CD151. Also, ligation of the CD53 antigen could transiently activate c‐jun NH2‐terminal kinase (JNK) and c‐jun‐dependent transcription in different cell types.( 50 ) In renal mesangial cells the ligation of CD53 induces a proliferative response mediated by the extracellular‐regulated kinases (ERK) ERK1 and ERK2.( 51 ) Also, CD53 and CD63 have been found to be associated with a tyrosine phosphatase (CD45) activity that dephosphorylates lymphocyte‐specific protein tyrosine kinase (lck),( 52 ) an important mediator of lymphocyte activation.( 53 , 54 ) All of the signaling molecules associated with tetraspanins have also been associated with different aspects of tumor biology in different systems. However, no systematic characterization of the response has been carried out.
Tetraspanin web
Palmitoylation determines the formation of cholesterol‐rich microdomains. Proteins associated with membranes can be modified covalently by palmitoylation, which can control their association and organization in different cellular membranes. The tetraspanins CD9, CD37, CD53, CD63, CD81, CD82 and CD151 are palmitoylated molecules.( 55 ) CD9, CD81 and CD82 palmitoylation is a requirement for their association with cholesterol complexes.( 13 ) The assembly of the tetraspanin web is started in the Golgi, where homodimers and heterodimers of CD9, CD81 and CD151 can be identified and isolated, and probably constitute intermediate building blocks in the assembly of the tetraspanin web.( 56 ) Furthermore, palmitoylated molecules appear to be important for assembly of the tetraspanin web, favoring association with other tetraspanins as well as other associated proteins.( 57 , 58 )
CD9 is palmitoylated intracellularly in juxtamembrane cysteine residues; it contributes to the association with other tetraspanins, CD81 and CD53, but is not affected by partitioning in detergents. Mutation of all CD9( 55 ) or CD151( 58 ) palmitoylation residues results in a more diffuse distribution of the proteins and prevents their association with cholesterol, suggesting that it plays a role in web stabilization. Although palmitoylated CD9 interacts with integrins, unpalmitoylated CD9 is freer and has enhanced binding to EWI‐2 and EWI‐F.( 59 ) The loss of palmitoylation does not affect spreading on the extracellular matrix, but these cells present a larger number of focal adhesions and an increase in adhesion‐induced phosphorylation of Akt, without affecting activation of focal adhesion kinase (FAK) or ERK1 and ERK2.( 57 )
Palmitoylation also controls the association between tetraspanins and intracellular signaling molecules. In these complexes, their ligation results in phosphorylation of signaling molecules such as vav, which is lost if cells are treated with cholesterol‐disrupting detergents.( 13 ) Interestingly, palmitoylation is regulated by the cellular redox state, and under oxidative stress palmitoylation is inhibited favoring signaling by 14‐3‐3 adaptor proteins.( 60 ) Thus, unpalmitoylated CD81 is bound constitutively to 14‐3‐3 protein, a serine–threonine binding protein.( 60 ) In B‐cells the coligation of the B‐cell receptor with CD19–CD21–CD81 promotes CD81 palmitoylation and stabilization of the complex within a cholesterol‐rich fraction.( 61 ) CD82 palmitoylation was shown to be necessary for mobility and invasion of PC13 cells, and loss of palmitoylation results not only in the abolishement of these properties, but also in regaining interaction with the p130 (CAS)–CrkII signaling complex.( 62 )
Palmitoylated tetraspanins (CD9, CD81, CD63) coexist and colocalize with palmitoylated integrin β4, which promotes the incorporation of CD151 into these tetraspanin complexes.( 63 ) Loss of integrin palmitoylation results in reduction of association among these tetraspanins, and in an increase in CD9 complexes, as well as signaling by p130 (CAS) when cells are grown on laminin.( 63 )
An overall picture regarding the role of tetraspanin palmitoylation emerges from these data (Fig. 2). There are two clearly different types of complexes: those with palmitoylated tetraspanins that favor their association with integrins and their integration into cholesterol‐rich fractions, and non‐palmitoylated tetraspanins that are accessible to binding with different signaling molecules, such as 14‐3‐3, p130 (CAS) or EWI proteins among others.
Figure 2.
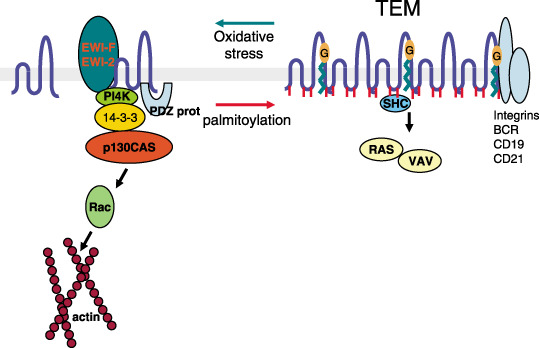
Organization and associations of tetraspanin proteins on the cell membrane in relation to cell signaling. The palmitoylated molecules are aggregated in the tetraspanin web, which is rich in cholesterol. The associated proteins differ depending on the state, as do the type of signals that they can modulate. The representation of signaling molecules in each state does not represent a direct interaction but their implication in the response. G, ganglioside that promotes tetraspanin interaction with integrins and affects signaling by growth factors; TEM, tetraspanin‐enriched microdomain.
Gangliosides are a regulatory component of tetraspanin complexes. Gangliosides are complex of glycolipids that contain a branched chain of as many as seven sugar residues. Specific interactions of gangliosides with tetraspanins have been reported in the case of several tetraspanins, and there appears to be some selectivity. Thus, the GM3 ganglioside interacts preferentially with CD9( 25 , 64 ) and CD81( 65 ) and reduces the mitogen‐activated protein kinase (MAPK) activation initiated by the interacting fibroblast growth factor receptor (FGFR).( 65 ) The GM2 ganglioside is mainly associated with CD82,( 36 , 66 ) and GD2 and GD3 gangliosides interact with CD151.( 67 ) Also, gangliosides such as GM3 promote the association of CD9 with the α3 integrin( 25 ) or α5 and inhibit cell motility.( 68 )
Dynamics of tetraspanin microdomains: Cell‐specific complexes. The web assembly of tetraspanins and their specific interactions can be interpreted within the general framework of the dynamics of a complex with the potential for a very high functional heterogeneity (Fig. 3). In this system there is a double level to increase complexity: the participation of the tetraspanins in the core, and the type and number of associated proteins. Thus variability in the combination of proteins can confer great flexibility to allow for specificity and functional differences depending on cell type. Therefore, tetraspanin complexes in specific cell types can be very different despite sharing several of their components. In that way the association of tetraspanin with membrane receptors may exist as an isolated complex on the cell membrane, or form part of a larger tetraspanin‐core complex. Obviously the association–dissociation kinetics represent an additional level of regulation, where palmitoylation and redox state play an important role (Fig. 2), but these possibilities have not yet been explored in any system.
Figure 3.
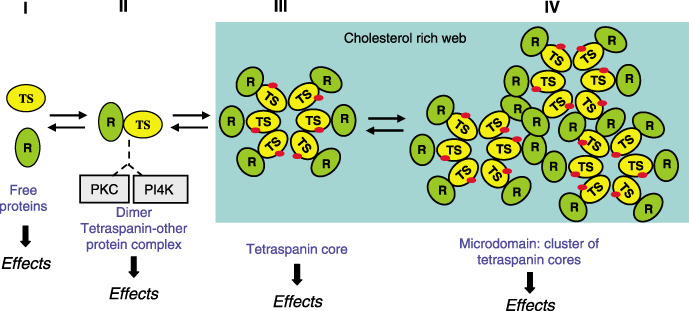
Dynamics of the formation of a tetraspanin microdomain. The tetraspanin protein binds to a receptor, which may be specific for each tetraspanin, on the plasma membrane. This heterodimer can be incorporated into a larger complex, where the tetraspanin proteins form the core. The participating tetraspanins are determined by their pattern and level of expression in a given cell type, as well as by the presence of their partners, which may also vary from cell to cell. These variations confer a great number of combinations that can be reflected in functional differences. The biological effects will depend on the situation of a specific tetraspanin at the time of stimulation. PI4K, phosphatidylinositol 4‐kinase; PKC, protein kinase C; R, receptor or membrane protein (some of which are listed in Table 1) that interacts with tetraspanins; TS, any of the 33 tetraspanin proteins. The red spots indicate palmitoylated molecules.
Individual associations of tetraspanin with other proteins may take place either in intracellular vesicles before moving to the membrane( 56 ) or in the membrane before incorporation into the complex, and might even associate with partner proteins during early biosynthesis.( 69 ) It is not known if the signals are different when a specific protein–tetraspanin is incorporated into a larger complex. However, initiation of signaling is very likely to be different if activated by either free molecules, in heterodimers, or a larger complex, such as the tetraspanin web.
Phenotypes associated with tetraspanin deficiencies
Mutation or loss of human tetraspanins has been detected with different phenotypes. In all cases there is a defect of cell adhesion properties and tissue organization. A rare single nucleotide insertion (G383) in CD151 exon 5 introduces a frameshift and stop signal at codon 140. The truncated protein lacks its integrin‐binding domain and causes skin blistering, deafness, β‐thalasemia and nephritis,( 70 ) but only the latter phenotype was reproduced in one of three knockout mice.( 71 ) Mutations in the specific tetraspanin A15 gene causes X‐linked mental retardation,( 72 ) and patients with mutations in the peripherin–RDS or ROM1 genes have retinal degeneration resulting from disorganization in the layered structure of photoreceptor cells.( 73 ) These phenotypes may reflect a defective interaction with the extracellular matrix; however, the reason why these are restricted to specific cell types when CD151 is expressed in most tissues is not known, but it is likely to be conditioned by their protein partners in each cell type.
The loss of surface CD53 antigen expression has been reported in one family, although the genetic cause underlying this loss of surface expression is not known. These patients had less than 10% of the expected surface expression of CD53 in B cells, T cells and neutrophils; therefore, the patients presented with a syndrome composed of multiple recurrent infectious diseases, very heterogeneous in nature from bacteria to viruses,( 74 ) which is very similar to chronic granulomatosis that shows defects in leukocyte adhesion and motility.
Several knockouts of murine tetraspanin genes also suggest a role in adhesion and motility. CD9( 75 ) and CD81( 76 ) knockout mice have reduced fertility as a consequence of defective sperm–egg fusion. CD151 knockout mice have defective organization of laminin, disrupting laminin–integrin binding( 77 ) and leading to defective reepitelization during wound healing.( 78 ) These animals had alterations in angiogenesis related to cell adhesion properties, but altered angiogenic properties were only detected when cells were grown in culture.( 79 ) These defects are also associated with defective signaling by protein kinase B (PKB)/c‐Akt, e‐nitric oxide synthase (NOS), ras‐related C3 botulinum toxin substrate 1 (rho family, small GTP‐binding protein Rac1) (Rac) and cell division cycle (Cdc)42 that was diminished.( 77 ) However, of the three CD151 knockout (ko) mice only one reproduces some of the phenotypes identified in humans. In addition, mice defective in CD81, CD9 or CD37 also show hyperproliferation of T cells that may be a consequence of lck hyperactivity.( 79 ) None of the murine models has been used to study tetraspanin implications in tumor susceptibility, formation or progression, and no predisposition to tumor formation has yet been reported in these animals, suggesting that tetraspanins are not likely to be implicated in, or play a minor role in, the initiation of tumors.
In Drosophila melanogaster, the loss of the tetraspanin late bloomer results in formation of defective neuromuscular junctions.( 80 ) Other tetraspanins have mild defects, which may reflect a functional redundancy. In Caenorhabditis elegans inactivation of the tetraspanin tsp‐15 (a likely ortholog of mammalian CD151) results in skin degeneration and blister formation indicating that it is required to maintain the integrity of the epithelium;( 81 ) a similar phenotype has been detected in animal models and some human diseases. This phenotype might indirectly reflect defective adhesion to the extracellular matrix.
Contribution of individual tetraspanins to relevant phenotypes in tumor biology: Cell adhesion and motility
CD151. In endothelial cells CD151 is localized mainly to the basal and lateral junctions,( 82 ) and blocking antibodies increase endothelial cell adhesion to the extracellular matrix, reducing their rate of invasion in collagen gels.( 83 ) Keratynocyte motility in wound healing assays is also reduced.( 84 ) In migrating ketarinocytes and breast cancer cells CD9, CD81 and CD151 are involved in transient interactions with the substrate before a more stable interaction is formed by attachment structures, such as in protrusions by lamellopodia formation. These structures are both dependent and independent of ligand binding, do not contain elements of the cytoskeleton, and colocalize with myristoylated alanine‐rich protein kinase C substrate (MARCKS), which are substrates of PKC that regulate cytoskeleton reorganization during migration.( 82 ) In human skin CD151 was detected clustered at the basal cell colocalizing with laminin 5( 70 ) and connecting focal adhesion with actin filaments as part of hemidesmosomes, which did not contain CD9 or CD81.( 85 ) In polarized epithelial cells, CD9, CD151 and CD81 also localize at lateral cell–cell contacts with a distribution similar to cadherins, although the effect of antibodies is cadherin independent. These three tetraspanins have been detected at heterotypic interactions between melanoma and endothelial cells.( 86 ) Overexpression of CD151 results in increased motility, and enhanced expression of matrix metalloproteinase‐9 and invasiveness.( 87 ) This probably occurs as a result of activating pathways mediated by small GTPases, which increase GTP binding to Cdc42 and rac, organizers of the cell cytoskeleton.( 88 ) The reduction in CD151 levels caused by small interfering RNA (siRNA) in primary melanocytes results in the loss of motility( 89 ) but has little effect on the steady‐state levels of integrins. However, these adhesion and motility alterations could be reversed by CD151 reexpression.( 90 ) In carcinoma cells CD151 forms complexes with the major laminin‐5 receptors, α3β1 and α6β4 integrins, in lateral cell surface membranes. The dissociation of CD151 from laminin‐binding integrins permits the migration of epithelial cells.( 91 ) These data on signaling and motility clearly relate CD151 to adhesion and dissemination of cells, but it has not been characterized in this context in human cancer.
CD82. The tetraspanin CD82 functions as a link between the actin cytoskeleton and membrane raft domains, inducing stable adhesion, spreading and development of membrane extensions. Signaling by CD82 involves its translocation from detergent‐resistant membranes to cytoskeleton pellets, and all of its effects are severely affected by cholesterol depletion.( 92 ) This CD82‐related actin polymerization involves src kinases including Vav1 and p56 lck and depends on the ability of CD82 to associate with the extracellular matrix.( 93 , 94 ) The depletion of gangliosides also destabilizes CD82 complexes,( 95 ) reducing the interaction with CD151 and increasing the interaction with EGFR( 66 ) and c‐Met,( 36 ) which modulates their signaling. Overexpression of CD82 might contribute to tumor invasion by inhibiting the cross‐talk between integrins, Met and src, and activation and phosphorylation of Src and its downstream targets p130 (CAS) and FAK.( 35 , 96 ) Inhibition of c‐Met or Src reduces invasion to the same extent as CD82 expression in PC3 cells.( 35 ) High levels of CD82 correlate with a lower invasion potential in prostate,( 96 ) lung( 97 ) and esophageal cancer( 98 ) and with its overexpression in multiple myeloma cell lines.( 99 )
The surface expression of CD82 antigen is upregulated by cytokines such as IL‐1β, IL‐4, IL‐6, IL‐13, interferon‐γ and tumor necrosis factor‐α.( 100 ) In lymphoid cell lines the coligation of CD82 and Fc receptors induces an increase in intracellular calcium level mediated by phospholipase C‐induced phosphatidyl inositol (PtdIns)(1,4,5)P3 second messengers followed by a more stable change, linked to extracellular calcium entry.( 100 ) In Jurkat cells stimulated with anti‐CD82 and anti‐CD3 monoclonal antibodies, implicates different transcription factors such as NF‐AT, AP‐1, and NF‐κB,( 101 ) resulting in an increased production of IL‐2. Morphologically, cells become strongly adherent and develop dendritic extensions, changes associated with the arrest of cell proliferation.( 102 )
Recently using a yeast two‐hybrid approach, duffy antigen receptor for chemokines (DARC), a membrane protein of endothelial cells, was identified as a binding partner of CD82 on prostate cells.( 103 ) This interaction makes cells enter senescence.( 104 ) Thus, in tumors losing this interaction, caused by CD82 loss, there would be a reduction in senescence, which is an antioncogenic mechanism, permitting tumor cell growth.( 105 )
CD9. CD9 can associate with the transmembrane region of TGF‐α, which results in the induction of EGFR activation and cellular proliferation.( 38 ) The metalloprotease ADAM10 promotes the association of CD9 with HB‐EGF.( 106 ) But more recently CD9 has been associated with control of cell motility. Functionally, anti‐CD9 antibodies inhibited transmigration of melanoma cells.( 86 ) The ectopic expression of CD9 can suppress cellular outgrowth by a neurite‐like process and promotes apoptotic death of cells that arre adherent to fibronectin.( 107 ) This reduction in adhesion correlates with the reduction in adhesion‐dependent phosphorylation of Akt but not that of FAK or JNK.( 107 ) However, the opposite effect is seen in primary melanocytes where reduction of CD9 by siRNA results in loss of motility.( 89 ) Motility dependent on binding to laminin‐5 is inhibited by the interaction of CD9 with the GM3 ganglioside.( 25 ) All of these data suggest that loss of CD9 and CD81 is likely to promote tumor dissemination. In agreement with this potential role, an inverse correlation between CD9 level and invasiveness has been reported in melanoma,( 108 ) breast cancer,( 109 ) oral squamous cell carcinoma,( 110 ) ovarian carcinoma( 111 ) and cervical carcinoma.( 112 )
It is interesting to note that CD9 knockout mice have reduced fertility as a consequence of defective sperm–egg fusion.( 75 ) More recently PSG17 has been identified as an essential CD9 ligand for sperm–egg fusion.( 113 , 114 ) PSG17, a placenta‐secreted pregnancy‐specific glycoprotein, upon binding to CD9 in monocytes induces the secretion of anti‐inflammatory cytokines such as IL‐10, IL‐6 and TGF‐β1, contributing to avoiding rejection.( 115 , 116 ) If a similar situation occured in a tumor environment, the loss of CD9 would prevent this response and probably contribute to tumor growth in an inflammatory environment.
CD81. The CD81 antigen was originally described as the TAPA1 antigen because it was the target of an antibody that has an antiproliferative effect and regulated the intracellular thiol levels.( 117 ) Later it was found to be associated with major histocompatibility complex (MHC) antigens( 118 ) and other tetraspanins,( 119 , 120 ) forming part of the CD19–CD21 signaling complex on B cells.( 121 ) CD81 and CD151 tetraspanin molecules are components of the endothelial lateral junctions implicated in the regulation of cell motility, either directly or by modulation of the function of the associated integrin heterodimers;( 83 ) CD81 was soon associated with adhesion and motility in lymphocytes.( 122 ) The link between CD81 and the actin cytoskeleton seems to be mediated by EWI‐2 and EWI‐F.( 123 ) CD81 overexpression reduces viability and motility in multiple myeloma cell lines.( 99 )
CD63. Most of the CD63 antigen is usually detected at a very high concentration in endosomal particles; therefore, it has attracted limited attention regarding its cell signaling characteristics. The C‐terminal intracellular region of CD63 interacts with the PDZ domain of syntenin‐1, a molecule that is implicated in the regulation of endocytosis and slows down CD63 internalization.( 124 ) However, CD63 is also expressed on the cell surface and is a clear marker of biological functions, particularly affecting tumor dissemination. CD63 was the first tetraspanin to be related to a very high invasive potential in melanomas, where an inverse correlation between CD63 levels and metastatic potential was identified with prognostic value( 125 , 126 ) and its reexpression in melanoma cells inhibits its invasion potential.( 127 ) A similar correlation has been detected in a colon carcinoma cell line.( 128 ) Despite this role CD63 expression in other tumors has not been studied.
CD53: Radioresistance and effects on redox state. The CD53 antigen has been mainly studied in rat and human cells. Ligation with specific antibodies stimulates several processes, such as the induction of nitric oxide synthase( 46 ) in macrophages, homotypic cell adhesion in B‐cell lymphomas( 48 ) and DNA synthesis in mesangial cells,( 51 ) all of which are dependent on PKC activation. Part of the signal is also transmitted by the JNK system and has a protective effect on apoptosis.( 129 )
Some tetraspanin proteins, CD53, CD82 and CD81, as well as their associated proteins CD19 and CD21, have been shown to interact with γ‐glutamyl transpeptidase, which regulates the intracellular redox state by modulating the level of glutathione.( 130 ) Interestingly the mechanism by which radiation therapy kills cells is based on its ability to generate free radicals within the cell and is therefore dependent on the intracellular redox state. The microarray analysis of gene expression in radioresistant LY cells has identified a very high overexpression of CD53 antigen as one of the main markers of these resistant populations,( 131 ) resulting in an increase in the intracellular level of glutathione, which has antioxidant properties and thus counteracts the radiation effect, facilitating cell survival.( 131 )
Expression of the tetraspanin protein family in normal and stem cells
Surface expression of tetraspanins in hematopoietic cells. The most complete study of tetraspanin protein expression was carried out during human B‐cell maturation.( 132 ) The relative surface levels of five tetraspanins (CD9, CD37, CD53, CD63 and CD81), and their interacting proteins (CD19, CD21 and HLA‐DR) revealed three different combinations of tetraspanin antigen expression (Fig. 4). In early bone marrow (BM) CD10+ B‐cell precursors there is high expression of CD81 and CD9 and a relatively low level of CD53, and these cells are negative for CD37 (Fig. 4a). In mature and peripheral B lymphocytes (CD10−) there is downregulation of CD9 and CD81 and upregulation of CD53 and CD37 (Fig. 4b). In plasma cells that have passed through secondary lymphoid tissues and reentered the BM there is CD9 reexpression and CD37 downregulation, but CD53 expression is maintained (Fig. 4c). These distinct patterns of tetraspanin expression may reflect the occurrence of different cellular interactions and homing properties during B‐cell maturation. These patterns in normal B‐cell maturation identify two switches in tetraspanin antigen expression occurring in the BM, in the transition from CD10+ B‐cell precursors to CD10− B lymphocytes, and later on during terminal differentiation into antibody‐secreting BM plasma cells.( 132 )
Figure 4.
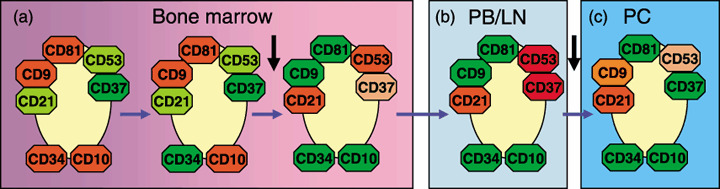
Expression of four tetraspanin antigens during B‐cell maturation. The relative levels of expression of four tetraspanins (CD9, CD37, CD53 and CD81) are shown along the maturation pathway of normal human B cells. Red indicates maximum expression; orange indicates medium levels of expression; green indicates negativity. Expression of other maturation markers is also included. Based on Barrena et al.( 132 ) LN, lymph node; PB, peripheral blood; PC, plasma cells. The horizontal arrows indicate the B‐cell maturation pathway. The black vertical arrows indicate the two main tetraspanin switchs.
Individual tetraspanin antigen expression was initially determined in some hematopoietic cells. CD9 was detected in platelets( 133 ) and in pre‐B‐cell lines.( 134 ) CD81 is an antiproliferative antigen in T cells,( 135 ) as is CD63 in melanoma cells.( 126 ) But soon it was realized that they were forming clusters.( 136 ) Increases in surface levels of CD9 and CD53 were detected in activated platelets and B cells, respectively. CD82 was also increased in response to several cytokines, and CD81 increased in eosinophils reacting to parasitic infections. All of these effects represent a role for tetraspanins as part of the cellular response.
The location of tetraspanins, and their interaction with β1 integrin, suggests that they might have a role in attachment to specific microenvironments, which may be niches of stem cells. Although precursors of lymphoid cells express relatively higher levels of CD9, they also express CD53 and CD63 in the endosomes. CD53 and CD63 are two proteins that are distributed asymmetrically in the CD133+ stem cell population,( 137 ) which is a hallmark of stem cells, but its significance is unknown.
Expression of tetraspanin proteins in epithelial cells. There are few reports on the expression of multiple tetraspanins in epithelial cells, where they might be part of different complexes depending on their location. In Madin–Darby canine kidney cells CD9, CD81 and CD151 are localized at lateral contact sites in membranes with a similar distribution to that of E‐cadherin. However, free CD9 was also detected in the apical surface of these cells.( 31 )
A systematic approach to detect six tetraspanin proteins, CD9, CD37, CD53, CD63, CD81 and CD82, in the gastrointestinal tract has been carried out. Two of these proteins, CD9 and CD82, were expressed at similarly high levels throughout the gastrointestinal tract, from the esophagus to the colon. CD63 expression was more restricted, ranging from the distal stomach to the colon. CD81 was detected only in basal layers of the esophagus. CD53 was barely detected and no expression of CD37 was observed.( 138 ) These differences clearly suggest that the organization of the tetraspanin web and its components could present significant differences depending on their localization in the gastrointestinal tract; however, their functional significance is not known. In the gastrointestinal tract some antigens, such as Co‐029, have been identified in colon tumors( 139 ) but have received no attention so far.
Tetraspanin proteins have been used to attempt to identify epithelial precursor cells in airway tract that is severely damaged in diseases such as cystic fibrosis. In the basal cells of the trachea there is a compartment of stem and transit amplifying cells. A marker of early stages is the detection of CD151 in combination with tissue factor antigens. Therefore, tracheal cells positive for CD151 and tissue factor were able to proliferate and reconstitute a fully differentiated epithelium in rat with epithelium‐denuded trachea. The cells were also positive for telomerase activity, considered a marker for the transit amplifying population. Cells of the columnar epithelium that were CD151 and tissue factor negative were unable to proliferate or reconstitute the epithelium and had no telomerase activity. These data suggest that CD151‐positive adult basal cells are present at least in the amplification compartment of the epithelium and have regenerative potential.( 140 ) It is not known if other epithelial cell markers or tetraspanins have the same effects.
In the squamous cervical epithelium there is a very high expression of CD9 in cells of the basal layer. This expression is downregulated in squamous cervical carcinomas and correlates with stage, but curiously as the tumor progresses there is a re‐expression of CD9 as it becomes more vascularized. This might be an important element in permitting tumor dissemination( 112 ) and might reflect an interaction with vascular cells not yet identified. In patients with scleroderma pigmentosum there is increased expression of CD151, CD53 and CD37,( 141 ) of which the latter two are not expressed in normal skin and therefore may give rise to skin alterations because they affect interactions with other cells or the stroma.( 142 )
Regulation of human tetraspanin gene expression. To understand cell‐specific expression of tetraspanin antigen genes it is necessary to characterizate their promoters. However, only two human CD9( 143 ) and CD53( 144 ) promoters have been partially studied, without any apparent overlap among the regulatory modules identified, suggesting that they are likely to have different regulation, consistent with their inverse expression in different cell types. The requirements for regulation of CD53 gene expression were determined in three different lymphoid cell lines. It is striking to note that the contribution of DNA response elements specific for different transcription factors varies depending on cell type, with some having a common effect and others having the opposite effect. Thus DNA response elements to v‐ets erythroblastosis virus E26 oncogene homology 1 (ets1), ETS domain‐containing protein Elk‐1 (elk1), C‐myc purine‐binding transcription factor (PuF), GATA1 and E4BP4 have different effects depending on cell type (Fig. 5). These data reflect the importance of regulatory sequence modularity in the control of cell‐specific gene expression.( 145 )
Figure 5.
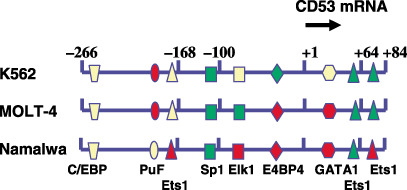
Differential regulation of CD53 gene expression depending on tumor cell type. The transcription factor requirement for human CD53 expression was studied in three lymphoid tumor cell lines of different origin, Namalwa (B‐cell tumor, Burkitt lymphoma), K562 (erythroleukemia) and Molt‐4 (T‐cell lymphoma). The positive or negative effect of transcription factors analyzed by mutagenesis of the CD53 gene promoter( 144 ) is indicated by a color‐coded summary of the results. Green (activation), red (inhibition), yellow (no effect).
Expression of tetraspanin proteins in human cancer
Most of the information on expression of multiple tetraspanin proteins is a consequence of the information obtained using expression microarrays in the analysis of different types of human cancer. These studies provide information on two aspects: the antigens expressed based on a given phenotype, and the relative change in the pattern as a function of differentiation or tumor phenotype, which indicates that they are functionally different.
Expression of tetraspanin proteins in B‐cell malignancies. The analysis of tetraspanin gene expression in a panel of 21 Burkitt lymphoma cell lines suggested that their phenotype corresponds to an intermediate stage of B‐cell development, and that the origin of these tumors is likely to be represented by a stage in which there is a switch in the expression of CD37 and CD9,( 146 , 147 ) a switch later identified in normal B‐cell development.( 132 )
The cell surface expression of four tetraspanin antigens and associated proteins has been studied in 67 cases of B‐cell malignancies.( 132 ) Hierarchical clustering analysis of flow cytometry immunophenotypic data showed a good correlation between tumor differentiation stage and the pattern of tetraspanin expression.( 132 ) Mature and peripheral B‐cell leukemias and lymphomas express high levels of CD37 and CD53, whereas those derived from B‐cell precursor (BCP)‐acute lymphocytic leukemia and clonal plasma cells coexpress CD9 with either CD81 or CD53, respectively. Despite these phenotypic similarities, variable levels of expression of one or more of these proteins were detected frequently and of these, phenotypic aberrations were common to most patients within a specific disease group. The differences in the pattern of tetraspanin surface expression can be used to discriminate two different lymphomas in an individual patient.( 148 ) In a well‐characterized diagnostic entity such as B‐chronic lymphocytic leukemia (CLL), the expression pattern of the CD9 and CD53 tetraspanins is associated with the pattern of in vivo tissue involvement. Thus, abnormally high reactivity to CD53 is associated with greater peripheral blood and lymph node infiltration. Previously, it was demonstrated that ligation of CD53 antigen protects lymphoid cells from apoptosis,( 129 ) an important property for mature and peripheral memory lymphocytes. Therefore, overexpression of CD53 could render B‐CLL cells more adapted to survive in peripheral blood and lymph nodes. In contrast, reduced CD9 expression on B‐CLL cells has been associated with a higher BM involvement. CD9 also functions as a motility or migration brake,( 95 ) which could help to explain the relationship observed between CD9 expression and the pattern of BM involvement.
Expression of tetraspanin proteins in carcinomas. Tetraspanins have received little attention in the study of human carcinomas but some information is emerging from three different lines of work. Information on expression is derived mostly from determining few tetraspanin antigens or from expression arrays, and functional information has been obtained experimentally using tumor cell lines. The alternative expression of some tetraspanin antigens permits the identification of subgroups in different types of human cancer.
The coordinated expression of tetraspanin initially identified in B cells (previous section) appears to be more general. In kidney cancer, high expression of CD53 and CD37 correlates inversely with respect to CD9 expression (low) in conventional renal cell carcinomas of clear cells and in papillary carcinomas, but not in other types such as granular carcinomas or oncocytomas in which CD9 expression is very high.( 149 ) Also, in melanomas there is no CD53 and CD37 expression and they express varying levels of CD9; the latter is inversely correlated with metastatic potential,( 108 ) a pattern consistent with a higher probability of infiltration in lymph nodes.( 132 )
The loss of CD9 has been correlated with higher motility and metastatic potential of tumor cells from lung,( 150 ) esophageal,( 98 ) oral,( 110 ) ovarian,( 111 ) cervical( 112 ) and gastric( 151 ) carcinomas. In the basal layer of the normal squamous epithelium of the uterine cervix CD9 is strongly expressed, but in invasive carcinomas it is downregulated. However, in some areas there is a re‐expression of CD9 that is correlated with lymphangiosis. This cluster of C9 might be an indicator of high risk of recurrence as CD9 has a role in transendothelial migration.( 112 ) In bladder cancer cell lines GM3 in glycosynapse 3 has a dual functional role. The first one modulates the interaction between α3 integrin and CD9. The second role is to activate or inhibit the activity of c‐src. Functionally high levels of GM3 reduce motility and invasiveness, whereas low levels have the opposite effect.( 64 ) CD63, one of the original tetraspanins, was identified in melanomas where its levels were inversely correlated with metastatic potential;( 126 ) in cell lines its overexpression suppresses the malignant phenotype.( 152 )
The expression of four tetraspanins, CD9, CD63, CD82 and CD151, has been studied in breast carcinoma cell lines with different invasive capabilities in in vitro assays. Low‐level expression clearly predicts their invasive potential, particularly for CD63, and the expression of CD9, CD63 and CD151 appeared to be coordinated by a common mechanism.( 153 )
In pancreatic carcinomas, overexpression of the little‐characterized Co‐029 protein has been associated with induction of angiogenic factor transcription activity, which includes increased matrix metalloproteinase and urokinase‐type plasminogen activator secretion, pronounced vascular endothelial growth factor expression in fibroblasts and vascular endothelial growth factor receptor expression.( 154 ) These effects can be blocked with an anti‐Co‐029 antibody.( 154 )
Tetraspanin expression has been studied in a model of colon carcinoma with cell lines derived from primary tumors and two from metastasis in colon carcinomas. The proteins associated with CD9 were determined by a proteomic approach. In this system 32 proteins were detected, including integrins, proteins with immunoglobulin domains, CD44, epithelial cell adhesion molecule, membrane proteases (ADAM10, TADG‐15 and CD26/dipeptidyl peptidase IV) and signaling proteins (heterotrimeric G proteins). Also, some differences were identified, particularly the Co‐029 tetraspanin antigen in metastasis, which was almost absent in primary tumors but very high in normal colon.( 17 )
In thyroid tumors CD82 is highly expressed in benign goiter, but expression at both the RNA and protein levels was significantly reduced in carcinomas, and was reduced even further in metastasic cells. There were no changes in the expression of CD63 in these tumors.( 155 )
In some prostate carcinomas with high levels of CD151, CD9 and CD53 are downregulated. CD9 and CD53 are upregulated in another group of prostate carcinomas.( 156 ) In these prostate carcinomas, CD9 and CD53 expression seems to be positively coordinated, which is the opposite of what has been detected in other cell types. In low‐grade prostate cancer the survival rate was higher in those cases in which CD151 levels were lower, and CD151 had a better prediction value than histological (Gleason) grade.( 157 ) Murine prostate cancer cells can attach to vascular endothelial cells through DARC, and this interaction inhibits proliferation and induces senescence by expression of T‐box 2 (TBX)2 and p21.( 103 ) The role of CD82 as a metastasis suppressor is compromised in DARC knockout mice. All of these data suggest that the interaction between DARC and CD82 is essential for the metastasis‐suppressor role of CD82.( 103 ) It would be interesting to know if a similar interaction is found for other tetraspanins that also may function as metastasis suppressors.
Bladder cancer represents a unique entity due to its characteristics. The expression of uroplakin, an antigen that seals the bladder epithelium, can be used to identify the type of tumor and its severity. Uroplakin II is mainly expressed in transitional bladder carcinomas but not in squamous cell carcinomas and can be used to monitor circulating cancer cells and metastasis.( 158 , 159 ) In this type of tumor the expression of the most common tetraspanins has not been characterized.
Conclusions and future directions
Despite the dispersed knowledge of tetraspanin proteins in relation to cancer, and that they have been mainly studied in an individual context rather than as complexes it is very likely that patterns of expression of these proteins will affect the behavior of tumor cells with respect to signaling by growth factors, cell motility and sensitivity to therapy, which will be identified when these studies are carried out. The regulatory role played by palmitoylation and gangliosides requires further characterization as they modulate cell signaling by associated growth factor receptors. It is expected that in the future these proteins will attract more attention and be studied in the proper context within tumor biology, as they are likely to play an important role in conferring specificity to many biological effects.
Although the basic processes in which tetraspanin proteins play a role have already been outlined, their specific participation in these processes and in different cell types will require a systematic approach where the multiplicity of components, their relatives levels and localization are taken simultaneously into account, thus generating a specific cell behavior.
Acknowledgments
This work was funded by grants from the Ministerio de Educación y Ciencia (SAF2004‐02900; SAF2007‐60242), Fundación de Investigación Médica MM and Federación de Cajas de Ahorro de Castilla y León.
References
- 1. Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol 2003; 19: 397–422. [DOI] [PubMed] [Google Scholar]
- 2. Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 2005; 6: 801–11. [DOI] [PubMed] [Google Scholar]
- 3. Boucheix C, Duc GHT, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert Rev Mol Med 2001; 2001: 1–17. [DOI] [PubMed] [Google Scholar]
- 4. Tarrant JM, Robb L, Van Spriel AB, Wright MD. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol 2003; 24: 610–17. [DOI] [PubMed] [Google Scholar]
- 5. Shoham T, Rajapaksa R, Kuo CC, Haimovich J, Levy S. Building of the tetraspanin web: distinct structural domains of CD81 function in different cellular compartments. Mol Cell Biol 2006; 26: 1373–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seigneuret M, Delaguillaumie A, Lagaudriere‐Gesbert C, Conjeaud H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J Biol Chem 2001; 276: 40 055–64. [DOI] [PubMed] [Google Scholar]
- 7. Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci 2003; 28: 106–12. [DOI] [PubMed] [Google Scholar]
- 8. Yunta M, Lazo PA. Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell Signal 2003; 15: 559–64. [DOI] [PubMed] [Google Scholar]
- 9. Wright MD, Moseley GW, Van Spriel AB. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens 2004; 64: 533–42. [DOI] [PubMed] [Google Scholar]
- 10. Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J 1997; 11: 428–42. [PubMed] [Google Scholar]
- 11. Min G, Wang H, Sun TT, Kong XP. Structural basis for tetraspanin functions as revealed by the cryo‐EM structure of uroplakin complexes at 6‐A resolution. J Cell Biol 2006; 173: 975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ritter LM, Boesze‐Battaglia K, Tam BM et al . Uncoupling of photoreceptor peripherin/rds fusogenic activity from biosynthesis, subunit assembly, and targeting: a potential mechanism for pathogenic effects. J Biol Chem 2004; 279: 39 958–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charrin S, Manie S, Thiele C et al . A physical and functional link between cholesterol and tetraspanins. European J Immunol 2003; 33: 2479–89. [DOI] [PubMed] [Google Scholar]
- 14. Levy S, Shoham T. Protein–protein interactions in the tetraspanin web. Physiology (Bethesda) 2005; 20: 218–24. [DOI] [PubMed] [Google Scholar]
- 15. Recio JA, Merlino G. Hepatocyte growth factor/scatter factor activates proliferation in melanoma cells through p38 MAPK, ATF‐2 and cyclin D1. Oncogene 2002; 21: 1000–8. [DOI] [PubMed] [Google Scholar]
- 16. Andre M, Le Caer JP, Greco C et al . Proteomic analysis of the tetraspanin web using LC‐ESI‐MS/MS and MALDI‐FTICR‐MS. Proteomics 2006; 6: 1437–49. [DOI] [PubMed] [Google Scholar]
- 17. Le Naour F, Andre M, Greco C et al . Profiling of the tetraspanin web of human colon cancer cells. Mol Cell Proteomics 2006; 5: 845–57. [DOI] [PubMed] [Google Scholar]
- 18. Watt FM. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J 2002; 21: 3919–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer 2002; 2: 91–100. [DOI] [PubMed] [Google Scholar]
- 20. Ridley AJ, Schwartz MA, Burridge K et al . Cell migration: integrating signals from front to back. Science 2003; 302: 1704–9. [DOI] [PubMed] [Google Scholar]
- 21. Janes SM, Watt FM. New roles for integrins in squamous‐cell carcinoma. Nat Rev Cancer 2006; 6: 175–83. [DOI] [PubMed] [Google Scholar]
- 22. Hemler ME, Lobb RR. The leukocyte beta 1 integrins. Curr Opin Hematol 1995; 2: 61–7. [DOI] [PubMed] [Google Scholar]
- 23. Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci 2001; 114: 4143–51. [DOI] [PubMed] [Google Scholar]
- 24. Yauch RL, Kazarov AR, Desai B, Lee RT, Hemler ME. Direct extracellular contact between integrin alpha (3) beta (1) and TM4SF protein CD151. J Biol Chem 2000; 275: 9230–8. [DOI] [PubMed] [Google Scholar]
- 25. Kawakami Y, Kawakami K, Steelant WF et al . Tetraspanin CD9 is a ‘proteolipid’, and its interaction with alpha3 integrin in microdomain is promoted by GM3 ganglioside, leading to inhibition of laminin‐5‐dependent cell motility. J Biol Chem 2002; 277: 34 349–58. [DOI] [PubMed] [Google Scholar]
- 26. Ono M, Handa K, Sonnino S, Withers DA, Nagai H, Hakomori S. GM3 ganglioside inhibits CD9‐facilitated haptotactic cell motility: coexpression of GM3 and CD9 is essential in the downregulation of tumor cell motility and malignancy. Biochemistry 2001; 40: 6414–21. [DOI] [PubMed] [Google Scholar]
- 27. Berditchevski F, Zutter MM, Hemler ME. Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Mol Biol Cell 1996; 7: 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishiuchi R, Sanzen N, Nada S et al . Potentiation of the ligand‐binding activity of integrin α3β1 via association with tetraspanin CD151. Proc Natl Acad Sci USA 2005; 102: 1939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hannigan GE, Leung‐Hagesteijn C, Fitz‐Gibbon L et al . Regulation of cell adhesion and anchorage‐dependent growth by a new beta 1‐integrin‐linked protein kinase. Nature 1996; 379: 91–6. [DOI] [PubMed] [Google Scholar]
- 30. Dedhar S. Cell–substrate interactions and signaling through ILK. Curr Opin Cell Biol 2000; 12: 250–6. [DOI] [PubMed] [Google Scholar]
- 31. Yanez‐Mo M, Tejedor R, Rousselle P, Sanchez‐Madrid F. Tetraspanins in intercellular adhesion of polarized epithelial cells: spatial and functional relationship to integrins and cadherins. J Cell Sci 2001; 114: 577–87. [DOI] [PubMed] [Google Scholar]
- 32. Stipp CS, Kolesnikova TV, Hemler ME. EWI‐2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem 2001; 276: 40 545–54. [DOI] [PubMed] [Google Scholar]
- 33. Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res 2003; 63: 2665–74. [PubMed] [Google Scholar]
- 34. Kolesnikova TV, Stipp CS, Rao RM, Lane WS, Luscinskas FW, Hemler ME. EWI‐2 modulates lymphocyte integrin α4β1 functions. Blood 2004; 103: 3013–19. [DOI] [PubMed] [Google Scholar]
- 35. Sridhar SC, Miranti CK. Tetraspanin KAI1/CD82 suppresses invasion by inhibiting integrin‐dependent crosstalk with c‐Met receptor and Src kinases. Oncogene 2006; 25: 2367–78. [DOI] [PubMed] [Google Scholar]
- 36. Todeschini AR, Dos Santos JN, Handa K, Hakomori SI. Ganglioside GM2/tetraspanin CD82 complex inhibits Met activation, and its cross‐talk with integrins: Basis for control of cell motility through glycosynapse. J Biol Chem 2007; 282: 8123–33. [DOI] [PubMed] [Google Scholar]
- 37. Trusolino L, Comoglio PM. Scatter‐factor and semaphorin receptors: cell signalling for invasive growth. Nature Rev Cancer 2002; 2: 289–300. [DOI] [PubMed] [Google Scholar]
- 38. Shi W, Fan H, Shum L, Derynck R. The tetraspanin CD9 associates with transmembrane TGF‐α and regulates TGF‐α‐induced EGF receptor activation and cell proliferation. J Cell Biol 2000; 148: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meyer‐Wentrup F, Figdor CG, Ansems M et al . Dectin‐1 interaction with tetraspanin CD37 inhibits IL‐6 production. J Immunol 2007; 178: 154–62. [DOI] [PubMed] [Google Scholar]
- 40. Szollosi J, Horejsi V, Bene L, Angelisova P, Damjanovich S. Supramolecular complexes of MHC class I, MHC class II, CD20, and tetraspan molecules (CD53, CD81, and CD82) at the surface of a B cell line JY. J Immunol 1996; 157: 2939–46. [PubMed] [Google Scholar]
- 41. Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic 2007; 8: 89–96. [DOI] [PubMed] [Google Scholar]
- 42. Unternaehrer JJ, Chow A, Pypaert M, Inaba K, Mellman I. The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc Natl Acad Sci USA 2007; 104: 234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci 2001; 58: 1189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berditchevski F, Tolias KF, Wong K, Carpenter CL, Hemler ME. A novel link between integrins, transmembrane‐4‐superfamily proteins (CD63 and CD81), and phosphatidylinositol 4‐kinase. J Biol Chem 1997; 272: 2595–8. [DOI] [PubMed] [Google Scholar]
- 45. Claas C, Stipp CS, Hemler ME. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J Biol Chem 2001; 276: 7974–84. [DOI] [PubMed] [Google Scholar]
- 46. Bosca L, Lazo PA. Induction of nitric oxide release by MRC OX‐44 (anti‐CD53) through a protein kinase C‐dependent pathway in rat macrophages. J Exp Med 1994; 179: 1119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barcia R, Garcia‐Vargas S, Bosca L, Lazo PA. CD53 antigen and epidermal growth factor induce similar changes in the pattern of phorbol ester binding in a B cell lymphoma. Cell Immunol 1996; 169: 107–12. [DOI] [PubMed] [Google Scholar]
- 48. Lazo PA, Cuevas L, Gutierrez del Arroyo A, Orue E. Ligation of CD53/OX44, a tetraspan antigen, induces homotypic adhesion mediated by specific cell–cell interactions. Cell Immunol 1997; 178: 132–40. [DOI] [PubMed] [Google Scholar]
- 49. Zhang XA, Bontrager AL, Hemler ME. Transmembrane‐4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta (1) integrins. J Biol Chem 2001; 276: 25 005–13. [DOI] [PubMed] [Google Scholar]
- 50. Yunta M, Oliva JL, Barcia R, Horejsi V, Angelisova P, Lazo PA. Transient activation of the c‐Jun N‐terminal kinase (JNK) activity by ligation of the tetraspan CD53 antigen in different cell types. Eur J Biochem 2002; 269: 1012–21. [DOI] [PubMed] [Google Scholar]
- 51. Yunta M, Rodriguez‐Barbero A, Arevalo MA, Lopez‐Novoa JM, Lazo PA. Induction of DNA synthesis by ligation of the CD53 tetraspanin antigen in primary cultures of mesangial cells. Kidney Int 2003; 63: 534–42. [DOI] [PubMed] [Google Scholar]
- 52. Carmo AM, Wright MD. Association of the transmembrane 4 superfamily molecule CD53 with a tyrosine phosphatase activity. Eur J Immunol 1995; 25: 2090–5. [DOI] [PubMed] [Google Scholar]
- 53. Penninger JM, Wallace VA, Kishihara K, Mak TW. The role of p56lck and p59fyn tyrosine kinases and CD45 protein tyrosine phosphatase in T‐cell development and clonal selection. Immunol Rev 1993; 135: 183–214. [DOI] [PubMed] [Google Scholar]
- 54. Sefton BM, Taddie JA. Role of tyrosine kinases in lymphocyte activation. Curr Opin Immunol 1994; 6: 372–9. [DOI] [PubMed] [Google Scholar]
- 55. Charrin S, Manie S, Oualid M, Billard M, Boucheix C, Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett 2002; 516: 139–44. [DOI] [PubMed] [Google Scholar]
- 56. Kovalenko OV, Yang X, Kolesnikova TV, Hemler ME. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross‐linking. Biochem J 2004; 377: 407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Berditchevski F, Odintsova E, Sawada S, Gilbert E. Expression of the palmitoylation‐deficient CD151 weakens the association of α3β1 integrin with the tetraspanin‐enriched microdomains and affects integrin‐dependent signaling. J Biol Chem 2002; 277: 36 991–7000. [DOI] [PubMed] [Google Scholar]
- 58. Yang X, Claas C, Kraeft SK et al . Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interacts, subcellular distribution, and integrin‐dependent cell morphology. Mol Biol Cell 2002; 13: 767–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang XH, Kovalenko OV, Kolesnikova TV et al . Contrasting effects of EWI proteins, integrins and protein palmitoylation on cell surface CD9 organization. J Biol Chem 2006; 281: 12 976–85. [DOI] [PubMed] [Google Scholar]
- 60. Clark KL, Oelke A, Johnson ME, Eilert KD, Simpson PC, Todd SC. CD81 associates with 14‐3‐3 in a redox‐regulated palmitoylation‐dependent manner. J Biol Chem 2004; 279: 19 401–6. [DOI] [PubMed] [Google Scholar]
- 61. Cherukuri A, Carter RH, Brooks S et al . B cell signaling is regulated by induced palmitoylation of CD81. J Biol Chem 2004; 279: 31 973–82. [DOI] [PubMed] [Google Scholar]
- 62. Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility‐ and invasiveness‐inhibitory activity. Cancer Res 2004; 64: 7455–63. [DOI] [PubMed] [Google Scholar]
- 63. Yang X, Kovalenko OV, Tang W, Claas C, Stipp CS, Hemler ME. Palmitoylation supports assembly and function of integrin‐tetraspanin complexes. J Cell Biol 2004; 167: 1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mitsuzuka K, Handa K, Satoh M, Arai Y, Hakomori S. A specific microdomain (‘glycosynapse 3’) controls phenotypic conversion and reversion of bladder cancer cells through GM3‐mediated interaction of α3β1 integrin with CD9. J Biol Chem 2005; 280: 35 545–53. [DOI] [PubMed] [Google Scholar]
- 65. Toledo MS, Suzuki E, Handa K, Hakomori S. Cell growth regulation through GM3‐enriched microdomain (glycosynapse) in human lung embryonal fibroblast WI38 and its oncogenic transformant VA13. J Biol Chem 2004; 279: 34 655–64. [DOI] [PubMed] [Google Scholar]
- 66. Odintsova E, Butters TD, Monti E, Sprong H, Van Meer G, Berditchevski F. Gangliosides play an important role in the organisation of CD82‐enriched microdomains. Biochem J 2006; 400: 315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thorne RF, Mhaidat NM, Ralston KJ, Burns GF. Shed gangliosides provide detergent‐independent evidence for type‐3 glycosynapses. Biochem Biophys Res Commun 2007; 356: 306–11. [DOI] [PubMed] [Google Scholar]
- 68. Miura Y, Kainuma M, Jiang H, Velasco H, Vogt PK, Hakomori S. Reversion of the Jun‐induced oncogenic phenotype by enhanced synthesis of sialosyllactosylceramide (GM3 ganglioside). Proc Natl Acad Sci USA 2004; 101: 16 204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yauch RL, Hemler ME. Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphoinositide 4‐kinase. Biochem J 2000; 351: 629–37. [PMC free article] [PubMed] [Google Scholar]
- 70. Karamatic Crew V, Burton N, Kagan A et al . CD151, the first member of the tetraspanin (TM4) superfamily detected on erythrocytes, is essential for the correct assembly of human basement membranes in kidney and skin. Blood 2004; 104: 2217–23. [DOI] [PubMed] [Google Scholar]
- 71. Sachs N, Kreft M, Van Den Bergh Weerman MA et al . Kidney failure in mice lacking the tetraspanin CD151. J Cell Biol 2006; 175: 33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zemni R, Bienvenu T, Vinet MC et al . A new gene involved in X‐linked mental retardation identified by analysis of an X;2 balanced translocation. Nat Genet 2000; 24: 167–70. [DOI] [PubMed] [Google Scholar]
- 73. Van Soest S, Westerveld A, De Jong PT, Bleeker‐Wagemakers EM, Bergen AA. Retinitis pigmentosa: defined from a molecular point of view. Surv Ophthalmol 1999; 43: 321–34. [DOI] [PubMed] [Google Scholar]
- 74. Mollinedo F, Fontan G, Barasoain I, Lazo PA. Recurrent infectious diseases in human CD53 deficiency. Clin Diagn Lab Immunol 1997; 4: 229–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9‐deficient mice. Science 2000; 287: 319–21. [DOI] [PubMed] [Google Scholar]
- 76. Rubinstein E, Ziyyat A, Prenant M et al . Reduced fertility of female mice lacking CD81. Dev Biol 2006; 290: 351–8. [DOI] [PubMed] [Google Scholar]
- 77. Takeda Y, Kazarov AR, Butterfield CE et al . Deletion of tetraspanin CD151 results in decreased pathological angiogenesis in vivo and in vitro . Blood 2006; 109: 1524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cowin AJ, Adams D, Geary SM, Wright MD, Jones JC, Ashman LK. Wound healing is defective in mice lacking tetraspanin CD151. J Invest Dermatol 2006; 126: 680–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Knobeloch KP, Wright MD, Ochsenbein AF et al . Targeted inactivation of the tetraspanin CD37 impairs T‐cell‐dependent B‐cell response under suboptimal costimulatory conditions. Mol Cell Biol 2000; 20: 5363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fradkin LG, Kamphorst JT, DiAntonio A, Goodman CS, Noordermeer JN. Genomewide analysis of the Drosophila tetraspanins reveals a subset with similar function in the formation of the embryonic synapse. Proc Natl Acad Sci USA 2002; 99: 13 663–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Moribe H, Yochem J, Yamada H, Tabuse Y, Fujimoto T, Mekada E. Tetraspanin protein (TSP‐15) is required for epidermal integrity in Caenorhabditis elegans . J Cell Sci 2004; 22: 5209–20. [DOI] [PubMed] [Google Scholar]
- 82. Berditchevski F, Odintsova E. Characterization of integrin–tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J Cell Biol 1999; 146: 477–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yanez‐Mo M, Alfranca A, Cabanas C et al . Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA‐1 and CD151/PETA‐3 with α3β1 integrin localized at endothelial lateral junctions. J Cell Biol 1998; 141: 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Penas PF, Garcia‐Diez A, Sanchez‐Madrid F, Yanez‐Mo M. Tetraspanins are localized at motility‐related structures and involved in normal human keratinocyte wound healing migration. J Invest Dermatol 2000; 114: 1126–35. [DOI] [PubMed] [Google Scholar]
- 85. Sterk LM, Geuijen CA, Oomen LC, Calafat J, Janssen H, Sonnenberg A. The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin α6β4 and may regulate the spatial organization of hemidesmosomes. J Cell Biol 2000; 149: 969–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Longo N, Yanez‐Mo M, Mittelbrunn M et al . Regulatory role of tetraspanin CD9 in tumor‐endothelial cell interaction during transendothelial invasion of melanoma cells. Blood 2001; 98: 3717–26. [DOI] [PubMed] [Google Scholar]
- 87. Hong I‐K, Jin Y‐J, Byun H‐J, Jeoung D‐I, Kim Y‐M, Lee H. Homophilic interactions of tetraspanin CD151 up‐regulate motility and matrix metalloproteinase‐9 expression of human melanoma cells through adhesion‐dependent c‐Jun activation signaling pathways. J Biol Chem 2006; 281: 24 279–92. [DOI] [PubMed] [Google Scholar]
- 88. Shigeta M, Sanzen N, Ozawa M, Gu J, Hasegawa H, Sekiguchi K. CD151 regulates epithelial cell–cell adhesion through PKC‐ and Cdc42‐dependent actin cytoskeletal reorganization. J Cell Biol 2003; 163: 165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Garcia‐Lopez MA, Barreiro O, Garcia‐Diez A, Sanchez‐Madrid F, Penas PF. Role of tetraspanins CD9 and CD151 in primary melanocyte motility. J Invest Dermatol 2005; 125: 1001–9. [DOI] [PubMed] [Google Scholar]
- 90. Winterwood NE, Varzavand A, Meland MN, Ashman LK, Stipp CS. A critical role for tetraspanin CD151 in α3β1 and α6β4 integrin‐dependent tumor cell functions on laminin‐5. Mol Biol Cell 2006; 17: 2707–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chometon G, Zhang ZG, Rubinstein E, Boucheix C, Mauch C, Aumailley M. Dissociation of the complex between CD151 and laminin‐binding integrins permits migration of epithelial cells. Exp Cell Res 2006; 312: 983–95. [DOI] [PubMed] [Google Scholar]
- 92. Delaguillaumie A, Harriague J, Kohanna S et al . Tetraspanin CD82 controls the association of cholesterol‐dependent microdomains with the actin cytoskeleton in T lymphocytes: relevance to co‐stimulation. J Cell Sci 2004; 117: 5269–82. [DOI] [PubMed] [Google Scholar]
- 93. Lagaudriere‐Gesbert C, Lebel‐Binay S, Hubeau C, Fradelizi D, Conjeaud H. Signaling through the tetraspanin CD82 triggers its association with the cytoskeleton leading to sustained morphological changes and T cell activation. Eur J Immunol 1998; 28: 4332–44. [DOI] [PubMed] [Google Scholar]
- 94. Delaguillaumie A, Lagaudriere‐Gesbert C, Popoff MR, Conjeaud H. Rho GTPases link cytoskeletal rearrangements and activation processes induced via the tetraspanin CD82 in T lymphocytes. J Cell Sci 2002; 115: 433–43. [DOI] [PubMed] [Google Scholar]
- 95. Ono M, Handa K, Withers DA, Hakomori S. Motility inhibition and apoptosis are induced by metastasis‐suppressing gene product CD82 and its analogue CD9, with concurrent glycosylation. Cancer Res 1999; 59: 2335–9. [PubMed] [Google Scholar]
- 96. Zhang XA, He B, Zhou B, Liu L. Requirement of the p130CAS‐Crk coupling for metastasis suppressor KAI1/CD82‐mediated inhibition of cell migration. J Biol Chem 2003; 278: 27 319–28. [DOI] [PubMed] [Google Scholar]
- 97. Adachi M, Taki T, Ieki Y, Huang CL, Higashiyama M, Miyake M. Correlation of KAI1/CD82 gene expression with good prognosis in patients with non‐small cell lung cancer. Cancer Res 1996; 56: 1751–5. [PubMed] [Google Scholar]
- 98. Uchida S, Shimada Y, Watanabe G et al . Motility‐related protein (MRP‐1/CD9) and KAI1/CD82 expression inversely correlate with lymph node metastasis in oesophageal squamous cell carcinoma. Br J Cancer 1999; 79: 1168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tohami T, Drucker L, Shapiro H, Radnay J, Lishner M. Overexpression of tetraspanins affects multiple myeloma cell survival and invasive potential. FASEB J 2007; 21: 691–9. [DOI] [PubMed] [Google Scholar]
- 100. Lebel‐Binay S, Lagaudriere C, Fradelizi D, Conjeaud H. CD82, tetra‐span‐transmembrane protein, is a regulated transducing molecule on U937 monocytic cell line. J Leukoc Biol 1995; 57: 956–63. [DOI] [PubMed] [Google Scholar]
- 101. Iwata S, Kobayashi H, Miyake‐Nishijima R et al . Distinctive signaling pathways through CD82 and beta1 integrins in human T cells. Eur J Immunol 2002; 32: 1328–37. [DOI] [PubMed] [Google Scholar]
- 102. Lebel‐Binay S, Lagaudriere C, Fradelizi D, Conjeaud H. CD82, member of the tetra‐span‐transmembrane protein family, is a costimulatory protein for T cell activation. J Immunol 1995; 155: 101–10. [PubMed] [Google Scholar]
- 103. Bandyopadhyay S, Zhan R, Chaudhuri A et al . Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nat Med 2006; 12: 933–8. [DOI] [PubMed] [Google Scholar]
- 104. Iiizumi M, Bandyopadhyay S, Watabe K. Interaction of Duffy antigen receptor for chemokines and KAI1: a critical step in metastasis suppression. Cancer Res 2007; 67: 1411–14. [DOI] [PubMed] [Google Scholar]
- 105. Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature 2004; 432: 307–15. [DOI] [PubMed] [Google Scholar]
- 106. Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein‐coupled receptors. J Cell Biol 2002; 158: 221–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Saito Y, Tachibana I, Takeda Y et al . Absence of CD9 enhances adhesion‐dependent morph differentiation, survival, matrix metalloproteinase‐2 production in small cell lung cancer cells. Cancer Res 2006; 66: 9557–65. [DOI] [PubMed] [Google Scholar]
- 108. Si Z, Hersey P. Expression of the neuroglandular antigen and analogues in melanoma. CD9 expression appears inversely related to metastatic potential of melanoma. Int J Cancer 1993; 54: 37–43. [DOI] [PubMed] [Google Scholar]
- 109. Huang CI, Kohno N, Ogawa E, Adachi M, Taki T, Miyake M. Correlation of reduction in MRP‐1/CD9 and KAI1/CD82 expression with recurrences in breast cancer patients. Am J Pathol 1998; 153: 973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kusukawa J, Ryu F, Kameyama T, Mekada E. Reduced expression of CD9 in oral squamous cell carcinoma: CD9 expression inversely related to high prevalence of lymph node metastasis. J Oral Pathol Med 2001; 30: 73–9. [DOI] [PubMed] [Google Scholar]
- 111. Houle CD, Ding XY, Foley JF, Afshari CA, Barrett JC, Davis BJ. Loss of expression and altered localization of KAI1 and CD9 protein are associated with epithelial ovarian cancer progression. Gynecol Oncol 2002; 86: 69–78. [DOI] [PubMed] [Google Scholar]
- 112. Sauer G, Windisch J, Kurzeder C, Heilmann V, Kreienberg R, Deissler H. Progression of cervical carcinomas is associated with down‐regulation of CD9 but strong local re‐expression at sites of transendothelial invasion. Clin Cancer Res 2003; 9: 6426–31. [PubMed] [Google Scholar]
- 113. Waterhouse R, Ha C, Dveksler GS. Murine CD9 is the receptor for pregnancy‐specific glycoprotein 17. J Exp Med 2002; 195: 277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ellerman DA, Ha C, Primakoff P, Myles DG, Dveksler GS. Direct binding of the ligand PSG17 to CD9 requires a CD9 site essential for sperm–egg fusion. Mol Biol Cell 2003; 14: 5090–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Snyder SK, Wessner DH, Wessells JL et al . Pregnancy‐specific glycoproteins function as immunomodulators by inducing secretion of IL‐10, IL‐6 and TGF‐β1 by human monocytes. Am J Reprod Immunol 2001; 45: 205–16. [DOI] [PubMed] [Google Scholar]
- 116. Ha CT, Waterhouse R, Wessells J, Wu JA, Dveksler GS. Binding of pregnancy‐specific glycoprotein 17 to CD9 on macrophages induces secretion of IL‐10, IL‐6, PGE2, and TGF‐β1. J Leukoc Biol 2005; 77: 948–57. [DOI] [PubMed] [Google Scholar]
- 117. Schick MR, Nguyen VQ, Levy S. Anti‐TAPA‐1 antibodies induce protein tyrosine phosphorylation that is prevented by increasing intracellular thiol levels. J Immunol 1993; 151: 1918–25. [PubMed] [Google Scholar]
- 118. Schick MR, Levy S. The TAPA‐1 molecule is associated on the surface of B cells with HLA‐DR molecules. J Immunol 1993; 151: 4090–7. [PubMed] [Google Scholar]
- 119. Mannion BA, Berditchevski F, Kraeft SK, Chen LB, Hemler ME. Transmembrane‐4 superfamily proteins CD81 (TAPA‐1), CD82, CD63, and CD53 specifically associated with integrin alpha 4 beta 1 (CD49d/CD29). J Immunol 1996; 157: 2039–47. [PubMed] [Google Scholar]
- 120. Rubinstein E, Le Naour F, Lagaudriere‐Gesbert C, Billard M, Conjeaud H, Boucheix C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA‐DR and VLA integrins. Eur J Immunol 1996; 26: 2657–65. [DOI] [PubMed] [Google Scholar]
- 121. Behr S, Schriever F. Engaging CD19 or target of an antiproliferative antibody 1 on human B lymphocytes induces binding of B cells to the interfollicular stroma of human tonsils via integrin a4/b1 and fibronectin. J Exp Med 1995; 182: 1191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Levy S, Todd SC, Maecker HT. CD81 (TAPA‐1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol 1998; 16: 89–109. [DOI] [PubMed] [Google Scholar]
- 123. Sala‐Valdes M, Ursa A, Charrin S et al . EWI‐2 and EWI‐F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin‐radixin‐moesin proteins. J Biol Chem 2006; 281: 19 665–75. [DOI] [PubMed] [Google Scholar]
- 124. Latysheva N, Muratov G, Rajesh S et al . Syntenin‐1 is a new component of tetraspanin‐enriched microdomains: mechanisms and consequences of the interaction of syntenin‐1 with CD63. Mol Cell Bio 2006; 24: 7707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hotta H, Takahashi N, Homma M. Transcriptional enhancement of the human gene encoding for a melanoma‐associated antigen (ME491) in association with malignant transformation. Jpn J Cancer Res 1989; 80: 1186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Hotta H, Ross AH, Huebner K et al . Molecular cloning and characterization of an antigen associated with early stages of melanoma tumor progression. Cancer Res 1988; 48: 2955–62. [PubMed] [Google Scholar]
- 127. Radford KJ, Mallesch J, Hersey P. Suppression of human melanoma cell growth and metastasis by the melanoma‐associated antigen CD63 (ME491). Int J Cancer 1995; 62: 631–5. [DOI] [PubMed] [Google Scholar]
- 128. Sordat I, Decraene C, Silvestre T et al . Complementary DNA arrays identify CD63 tetraspanin and α3 integrin chain as differentially expressed in low and high metastatic human colon carcinoma cells. Lab Invest 2002; 82: 1715–24. [DOI] [PubMed] [Google Scholar]
- 129. Yunta M, Lazo PA. Apoptosis protection and survival signal by the CD53 tetraspanin antigen. Oncogene 2003; 22: 1219–24. [DOI] [PubMed] [Google Scholar]
- 130. Nichols TC, Guthridge JM, Karp DR, Molina H, Fletcher DR, Holers VM. Gamma‐glutamyl transpeptidase, an ecto‐enzyme regulator of intracellular redox potential, is a component of TM4 signal transduction complexes. Eur J Immunol 1998; 28: 4123–9. [DOI] [PubMed] [Google Scholar]
- 131. Voehringer DW, Hirschberg DL, Xiao J et al . Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc Natl Acad Sci USA 2000; 97: 2680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Barrena S, Almeida J, Yunta M et al . Aberrant expression of tetraspanin molecules in B‐cell chronic lymphoproliferative disorders and its correlation with normal B‐cell maturation. Leukemia 2005; 19: 1376–83. [DOI] [PubMed] [Google Scholar]
- 133. Slupsky JR, Seehafer JG, Tang S‐C, Masellis‐Smith A, Shaw ARE. Evidence that monoclonal antibodies against CD9 antigen induce specific association between CD9 and the platelet glycoprotein IIb‐IIIa complex. Jbiolchem 1989; 264: 12 289–93. [PubMed] [Google Scholar]
- 134. Masellis‐Smith A, Jensen GS, Seehafer JG, Slupsky JR, Shaw AR. Anti‐CD9 monoclonal antibodies induce homotypic adhesion of pre‐B cell lines by a novel mechanism. J Immunol 1990; 144: 1607–13. [PubMed] [Google Scholar]
- 135. Takahashi S, Doss C, Levy S, Levy R. TAPA‐1, the target of an antiproliferative antibody, is associated on the cell surface with the Leu‐13 antigen. J Immunol 1990; 145: 2207–13. [PubMed] [Google Scholar]
- 136. Horejsi V, Vlcek C. Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett 1991; 288: 1–4. [DOI] [PubMed] [Google Scholar]
- 137. Beckmann J, Scheitza S, Wernet P, Fischer JC, Giebel B. Asymmetric cell division within the human hematopoietic stem and progenitor cell compartment: Identification of asymmetrically segregating proteins. Blood 2007; 109: 5494–501. [DOI] [PubMed] [Google Scholar]
- 138. Okochi H, Mine T, Nashiro K, Suzuki J, Fujita T, Furue M. Expression of tetraspans transmembrane family in the epithelium of the gastrointestinal tract. J Clin Gastroenterol 1999; 29: 63–7. [DOI] [PubMed] [Google Scholar]
- 139. Szala S, Kasai Y, Steplewski Z, Rodeck U, Koprowski H, Linnenbach AJ. Molecular cloning of cDNA for the human tumor‐associated antigen CO‐029 and identification of related transmembrane antigens. Proc Natl Acad Sci USA 1990; 87: 6833–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hajj R, Baranek T, Le Naour R, Lesimple P, Puchelle E, Coraux C. Basal cells of the human adult airway surface epithelium retain transit‐amplifying cell properties. Stem Cells 2006; 25: 139–48. [DOI] [PubMed] [Google Scholar]
- 141. Whitfield ML, Finlay DR, Murray JI et al . Systemic and cell type‐specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci USA 2003; 100: 12 319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Okochi H, Kato M, Nashiro K, Yoshie O, Miyazono K, Furue M. Expression of tetra‐spans transmembrane family (CD9, CD37, CD53, CD63, CD81 and CD82) in normal and neoplastic human keratinocytes: an association of CD9 with alpha 3 beta 1 integrin. Br J Dermatol 1997; 137: 856–63. [PubMed] [Google Scholar]
- 143. Le Naour F, Prenant M, Francastel C, Rubinstein E, Uzan G, Boucheix C. Transcriptional regulation of the human CD9 gene: characterization of the 5′‐flanking region. Oncogene 1996; 13: 481–6. [PubMed] [Google Scholar]
- 144. Hernandez‐Torres J, Yunta M, Lazo PA. Differential cooperation between regulatory sequences required for human CD53 gene expression. J Biol Chem 2001; 276: 35 405–13. [DOI] [PubMed] [Google Scholar]
- 145. Davidson EH. The Regulatory Genome. Gene Regulatory Networks in Development and Evolution. San Diego, CA: Academic Press, 2006. [Google Scholar]
- 146. Ferrer M, Yunta M, Lazo PA. Tetraspan transmembrane antigen levels in Burkitt lymphoma cell lines. Leukemia 1998; 12: 773. [DOI] [PubMed] [Google Scholar]
- 147. Ferrer M, Yunta M, Lazo PA. Pattern of expression of tetraspanin antigen genes in Burkitt lymphoma cell lines. Clin Exp Immunol 1998; 113: 346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Barrena S, Almeida J, Yunta M et al . Discrimination of biclonal B‐cell chronic lymphoproliferative neoplasias by tetraspanin antigen expression. Leukemia 2005; 19: 1708–9. [DOI] [PubMed] [Google Scholar]
- 149. Higgins JP, Shinghal R, Gill H et al . Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol 2003; 162: 925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Higashiyama M, Taki T, Ieki Y et al . Reduced motility related protein‐1 (MRP1/CD9) gene expression as a factor of poor prognosis in non‐small cell lung cancer. Cancer Res 1995; 55: 6040–4. [PubMed] [Google Scholar]
- 151. Hori H, Yano S, Koufuji K, Takeda J, Shirouzu K. CD9 expression in gastric cancer and its significance. J Surg Res 2004; 117: 208–15. [DOI] [PubMed] [Google Scholar]
- 152. Hotta H, Hara I, Miyamoto H, Homma M. Overexpression of the human melanoma‐associated antigen ME491 partially suppresses in vivo malignant phenotypes of H‐ras‐transformed NIH3T3 cells in athymic nude mice. Melanoma Res 1991; 1: 125–32. [DOI] [PubMed] [Google Scholar]
- 153. Sauer G, Kurzeder C, Grundmann R, Kreienberg R, Zeillinger R, Deissler H. Expression of tetraspanin adaptor proteins below defined threshold values is associated with in vitro invasiveness of mammary carcinoma cells. Oncol Report 2003; 10: 405–10. [PubMed] [Google Scholar]
- 154. Gesierich S, Berezovskiy I, Ryschich E, Zoller M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1a/CO‐029. Cancer Res 2006; 66: 7083–94. [DOI] [PubMed] [Google Scholar]
- 155. Chen Z, Mustafa T, Trojanowicz B et al . CD82, and CD63 in thyroid cancer. Int J Mol Med 2004; 14: 517–27. [PubMed] [Google Scholar]
- 156. Lapointe J, Li C, Higgins JP et al . Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA 2004; 101: 811–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Ang J, Lijovic M, Ashman LK, Kan K, Frauman AG. CD151 protein expression predicts the clinical outcome of low‐grade primary prostate cancer better than histologic grading: a new prognostic indicator? Cancer Epidemiol Biomarkers Prev 2004; 13: 1717–21. [PubMed] [Google Scholar]
- 158. Yuasa T, Yoshiki T, Isono T, Tanaka T, Hayashida H, Okada Y. Expression of transitional cell‐specific genes, uroplakin Ia and II, in bladder cancer: detection of circulating cancer cells in the peripheral blood of metastatic patients. Int J Urol 1999; 6: 286–92. [DOI] [PubMed] [Google Scholar]
- 159. Olsburgh J, Harnden P, Weeks R et al . Uroplakin gene expression in normal human tissues and locally advanced bladder cancer. J Pathol 2003; 199: 41–9. [DOI] [PubMed] [Google Scholar]


