Abstract
To identify novel methylation‐silenced genes in gastric cancers, we carried out a chemical genomic screening, a genome‐wide search for genes upregulated by treatment with a demethylating agent, 5‐aza‐2′‐deoxycytidine (5‐aza‐dC). After 5‐aza‐dC treatment of a gastric cancer cell line (AGS) 579 genes were upregulated 16‐fold or more, using an oligonucleotide microarray with 39 000 genes. From these genes, we selected 44 known genes on autosomes whose silencing in gastric cancer has not been reported. Thirty‐two of these had CpG islands (CGI) in their putative promoter regions, and all of the CGI were methylated in AGS, giving an estimated number of 421 ± 75 (95% confidence interval) methylation‐silenced genes. Additionally, we analyzed the methylation status of 16 potential tumor‐related genes with promoter CGI that were upregulated four‐fold or more, and 14 of these were methylated in AGS. Methylation status of the 32 randomly selected and 16 potential tumor‐related genes was analyzed in 10 primary gastric cancers, and 42 genes (ABHD9, ADFP, ALDH1A3, ANXA5, AREG, BDNF, BMP7, CAV1, CDH2, CLDN3, CTSL, EEF1A2, F2R, FADS1, FSD1, FST, FYN, GPR54, GREM1, IGFBP3, IGFBP7, IRS2, KISS1, MARK1, MLF1, MSX1, MTSS1, NT5E, PAX6, PLAGL1, PLAU, PPIC, RBP4, RORA, SCRN1, TBX3, TFAP2C, TNFSF9, ULBP2, WIF1, ZNF177 and ZNF559) were methylated in at least one primary gastric cancer. A metastasis suppressor gene, MTSS1, was located in a genomic region with frequent loss of heterozygosity (8q22), and was expressed abundantly in the normal gastric mucosa, suggesting its role in gastric carcinogenesis. (Cancer Sci 2006; 97: 64 –71)
Abbreviations:
- 5‐aza‐dC
5‐aza‐2′‐deoxycytidine
- CGI
CpG island
- LOH
loss of heterozygosity
- MS‐RDA
methylation sensitive‐representational analysis
- MSP
methylation‐specific PCR
- PCR
polymerase chain reaction.
Epigenetic alterations are involved in cancer development and progression, and methylation of promoter CGI leads to transcriptional silencing of their downstream genes.( 1 ) In various human cancers, silencing of tumor‐suppressor genes, such as CDKN2A (p16), CDH1 (E‐cadherin) and MLH1, is known to be one of the major mechanisms for their inactivation, along with mutations and LOH. To identify genes silenced by promoter methylation by genome‐wide screenings, various techniques have been developed.( 2 ) Most techniques are based on the methylation status of genomic DNA, including MS‐RDA and restriction landmark genomic scanning. In contrast, Suzuki et al. developed a technique that screens genes re‐expressed after treatment with a demethylating agent, 5‐aza‐dC, using a microarray.( 3 ) The chemical genomic screening technique is simple and is effective in identifying genes silenced in cell lines. It has been applied to colon, bladder, esophageal, pancreatic and prostate cancers.( 3 , 4 , 5 , 6 , 7 )
Gastric cancer is the second most common cause of cancer death in the world.( 8 ) As its molecular basis, deep involvement of aberrant DNA methylation has been indicated by the higher incidences of aberrant DNA methylation of known tumor‐suppressor genes than of mutations.( 9 ) We previously searched for genes silenced in MKN28 and MKN74 cell lines using MS‐RDA,( 10 ) and identified lysyl oxidase as a novel tumor‐suppressor gene.( 11 ) However, the entire picture of methylation‐silenced genes in gastric cancers is still unclear, and further searches for methylation‐silenced genes are necessary.
In the present study, we carried out a chemical genomic screening of methylation‐silenced genes in the human gastric cancer cell line AGS.
Materials and Methods
Tissue samples, cell lines and 5‐aza‐dC treatment
Ten primary gastric cancer samples (male/female = 7/3, aged 38–81 years) and two normal gastric mucosae were obtained from 10 patients undergoing gastrectomy at Aichi Cancer Center (Nagoya, Japan) with informed consent. These samples were frozen and stored at −80°C until extraction of DNA or RNA. Gastric cancer cell lines AGS, MKN28, MKN45 and KATOIII were obtained from the Japanese Collection of Research Bioresources (Tokyo, Japan) and American Type Culture Collection (Manassas, VA, USA). Two gastric cancer cell lines, HSC44 and HSC57, were gifted by Dr Kazuyoshi Yanagihara at the National Cancer Center Research Institute (Tokyo, Japan). AGS cells were seeded at a density of 3 × 105 cells/10 cm dish on day 0 and treated with freshly prepared 1 µM 5‐aza‐dC (Sigma) for 24 h on days 1, 3 and 5. After each treatment, the cells were placed in fresh medium and harvested on day 6. Genomic DNA was extracted by standard phenol/chloroform procedures. Total RNA was extracted using ISOGEN (Nippon Gene, Tokyo, Japan) and purified using an RNeasy Mini kit (Qiagen, Valencia, CA, USA).
Oligonucleotide microarray analysis
Oligonucleotide microarray analysis was carried out using GeneChip Human Genome 133 Plus 2.0 (Affymetrix, Santa Clara, CA, USA) with 54 000 probe sets and 47 400 transcripts from 39 000 genes. From 8 µg of total RNA, the first‐strand cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen, Groningen, the Netherlands) and a T7‐(dT)24 primer (Amersham Bioscience, Buckinghamshire, UK), and the double‐stranded cDNA was then synthesized. From the double‐stranded cDNA, biotin‐labeled cRNA was prepared using a BioArray HighYield RNA transcript labeling kit (Enzo, Farmingdale, NY, USA). Labeled cRNA (20 µg) was fragmented, and the GeneChips were hybridized. The arrays were stained and scanned according to the protocol from Affymetrix. The data were processed using GeneChip Operating Software. The signal intensities were normalized so that the average of all of the genes on a GeneChip would be 500. The P‐values for different expression (change P‐value) were calculated in each probe by statistical algorithms based on the Wilcoxon's signed rank test. The change P‐values of 0.003 and 0.997 were used as thresholds to define genes with increased and decreased expression, respectively. Expression data for the normal tissues using GeneChip were obtained from the database RefEXA (http://www.lsbm.org/site_e/database/index.html),( 12 ) with kind permission from Dr H. Aburatani.
Methylation‐specific polymerase chain reaction
DNA (1 µg) digested with BamHI was denatured in 0.3 M NaOH at 37°C for 15 min. Then, 3.6 M sodium bisulfite (pH 5.0) and 0.6 mM hydroquinone were added, and the sample underwent 15 cycles of 30‐s denaturation at 95°C and a 15‐min incubation at 50°C. The sample was desalted with the Wizard DNA Clean‐Up system (Promega, Madison, WI, USA) and desulfonated in 0.3 M NaOH. DNA was ethanol‐precipitated and dissolved in 40 µL of Tris‐EDTA buffer. MSP was carried out with a primer set specific to the methylated or unmethylated sequence (M or U set), using 0.5 µL of the sodium‐bisulfite‐treated DNA. A region 200 bp or less upstream of a putative transcriptional start site was analyzed, except for BDNF (−401 to −214). Primer sequences and PCR conditions are shown in Table 1. DNA methylated with SssI methylase was used to determine specific conditions of PCR for M sets.
Table 1.
Primers for methylation‐specific polymerase chain reaction
| Genes | M/U | Forward primer | Reverse primer | Annealing(°C) | No.cycles | ||
|---|---|---|---|---|---|---|---|
| Position † | Sequence | Position | Sequence | ||||
| ADFP | M | −169 | GGTCGGGTTTTCGTTCGGTTTTC | −36 | ACCCGAATATCACCCTCGAACACG | 55 | 34 |
| U | −170 | AGGTTGGGTTTTTGTTTGGTTTTT | −36 | ACCCAAATATCACCCTCAAACACA | 57 | 34 | |
| ALDH1A3 | M | −108 | TCGGTTTCGTAGTTAATTAGGC | −18 | GACTCGACCCGAACACTACGCA | 55 | 35 |
| U | −108 | TTGGTTTTGTAGTTAATTAGGT | −18 | CAACTCAACCCAAACACTACACA | 49 | 34 | |
| ANXA5 | M | −164 | TATTTAGGTTCGCGAGATTAGC | −48 | CCAAAACCCCAACCGCAAACCG | 57 | 34 |
| U | −154 | TGTGAGATTAGTGGGATAGTTT | −48 | ACCAAAACCCCAACCACAAACCA | 55 | 34 | |
| AREG | M | −173 | TTTTTAGCGAATTTTTACGTAC | −22 | ATAAAACGACGCGCACCTACCG | 55 | 35 |
| U | −165 | GAATTTTTATGTATGAGGGAGGT | −22 | ATAAAACAACACACACCTACCA | 55 | 34 | |
| BDNF | M | −401 | TACGTAAATAGCGAGGTTAGTC | −214 | AACTCCGACGAAACTAAATTCG | 55 | 34 |
| U | −411 | GTGAGTTGGTTATGTAAATAGT | −214 | AACTCCAACAAAACTAAATTCA | 52 | 34 | |
| CAV1 | M | −80 | TTTCGGGACGTTTTTCGGTGGT | −6 | TAAAAACGTTTCTCCCGCGCTA | 59 | 34 |
| U | −95 | GAAAATATTTGTTTTTTTTGGGAT | −3 | ACAAATAAAAACATTTCTCCCACA | 55 | 35 | |
| EEF1a2 | M | −248 | GTTTCGTTTTTCGGGTTCGTC | −28 | GCCCTACAACACGCCAATACG | 57 | 34 |
| U | −250 | TTGTTTTGTTTTTTGGGTTTGTT | −28 | ACACCCTACAACACACCAATACA | 58 | 34 | |
| F2R | M | −187 | TTAGGAGGGTCGAGACGGTCGC | −96 | TCCTCTAAACACCGTTAATTCG | 55 | 34 |
| U | −189 | TTTTAGGAGGGTTGAGATGGTTGT | −98 | TCCTCTAAACACCATTAATTCACA | 55 | 35 | |
| FADS1 | M | −234 | GTTCGTTTGACGTTAGGAAGTC | −34 | GCCCAAAACCAACCGCCTACG | 55 | 35 |
| U | −234 | GTTTGTTTGATGTTAGGAAGTT | −34 | CACCCAAAACCAACCACCTACA | 55 | 34 | |
| FSD1 | M | −159 | AGGGTTTTGGGCGAGGTTAGC | −25 | AAACTACCTTTACCGCGACCG | 56 | 34 |
| U | −158 | GGGTTTTGGGTGAGGTTAGTGT | −25 | CAAACTACCTTTACCACAACCA | 58 | 34 | |
| FST | M | −172 | TTTAGATTTAAAGCGCGGTTGC | −47 | ACGAATAACTCGAACGAACG | 55 | 34 |
| U | −173 | GTTTAGATTTAAAGTGTGGTTGT | −47 | ACAAATAACTCAAACAAACA | 55 | 34 | |
| GREM1 | M | −134 | CGTCGGTATTTAAACGGGAGAC | −35 | GAAACTCGACGCGAAATCAACG | 55 | 35 |
| U | −134 | TGTTGGTATTTAAATGGGAGAT | −35 | CAAAACTCAACACAAAATCAACA | 55 | 34 | |
| IGFBP7 | M | −195 | GGGTCGGTTACGTCGGGTGTTC | −18 | GACAAAAACGCGAATAAACCG | 55 | 35 |
| U | −197 | ATGGGTTGGTTATGTTGGGTGTTT | −18 | CAACAACAAAAACACAAATAAACCA | 60 | 35 | |
| IL6R | M | −117 | TTTTTATAGCGTAATTTCGTTTAC | 78 | AACCGAAACGAATAACGCAACA | 48 | 35 |
| U | −124 | GGTGTGTTTTTTATAGTGTAATTTT | 65 | TAACACAACAACCCCACACACCA | 60 | 34 | |
| IRS2 | M | −127 | GCGGCGTTAATGCGAGGTAGC | −29 | TAAATAACACATCGCGCACCG | 55 | 35 |
| U | −128 | TGTGGTGTTAATGTGAGGTAGT | −24 | CACACAATAAATAACACATCACA | 60 | 35 | |
| KISS1 | M | −198 | AAAGTTTCGTTTCGGAGGGTTC | −49 | CTTTTATAAAACCCGAAATAACG | 58 | 34 |
| U | −198 | AAAGTTTTGTTTTGGAGGGTTT | −49 | CCTTTTATAAAACCCAAAATAACA | 55 | 33 | |
| MARK1 | M | −268 | TTTAGACGATCGTAAATTTTGC | −26 | TCAAAAAAAACGACCCGAACCG | 52 | 34 |
| U | −216 | GGATAGGTGGGTAAGAGAGTGT | −32 | AAAACAACCCAAACCAACTACA | 55 | 34 | |
| MLF1 | M | −118 | GGGTAGCGGCGTATTGTTTTTC | −16 | CTCACTCGCCGCGACGCAAACG | 55 | 35 |
| U | −120 | TAGGGTAGTGGTGTATTGTTTTTT | −7 | ACAAACAACACCTCACTCACCACA | 60 | 35 | |
| MSX1 | M | −178 | CGTCGTTTGGGTTTTGTTTTGC | −18 | CCGACTCCGAACCCTACCG | 55 | 34 |
| U | −160 | TTGTGTGTTTTTAGGTTTAGTGT | −18 | CCAACTCCAAACCCTACCA | 58 | 34 | |
| MX1 | M | −178 | GGGTTCGGGTTCGAGAATTTGC | −21 | TTCGCCTCTTTCACCCCG | 55 | 34 |
| U | −179 | TGGGTTTGGGTTTGAGAATTTGT | −21 | ACTTCACCTCTTTCACCCCA | 55 | 36 | |
| NT5E | M | −183 | AGTCGATAGTCGCGTTAGGGTC | −36 | GAACAACTAAAACCGAAACTCG | 55 | 35 |
| U | −184 | TAGTTGATAGTTGTGTTAGGGTT | −41 | AACTAAAACCAAAACTCAATACC | 53 | 35 | |
| PLAGL1 | M | −195 | GTTCGGGTTTATTTGCGTTAGC | −47 | AACCCCTAACGAAAACGTCACG | 60 | 33 |
| U | −196 | GGTTTGGGTTTATTTGTGTTAGT | −47 | CCCCTAACAAAAACATCACA | 60 | 34 | |
| PPIC | M | −162 | GTTTTTCGTATTCGTTTAAGGC | −33 | AAAATAAAAATCGAACAATCCG | 55 | 35 |
| U | −165 | GGTGTTTTTTGTATTTGTTTAAGGT | −57 | AAAAACAAAAACCCAAAACACA | 55 | 34 | |
| PYCARD | M | −186 | CGGGGAATCGCGGAGGTTTC | −36 | AATAAAACCCGAAAAAAAACCG | 55 | 35 |
| U | −190 | GGTTTGGGGAATTGTGGAGGTTTT | −13 | ATCACACCCTCCAACTAACCTACA | 55 | 35 | |
| RBP4 | M | −32 | TTCGGGTTTCGGTGAGTTAGGGC | 69 | CCGCTACTTTATAACGCCG | 58 | 34 |
| U | −33 | GTTTGGGTTTTGGTGAGTTAGGGT | 69 | ACCCCACTACTTTATAACACCA | 60 | 33 | |
| RGS2 | M | −184 | ACGTTAGTAGCGTTTCGGTTTC | −37 | GTCGCAACATTTATAAAACCTCG | 55 | 35 |
| U | −185 | GATGTTAGTAGTGTTTTGGTTTT | −37 | CATCACAACATTTATAAAACCTCA | 60 | 34 | |
| SCRN1 | M | −106 | GAGGGTGGGTTCGCGGTTAC | −14 | CTACAATAACGAAAACGACCG | 55 | 35 |
| U | −106 | GAGGGTGGGTTTGTGGTTATGT | −21 | CAATAACAAAAACAACCACCAAACA | 60 | 35 | |
| TBX3 | M | −98 | TTGGTTCGAAAGCGTTAAAGAG | −22 | ACCGAACGTCTACTCGACGACT | 53 | 35 |
| U | −110 | GTAGTAATATAATTGGTTTGAAAGT | −33 | CTACTCAACAACTCTAAAAAATCA | 55 | 35 | |
| TFAP2C | M | −146 | GCGTTGCGTTAGGTTCGGGTGC | 40 | CGCGAATATCAAAACCGCTCCG | 55 | 35 |
| U | −148 | TGGTGTTGTGTTAGGTTTGGGTGT | 40 | ACCACAAATATCAAAACCACTCCA | 60 | 35 | |
| ULBP2 | M | −213 | TGAGTTTGTCGTGGAAGGAATC | −89 | GTCAAACGAATCATAACGTCACG | 55 | 35 |
| U | −193 | TTGTGTTTTGGTAGGAGTTGGGT | −71 | ATCAAACAAATCATAACATCACA | 52 | 34 | |
| WIF1 | M | −131 | CGTTCGCGTTTTATTTTTTTGC | −27 | AACGCGTCGCCTCCCGACCTAA | 53 | 35 |
| U | −126 | GTGTTTTATTTTTTTGTGTGATTT | −21 | AACAACTAAACACATCACCTCCCA | 55 | 34 | |
| ZNF559 | M | −147 | GGTTCGGGAATTCGAGGTTTC | −43 | TACCTCAAACGCCAACGAAAACG | 58 | 34 |
| U | −149 | TGGGTTTGGGAATTTGAGGTTTT | −70 | CTATTAAAATAACAACCATTATACA | 52 | 34 | |
| ABHD9 | M | −195 | CGTGAGTTATCGTATTCGGTTC | −115 | TCCTATACGAAACTTAAAACCG | 59 | 33 |
| U | −197 | GGTGTGAGTTATTGTATTTGGTTT | −102 | ACAAAACCTAACAAATCCTATACA | 55 | 34 | |
| BMP7 | M | −227 | GTTTTTTCGTTGTTTTTTCGGC | −82 | ATACTAACCCCGAACCCCTCG | 55 | 34 |
| U | −231 | GTTTGTTTTTTTGTTGTTTTTTTGGT | −82 | AATACTAACCCCAAACCCCTCA | 59 | 34 | |
| CDH2 | M | −217 | GCGGTGTCGTTATATAGTAGC | −128 | ACTCTAAACCTACGCCGCCG | 50 | 34 |
| U | −294 | GTAAAATTATGAGTTTGAAATTTTGT | −114 | AAAAAAAAACATATAAACATCTACA | 55 | 34 | |
| CDKN2D | M | −122 | GCGGTGTCGTTATATAGTAGC | −15 | ACTCTAAACCTACGCCGCCG | 55 | 34 |
| U | −164 | GGTTTTGTGGGTGGAATGTT | −15 | CTACTCTAAACCTACACCACCA | 55 | 34 | |
| CLDN3 | M | −111 | AGGTTTTGGAGAGCGCGGTTTC | −47 | ACCCTAAACTAAAACCGATACG | 50 | 34 |
| U | −105 | TGGAGAGTGTGGTTTTGTTTTTATT | −47 | CTAACCCTAAACTAAAACCAATACA | 55 | 34 | |
| CTSL | M | −182 | GATTTTATTTTGCGTCGTTTC | −40 | ACGCTACGATTAACTATACCG | 55 | 34 |
| U | −186 | GTTTGATTTTATTTTGTGTTGTTTT | −40 | ACTACACTACAATTAACTATACCA | 48 | 34 | |
| FYN | M | −228 | TCGTACGTATTTTGGGATGTTC | −141 | CTACGAACCGCAACCATTAACG | 55 | 34 |
| U | −229 | ATTGTATGTATTTTGGGATGTTT | −129 | ACCCCTTAAAAACTACAAACCA | 55 | 34 | |
| GPR54 | M | −200 | TTATAAACGTTCGGTCGTAGC | −54 | CAAAATTACGCCCTAACACCG | 52 | 34 |
| U | −206 | TATGGGTTATAAATGTTTGGTT | −54 | CAAAATTACACCCTAACACCA | 58 | 34 | |
| IGFBP3 | M | −99 | TTTCGGTTTTTATATAGCGGTC | −37 | AAAAAACGACTAATCCTCAACG | 55 | 34 |
| U | −102 | TTATTTTGGTTTTTATATAGTGGTT | −37 | AACAAAAAACAACTAATCCTCAACA | 48 | 34 | |
| MTSS1 | M | −130 | GAGAGCGCGTTTTCGTTTGGC | −32 | CGCCTCCTTTTCACTCCTACG | 59 | 34 |
| U | −130 | GAGAGTGTGTTTTTGTTTGGT | −32 | CCACCTCCTTTTCACTCCTACA | 55 | 34 | |
| PAX6 | M | −188 | AGGGAGTATTTAATCGGTTGGC | −47 | CTCCTACGCCTAAACCAAAACG | 59 | 34 |
| U | −138 | GTAATATTTTGTGTGAGAGTGAGT | −47 | TCCTCCTACACCTAAACCAAAACA | 55 | 34 | |
| PLAU | M | −177 | TTTGTGAGCGTTGCGGAAGTAC | −51 | ACGATCTCCGCACTATACTACG | 55 | 34 |
| U | −153 | GGGGTTTGGGTTGTTGAGT | −51 | CTACAATCTCCACACTATACTACA | 50 | 34 | |
| RORA | M | −213 | GGTTGGAGAAGTTTTCGTTAGC | −111 | GACGAACGAACAAACAAAAACG | 55 | 34 |
| U | −215 | TTGGTTGGAGAAGTTTTTGTTAGT | −123 | CAAACAAAAACACAAAAAAACACA | 55 | 34 | |
| SNAI1 | M | −155 | ATTTGTTCGGGGAGTGGTTTTC | −91 | AAAACGAAACCTTATCTACCACG | 55 | 34 |
| U | −213 | GGAGTTTTTGTTTGGGTTTTTATT | −91 | AAAAACAAAACCTTATCTACCACA | 55 | 34 | |
| TNFSF9 | M | −198 | GTCGAGTTTGGAAGGTCGGAAAC | −65 | AAAAAACCACGCCCCTCCG | 56 | 34 |
| U | −199 | GGTTGGAAATGGAAAGGAGAGT | −65 | AAAAAAAACCACACCCCTCCA | 55 | 34 | |
| ZNF177 | M | −119 | GTAGGAGTATTTGCGATGTTTC | −12 | AAAATAACGAAACGACGAACG | 58 | 34 |
| U | −97 | GTTTTTAAGTTTTTAGGGTGAATTT | −22 | AAACAACAAACACCCACTTCCA | 55 | 34 | |
Transcription start site = 0. All primers were designed on the top strand sequences. M, specific to methylated DNA; U, specific to unmethylated DNA.
Results
Oligonucleotide microarray analysis
AGS cells were treated with 1 µM of 5‐aza‐dC, which caused growth suppression at 49%, and upregulated genes were searched for using an oligonucleotide microarray. Among the 39 000 genes (54 000 probe sets) analyzed, 1430 genes (1747 probes) were upregulated four‐fold or more (signal log ratio > 2) and 579 genes (678 probes) were upregulated 16‐fold or more (signal log ratio > 4). To identify silenced genes with known functions from the 579 genes, we excluded genes on chromosome X (95 probes, 70 genes) and genes without known functions (i.e. FLJ genes, KIAA genes, LOC genes, MG genes and Orf genes [149 probes, 141 genes]).
Among the remaining 368 genes (434 probes), we found eight genes (14 probes) whose methylation‐silencing had been reported in gastric cancers (BNIP3,( 13 ) CDKN2A (p16),( 14 ) CHFR,( 15 ) ID4,( 16 ) RBP1,( 17 ) RUNX3,( 18 ) THBD,( 10 ) TIMP ( 19 )). The remaining 360 genes (420 probes) were considered as candidates for novel methylation‐silenced genes in gastric cancers (Table 1).
Methylation analysis of genes upregulated by 5‐aza‐dC treatment
From the 360 genes upregulated 16‐fold or more, we selected 44 genes randomly (Table 2). Among these 44 genes, 32 genes (73%) had CGI in their 5′ regions, which were considered as promoter regions (Table 2). To examine whether the induction of these genes by 5‐aza‐dC treatment was really due to promoter demethylation, the methylation status of these 5′‐CGIs were analyzed by MSP. For all the 32 genes, only methylated molecules were detected before 5‐aza‐dC treatment, and unmethylated DNA molecules were detected after the treatment in AGS, suggesting silencing of the 32 genes by methylation of their 5′‐CpG islands (representative results in Fig. 1).
Table 2.
Genes upregulated after 5‐aza‐dC treatment in the AGS cell line
| Probe set | Gene title | Symbol | Fold change | CGI |
|---|---|---|---|---|
| The 44 genes picked randomly from the genes showing greater than 16‐fold upregulation after 5‐aza‐dC treatment | ||||
| 209122_at | Adipose differentiation‐related protein | ADFP | 36.0 | Yes |
| 203180_at | Aldehyde dehydrogenase 1 family, member A3 | ALDH1A3 | 46.2 | Yes |
| 200782_at | Annexin A5 | ANXA5 | 32.5 | Yes |
| 205239_at | Amphiregulin (schwannoma‐derived growth factor) | AREG | 59.3 | Yes |
| 206382_s_at | Brain‐derived neurotrophic factor | BDNF | 29.2 | Yes |
| 203065_s_at | Caveolin 1, caveolae protein, 22 kDa | CAV1 | 37.2 | Yes |
| 210140_at | Cystatin F (leukocystatin) | CST7 | 22.1 | No |
| 219424_at | Epstein–Barr virus induced gene 3 | EBI3 | 18.5 | No |
| 204540_at | Eukaryotic translation elongation factor 1 alpha 2 | EEF1A2 | 25.0 | Yes |
| 203989_x_at | Coagulation factor II (thrombin) receptor | F2R | 59.3 | Yes |
| 208962_s_at | Fatty acid desaturase 1 | FADS1 | 23.0 | Yes |
| 203240_at | Fc fragment of IgG binding protein | FCGBP | 16.8 | No |
| 1570515_a_at | Filamin A interacting protein 1 | FILIP1 | 34.8 | No |
| 219170_at | Fibronectin type 3 and SPRY domain containing 1 | FSD1 | 16.0 | Yes |
| 226847_at | Follistatin | FST | 29.2 | Yes |
| 218469_at | Gremlin 1 homolog, cysteine knot superfamily | GREM1 | 59.3 | Yes |
| 213620_s_at | Intercellular adhesion molecule 2 | ICAM2 | 18.5 | No |
| 201162_at | Insulin‐like growth factor binding protein 7 | IGFBP7 | 38.4 | Yes |
| 205945_at | Interleukin 6 receptor | IL6R | 26.0 | Yes |
| 209185_s_at | Insulin receptor substrate 2 | IRS2 | 24.0 | Yes |
| 205563_at | KiSS‐1 metastasis‐suppressor | KISS1 | 54.8 | Yes |
| 221047_s_at | MAP/microtubule affinity‐regulating kinase 1 | MARK1 | 17.6 | Yes |
| 1552456_a_at | Methyl‐CpG binding domain protein 3‐like 2 | MBD3L2 | 38.4 | No |
| 206560_s_at | Melanoma inhibitory activity | MIA | 23.0 | No |
| 204784_s_at | Myeloid leukemia factor 1 | MLF1 | 27.0 | Yes |
| 205932_s_at | Msh homeo box homolog 1 (Drosophila) | MSX1 | 16.0 | Yes |
| 202086_at | Myxovirus (influenza virus) resistance 1 | MX1 | 18.5 | Yes |
| 205581_s_at | Nitric oxide synthase 3 (endothelial cell) | NOS3 | 18.5 | No |
| 203939_at | 5′‐nucleotidase, ecto (CD73) | NT5E | 53.3 | Yes |
| 207002_s_at | Pleiomorphic adenoma gene‐like 1 | PLAGL1 | 46.2 | Yes |
| 204517_at | Peptidylprolyl isomerase C (cyclophilin C) | PPIC | 23.0 | Yes |
| 221666_s_at | PYD and CARD domain containing | PYCARD | 25.0 | Yes |
| 219140_s_at | Retinol binding protein 4, plasma | RBP4 | 43.6 | Yes |
| 202388_at | Regulator of G‐protein signaling 2, 24 kDa | RGS2 | 33.6 | Yes |
| 201462_at | Secernin 1 | SCRN1 | 31.4 | Yes |
| 204614_at | Serine (or cysteine) proteinase inhibitor, clade B, member 2 | SERPINB2 | 36.0 | No |
| 202627_s_at | Serine (or cysteine) proteinase inhibitor, clade E, member 1 | SERPINE1 | 26.0 | No |
| 208539_x_at | Small proline‐rich protein 2 A | SPRR2A | 22.1 | No |
| 224167_at | Likely ortholog of mouse spermatogenic Zip 1 | SPZ1 | 18.5 | No |
| 219682_s_at | T‐box 3 | TBX3 | 36.0 | Yes |
| 205286_at | Transcription factor AP‐2 gamma | TFAP2C | 32.5 | Yes |
| 221291_at | UL16 binding protein 2 | ULBP2 | 29.2 | Yes |
| 204712_at | WNT inhibitory factor 1 | WIF1 | 31.4 | Yes |
| 224518_s_at | Zinc finger protein 559 | ZNF559 | 30.3 | Yes |
| Genes showing greater than four‐fold upregulation after 5‐aza‐dC treatment, having CpG islands, and having cancer related function or having chromosomal location in the region of frequent loss in gastric cancer. | ||||
| 220013_at | Abhydrolase domain containing 9 | ABHD9 | 8.0 | Yes |
| 209591_s_at | Bone morphogenetic protein 7 (osteogenic protein 1) | BMP7 | 13.9 | Yes |
| 203440_at | Cadherin 2, type 1, N‐cadherin (neuronal) | CDH2 | 26.0 | Yes |
| 210240_s_at | Cyclin‐dependent kinase inhibitor 2D (p19) | CDKN2D | 4.4 | Yes |
| 203953_s_at | Claudin 3 | CLDN3 | 45.3 | Yes |
| 202087_s_at | Cathepsin L | CTSL | 13.0 | Yes |
| 216033_s_at | FYN oncogene related to SRC, FGR, YES | FYN | 78.8 | Yes |
| 242517_at | G protein‐coupled receptor 54 | GPR54 | 6.3 | Yes |
| 210095_s_at | Insulin‐like growth factor binding protein 3 | IGFBP3 | 9.8 | Yes |
| 203037_s_at | Metastasis suppressor 1 | MTSS1 | 6.5 | Yes |
| 205646_s_at | Paired box gene 6 (aniridia, keratitis) | PAX6 | 18.4 | Yes |
| 205479_s_at | Plasminogen activator, urokinase | PLAU | 137.2 | Yes |
| 210479_s_at | RAR‐related orphan receptor A | RORA | 55.7 | Yes |
| 219480_at | Snail homolog 1 (Drosophila) | SNAI1 | 4.6 | Yes |
| 206907_at | Tumor necrosis factor superfamily, member 9 | TNFSF9 | 4.8 | Yes |
| 207417_s_at | Zinc finger protein 177 | ZNF177 | 4.4 | Yes |
| Genes reported as silenced genes in gastric cancer and showing greater than 16‐fold upregulation after 5‐aza‐dC treatment | ||||
| 201848_s_at | BCL2/adenovirus E1B 19 kDa interacting protein 3 | BNIP3 | 20.3 | Yes |
| 207039_at | Cyclin‐dependent kinase inhibitor 2 A (p16) | CDKN2A | 33.6 | Yes |
| 223931_s_at | Checkpoint with forkhead and ring finger domains | CHFR | 16.8 | Yes |
| 209291_at | Inhibitor of DNA binding 4 | ID4 | 57.8 | Yes |
| 203423_at | Retinol binding protein 1, cellular | RBP1 | 54.8 | Yes |
| 204198_s_at | Runt‐related transcription factor 3 | RUNX3 | 27.0 | Yes |
| 203888_at | Thrombomodulin | THBD | 29.2 | Yes |
| 201147_s_at | Tissue inhibitor of metalloproteinase 3 | TIMP3 | 65.6 | Yes |
Figure 1.
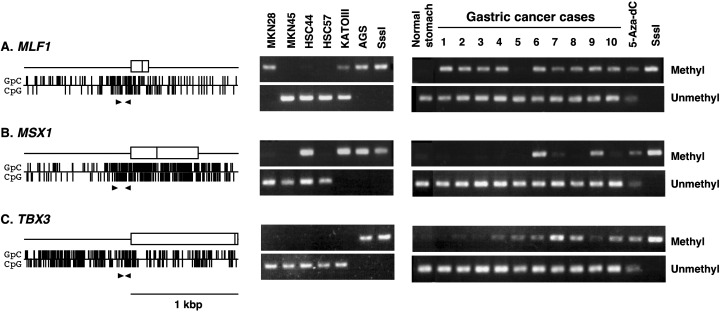
A representative result of methylation analysis. (A) MLF1; (B) MSX1; and (C) TBX3. The left sides of each panel represent the 5′ CpG islands and regions analyzed by methylation‐specific polymerase chain reaction (MSP). Vertical marks, individual GpC and CpG sites; Open boxes, non‐coding and coding exons; and arrowheads, positions of MSP primers (M sets). The right sides show the results of MSP in gastric cancer cell lines, normal gastric mucosa and primary gastric cancers. 5‐aza‐dC, AGS cells after treatment with 5‐aza‐2′‐deoxycytidine; SssI, genomic DNA methylated with SssI methylase.
Analysis of five additional gastric cancer cell lines (MKN28, MKN45, HSC44, HSC57, KATOIII) showed that five genes (ANXA5, AREG, CAV1, IL6R, TBX3) were methylated only in AGS, and 27 genes were methylated in multiple gastric cancer cell lines (Table 3). The microarray analysis of KATOIII and HSC57 showed that none of the 32 genes were expressed when unmethylated DNA molecules were not present.
Table 3.
Methylation profiles in gastric cancer
| Gene Symbol | Function | Chromo‐ somal location | Methylation status in gastric cancer cell line † | Expression (signal intensity of GeneChip) | Methylation status in gastric cancer ‡ | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MKN28 | MKN45 | HSC44 | HSC57 | KATOIII | AGS | HSC57 | KatoIII | +5‐aza‐dC | Stomach § | Case1 | Case2 | Case3 | Case4 | Case5 | Case6 | Case7 | Case8 | Case9 | Case10 | Normal | ||||
| AGS | AGS | (RefEXA) | tub1 | tub2 | por2 | por | tub2 | por1 | por2 | por | por2 | por2 | ||||||||||||
| 32 genes selected randomly and with GGI | ||||||||||||||||||||||||
| ADFP | Fatty acid transport | 9p22.1 | M | U | M/U | U | M | M | 2191 | 74 | 33 | 1186 | 154 | U | U | U | U | U | M | U | U | U | U | – |
| ALDH1A3 | Vitamin A metabolism | 15q26.3 | U | U | U | U | M | M | 322 | 69 | 16 | 1435 | 65 | U | U | U | U | U | M | M | U | M– | U | – |
| ANXA5 | Apoptosis | 4q28‐q32 | U | U | U | U | U | M | 1740 | 4933 | 144 | 6960 | 433 | U | U | U | U | U | M | U | U | U | U | – |
| AREG | Cell proliferation | 4q13‐q21 | U | U | U | U | U | M | 9568 | 9103 | 7 | 2677 | 137 | U | U | U | U | U | M | U | U | U | U | – |
| BDNF | Growth factor activity | 11p13 | M | M | M | M/U | U | M | 50 | 20 | 5 | 447 | 32 | M | U | M | M | U | M | M | M | M | M | – |
| CAV1 | Cell aging | 7q31.1 | U | U | U | U | U | M | 24 | 167 | 20 | 1411 | 102 | U | U | U | U | U | M | U | U | U | U | – |
| EEF1A2 | Translational elongation | 20q13.3 | U | U | M | U | M | M | 240 | 60 | 112 | 7234 | 58 | U | U | U | U | U | M | U | U | U | U | – |
| F2R | G‐protein signaling | 5q13 | U | U | M/U | U | M | M | 140 | 7 | 4 | 820 | 26 | U | U | U | U | U | M | M | U | U | U | – |
| FADS1 | Fatty acid desaturation | 11q12.2‐q13.1 | U | U | M/U | U | M | M | 375 | 19 | 22 | 542 | 40 | U | U | U | U | U | M | M– | M | M | U | – |
| FSD1 | Microtubule depolymerization | 19p13.3 | U | M/U | M/U | U | M | M | 49 | 25 | 31 | 362 | 5 | U | U | U | U | U | M | U | U | M | U | U |
| FST | Transcription factor | 5q11.2 | U | U | M | U | M | M | 6 | 4 | 7 | 278 | – | U | U | U | U | U | M | M | U | M | U | – |
| GREM1 | Signal transduction | 15q13–15q15 | M | M | M | U | M | M | 77 | 33 | 5 | 2128 | 184 | M | M | M | M | M | M | M | M | M | M | – |
| IGFBP7 | Angiogenesis | 4q12 | M | U | M | U | M | M | 13 | 11 | 9 | 934 | 483 | U | U | M– | U | U | M | M | U | M | M– | – |
| IL6R | Immune response | 1q21 | U | U | U | U | U | M | 124 | 227 | 12 | 574 | 72 | U | U | U | U | U | U | U | U | U | U | – |
| IRS2 | Signal transduction | 13q34 | U | U | U | U | M | M | 6602 | 57 | 30 | 1245 | 274 | U | U | U | U | U | U | U | U | M– | U | – |
| KISS1 | Metastasis suppressor | 1q32 | M/U | M | M | U | U | M | 19 | 26 | 21 | 3697 | 7 | U | M– | M– | M– | U | M– | M– | M– | M– | M– | U |
| MARK1 | Protein amino acid phosphorylation | 1q41 | U | U | U | U | M | M | 79 | 85 | 45 | 845 | 43 | U | U | U | U | U | M | U | M | U | U | – |
| MLF1 | Cell differentiation | 3q25.1 | M | U | U | U | M/U | M | 936 | 1078 | 22 | 709 | 42 | M | M | M | M | U | M | M | M | M | M | U |
| MSX1 | Transcription factor | 4p16.3‐p16.1 | U | U | M/U | U | M | M | 524 | 13 | 43 | 2675 | 32 | U | U | U | U | U | M | M– | U | M | U | U |
| MX1 | Response to virus | 21q22.3 | M | U | M | U | U | M | 856 | 1533 | 48 | 996 | 223 | U | U | U | U | U | U | U | U | U | U | – |
| NT5E | DNA metabolism | 6q14–6q21 | U | U | U | U | M/U | M | 115 | 394 | 4 | 821 | 31 | U | U | U | U | U | U | M– | U | U | U | – |
| PLAGL1 | Induction of apoptosis | 6q24–6q25 | M | M | M | M/U | M | M | 456 | 16 | 4 | 668 | 81 | M– | M | M | M | M | M | M | M | M | M | U |
| PPIC | Signal transduction | 5q23.2 | U | U | U | U | M | M | 2724 | 59 | 49 | 1621 | 186 | U | U | U | U | U | M | M– | U | U | U | – |
| PYCARD | Signal transduction | 16p12–16p11.2 | M | M | M | U | U | M | 2739 | 557 | 9 | 420 | 39 | U | U | U | U | U | U | U | U | U | U | – |
| RBP4 | Vitamin A metabolism | 10q23–10q24 | M | U | M | U | U | M | 86 | 643 | 10 | 880 | 11 | U | U | U | M– | M– | M | M | M– | U | M– | U |
| RGS2 | G‐protein signaling | 1q31 | U | U | U | U | M/U | M/U | 27 | 90 | 27 | 1727 | 575 | U | U | U | U | U | U | U | U | U | U | – |
| SCRN1 | Exocytosis | 7p14.3‐p14.1 | M/U | U | U | U | M | M | 1595 | 57 | 19 | 1306 | 127 | U | U | U | U | U | M | U | U | M | U | – |
| TBX3 | Morphogenesis | 12q24.1 | U | U | U | U | U | M | 1280 | 881 | 15 | 1714 | 38 | U | U | U | M | M | M | M | M | M– | M | U |
| TFAP2C | Transcription factor | 20q13.2 | U | U | M/U | U | M | M | 1486 | 26 | 27 | 2446 | 51 | U | M– | M | U | U | M | M | U | M | U | – |
| ULBP2 | T cell proliferation | 6q25 | U | U | U | U | M/U | M | 365 | 510 | 29 | 1286 | 8 | U | U | U | U | U | M | U | U | U | U | U |
| WIF1 | Wnt signaling | 12q14.3 | M | M | M | M/U | M | M | 9 | 41 | 7 | 241 | 2 | U | U | M | M | M– | M | M | M | M | M– | U |
| ZNF559 | Transcription factor | 19p13.2 | M | U | M/U | U | M | M | 609 | 7 | 7 | 532 | – | M– | M | M– | M– | U | U | U | M | M– | M– | U |
| 16 genes with potential tumor‐related functions | ||||||||||||||||||||||||
| ABHD9 | Response to chemical substance | 19p13.12 | M | M | M | U | M | M | 19 | 18 | 13 | 140 | 3 | U | M | U | M– | M | M | M | M | M | M– | – |
| BMP7 | Cell proliferation | 20q13 | U | U | M | U | M/U | M | 4166 | 19 | 61 | 1185 | 68 | U | U | U | M– | U | M | M | M– | M | M– | – |
| CDH2 | Cell adhesion | 18q11.2 | M | M | M | U | M | M | 13 | 13 | 9 | 487 | 57 | M | U | M | M | M– | M | M | M | M | M | – |
| CDKN2D | Cdk inhibitor | 19p13 | U | U | U | U | U | U | 47 | 339 | 119 | 489 | 28 | U | U | U | U | U | U | U | U | U | U | – |
| CLDN3 | Cell adhesion | 7q11.23 | U | U | M/U | U | U | M | 7584 | 69 | 7 | 359 | 1 | U | U | U | U | U | M | M | M– | M | U | U |
| CTSL | Protein processing | 9q21–9q22 | U | U | M | U | M/U | M | 1017 | 102 | 50 | 1753 | 118 | U | U | U | U | M– | M | M | M | U | M | – |
| FYN | Proto‐oncogene | 6q21 | U | U | U | U | U | M | 341 | 1046 | 5 | 797 | 64 | U | U | U | U | U | M | U | U | U | U | – |
| GPR54 | G‐protein signaling | 19p13.3 | U | U | M | U | M | M | 89 | 85 | 35 | 210 | – | U | U | U | U | U | M | U | U | U | U | – |
| IGFBP3 | Induction of apoptosis | 7p13–7p12 | M/U | M/U | M/U | U | M | M | 106 | 15 | 31 | 277 | 422 | M | U | M | M | M– | M | M | M | M | M | – |
| MTSS1 | Cytoskeletal organization | 8p22 | M/U | U | M | U | U | M | 17 | 694 | 73 | 625 | 400 | M | U | U | U | U | M | U | M | U | U | – |
| PAX6 | Transcription factor | 11p13 | M | U | M/U | U | M | M | 17 | 21 | 29 | 417 | 6 | U | U | M | M | U | M | M | M | M | M– | U |
| PLAU | Angiogenesis | 10q24 | M | U | U | U | M | M | 1060 | 168 | 11 | 2950 | 80 | U | U | U | U | U | M– | U | U | U | U | – |
| RORA | Signal transduction | 15q21‐q22 | U | U | M | U | M | M | 195 | 49 | 2 | 340 | 20 | M | M | U | U | M | M– | M | M | M | M | U |
| SNAI1 | Transcriptional repressor | 20q13.1‐q13.2 | U | U | U | U | U | U | 235 | 40 | 52 | 319 | 8 | U | U | U | U | U | U | U | U | U | U | – |
| TNFSF9 | Apoptosis | 19p13.3 | U | U | U | U | U | M | 615 | 1133 | 84 | 916 | 23 | U | U | U | U | U | M | U | U | U | U | – |
| ZNF177 | Transcription factor | 19p13.2 | M | M | M | M | U | M | 76 | 59 | 47 | 222 | 14 | M | M | M | M | M | M | M | M | M | M | U |
From genes upregulated 16‐fold or more, 44 genes were selected randomly, and the methylation status of 32 genes with CGI were analyzed. In addition, 16 genes with potential tumor‐related functions and CGI were selected for methylation analysis. †In cancer cell lines, M: only methylated molecules detected, U: only unmethylated molecules detected, M/U: both methylated and unmethylated molecules detected. ‡In primary cancer samples, ‘M’ and ‘U’ indicate detection and absence, respectively, of methylated molecules. ‘M–’ indicates slight detection of methylated molecules. §Obtained from the RefEXA database. Underlined chromosomal locations, regions of frequent loss in gastric cancer.
We next selected 16 potential tumor‐related genes with promoter CGI and four‐fold or greater upregulation as the above analysis suggested that a considerable number of silenced genes were still present among the genes with upregulation of 16‐fold or less (Table 2). The potential tumor‐related genes were selected based on their tumor‐related function and location in genomic regions with frequent LOH (5q21‐23,( 20 , 21 ) 8p22,( 20 , 21 ) 9p12‐24( 20 , 21 , 22 )) or with DNA loss by comparative genomic hybridization (19p13.12‐p13.3( 23 )) in gastric cancers. MSP showed that 14 of these 16 genes were methylated in AGS before 5‐aza‐dC treatment (Table 3). CDKN2D and SNAI1 were not methylated even before 5‐aza‐dC treatment, suggesting that they were induced as a stress response by 5‐aza‐dC treatment.
Presence of methylation in primary gastric cancers
The methylation status of the above 48 genes (32 selected randomly and 16 tumor‐related genes) were examined in 10 primary gastric cancers. It was shown that 42 genes (ABHD9, ADFP, ALDH1A3, ANXA5, AREG, BDNF, BMP7, CAV1, CDH2, CLDN3, CTSL, EEF1A2, F2R, FADS1, FSD1, FST, FYN, GPR54, GREM1, IGFBP3, IGFBP7, IRS2, KISS1, MARK1, MLF1, MSX1, MTSS1, NT5E, PAX6, PLAGL1, PLAU, PPIC, RBP4, RORA, SCRN1, TBX3, TFAP2C, TNFSF9, ULBP2, WIF1, ZNF177 and ZNF559) were methylated in at least one gastric cancer (Table 3). The numbers of methylated genes in each case ranged from one to 10. Case 6 had a large number of methylated genes, which was similar to AGS (Table 3). The expression levels of the 48 genes in the normal gastric mucosae were obtained from the RefEXA database (Table 3).
There remained a possibility that these silenced genes were normally methylated or were methylated tissue‐specifically. Therefore, we selected 11 genes with relatively high chances of having methylated CGI, based on their low expression in the normal gastric mucosae (CLDN3, FADS1, KISS1, PAX6, PLAGL1, RBP4, RORA, ULBP2, WIF1, ZNF177 and ZNF559). Along with three additional genes (MLF1, MSX1 and TBX3), their methylation status was examined in the normal gastric mucosae. However, none of the 14 genes were methylated.
Discussion
Chemical genomic screening revealed that a considerable number of genes were methylation‐silenced in the AGS gastric cancer cell line. After 5‐aza‐dC treatment of AGS, 579 genes were upregulated 16‐fold or more. When we analyzed 44 selected genes, 32 of them had CGI in their promoter regions, and all of the 32 genes turned out to be methylation‐silenced. Because 32 of the 44 genes selected from 579 genes were silenced, it was estimated that 421 ± 75 (95% confidence interval) genes were silenced in AGS. To avoid overestimation, we randomly selected 44 genes from the 360 genes after excluding: (i) genes on chromosome X, which harbors many normally methylated genes like MAGE; (ii) genes that have not been characterized yet; and (iii) genes whose methylation‐associated silencing was already known in gastric cancers. Among the 16 potential tumor‐related genes, 10 were upregulated 16‐fold or less, and eight of the 10 genes were found to be methylation‐silenced. If genes with relatively small upregulation were analyzed, the number of silenced genes in AGS was expected to be larger.
As for the number of methylation‐silenced genes in a cancer, Costello et al. estimated that an average of 600 CGI in the whole genome were methylated aberrantly in the tumors.( 24 ) However, the number was calculated by analyzing CGI in any location against a gene, and the number of genes silenced, for which methylation of promoter CGI is necessary, was not determined. Using chemical genomic screening, Sato et al. estimated that an average of 140 genes would be methylated aberrantly in pancreatic cancers.( 6 ) Compared with this number, the number of genes silenced in the AGS cell line was considered to be much larger. We recently found that AGS had an increased rate of de novo methylation,( 25 ) and this could be one of the mechanisms.
By methylation analysis of 48 genes (Table 3), 46 genes were found to be methylated in AGS, and 42 genes were methylated in at least one primary gastric cancer. Among the 42 genes, eight genes (CAV1,( 26 , 27 ) IGFBP3,( 28 ) IGFBP7[MAC25/IGFBP‐rP1],( 29 ) PAX6,( 30 ) PLAGL1[ZAC/LOT1],( 31 , 32 ) PLAU[uPA],( 33 , 34 ) RBP4 ( 35 ) and WIF1 ( 36 )) were reported to be silenced with functional relevance in cancers other than gastric cancers. In addition, two genes (CDH2 ( 37 ) and FYN ( 38 )) were reported to be methylated in some cancers, but their functional significance needs clarification.
Also among the 32 genes whose silencing was novel, we were able to find potential tumor‐related genes. To achieve this, some genes were selected based on (i) antioncogenic cellular functions or (ii) location in genomic regions with frequent LOH in gastric cancers. Candidate tumor‐related genes were further selected based on (iii) the presence of methylation of promoter CGI in primary gastric cancers, and (iv) expression in normal gastric mucosae when various tissues were compared. MTSS1/MIM/BEG4 met all of these criteria, and was a good candidate for a novel tumor‐related gene. It mediates Sonic hedgehog signaling by potentiating Gli‐dependent transcription,( 39 ) and is known as a metastasis suppressor gene in bladder cancers.( 40 ) Although LOH was not frequent in their locations, ANXA5, AREG, GREM, IGFBP7, IRS2, BMP7, CTSL and IGFBP3 were expressed in the normal gastric mucosae and had potential antioncogenic functions, such as mediation of SMAD signaling (BMP7)( 41 ) and induction of apoptosis (IGFBP3).( 42 ) There is a possibility that silencing of these genes is causally related to development and progression of gastric cancers. However, considering the large number of methylation‐silenced genes, it was likely that the majority of the genes silenced in AGS did not have causal roles in gastric carcinogenesis.
In summary, we found a considerable number of methylation‐silenced genes in a gastric cancer cell line AGS. Potential tumor‐related genes were selected based on their known functions, chromosomal locations, methylation in primary samples and expression in normal gastric mucosae. The usefulness of chemical genomic screening was confirmed.
Acknowledgments
This work was supported by a Grant‐in‐Aid on Priority Area from the Ministry of Education, Sciences, Culture and Sports (MEXT), Japan.
References
- 1. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3: 415–28. [DOI] [PubMed] [Google Scholar]
- 2. Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer 2005; 5: 223–31. [DOI] [PubMed] [Google Scholar]
- 3. Suzuki H, Gabrielson E, Chen W et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet 2002; 31: 141–19. [DOI] [PubMed] [Google Scholar]
- 4. Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5‐aza‐2′‐deoxycytidine. Cancer Res 2002; 62: 961–6. [PubMed] [Google Scholar]
- 5. Yamashita K, Upadhyay S, Osada M et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell 2002; 2: 485–95. [DOI] [PubMed] [Google Scholar]
- 6. Sato N, Fukushima N, Maitra A et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high‐throughput microarrays. Cancer Res 2003; 63: 3735–42. [PubMed] [Google Scholar]
- 7. Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res 2005; 65: 4218–27. [DOI] [PubMed] [Google Scholar]
- 8. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 9. Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell 2004; 5: 121–5. [DOI] [PubMed] [Google Scholar]
- 10. Kaneda A, Kaminishi M, Yanagihara K, Sugimura T, Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res 2002; 62: 6645–50. [PubMed] [Google Scholar]
- 11. Kaneda A, Wakazono K, Tsukamoto T et al. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res 2004; 64: 6410–15. [DOI] [PubMed] [Google Scholar]
- 12. Ge X, Yamamoto S, Tsutsumi S et al. Interpreting expression profiles of cancers by genome‐wide survey of breadth of expression in normal tissues. Genomics 2005; 86: 127–41. [DOI] [PubMed] [Google Scholar]
- 13. Murai M, Toyota M, Suzuki H et al. Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin Cancer Res 2005; 11: 1021–7. [PubMed] [Google Scholar]
- 14. Lee YY, Kang SH, Seo JY et al. Alterations of p16INK4A and p15INK4B genes in gastric carcinomas. Cancer 1997; 80: 1889–96. [DOI] [PubMed] [Google Scholar]
- 15. Satoh A, Toyota M, Itoh F et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res 2003; 63: 8606–13. [PubMed] [Google Scholar]
- 16. Chan AS, Tsui WY, Chen X et al. Downregulation of ID4 by promoter hypermethylation in gastric adenocarcinoma. Oncogene 2003; 22: 6946–53. [DOI] [PubMed] [Google Scholar]
- 17. Esteller M, Guo M, Moreno V et al. Hypermethylation‐associated inactivation of the cellular retinol‐binding‐protein 1 gene in human cancer. Cancer Res 2002; 62: 5902–5. [PubMed] [Google Scholar]
- 18. Li QL, Ito K, Sakakura C et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002; 109: 113–24. [DOI] [PubMed] [Google Scholar]
- 19. Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along the multistep pathway of gastric carcinogenesis. Lab Invest 2003; 83: 635–41. [DOI] [PubMed] [Google Scholar]
- 20. Yustein AS, Harper JC, Petroni GR, Cummings OW, Moskaluk CA, Powell SM. Allelotype of gastric adenocarcinoma. Cancer Res 1999; 59: 1437–41. [PubMed] [Google Scholar]
- 21. Kim KM, Kwon MS, Hong SJ et al. Genetic classification of intestinal‐type and diffuse‐type gastric cancers based on chromosomal loss and microsatellite instability. Virchows Arch 2003; 443: 491–500. [DOI] [PubMed] [Google Scholar]
- 22. Choi SW, Park SW, Lee KY, Kim KM, Chung YJ, Rhyu MG. Fractional allelic loss in gastric carcinoma correlates with growth patterns. Oncogene 1998; 17: 2655–9. [DOI] [PubMed] [Google Scholar]
- 23. Weiss MM, Kuipers EJ, Postma C et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol 2004; 26: 307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Costello JF, Fruhwald MC, Smiraglia DJ et al. Aberrant CpG‐island methylation has non‐random and tumour‐type‐specific patterns. Nat Genet 2000; 24: 132–8. [DOI] [PubMed] [Google Scholar]
- 25. Ushijima T, Watanabe N, Shimizu K, Miyamoto K, Sugimura T, Kaneda A. Decreased fidelity in replicating CpG methylation patterns in cancer cells. Cancer Res 2005; 65: 11–17. [PubMed] [Google Scholar]
- 26. Sunaga N, Miyajima K, Suzuki M et al. Different roles for caveolin‐1 in the development of non‐small cell lung cancer versus small cell lung cancer. Cancer Res 2004; 64: 4277–85. [DOI] [PubMed] [Google Scholar]
- 27. Wiechen K, Diatchenko L, Agoulnik A et al. Caveolin‐1 is down‐regulated in human ovarian carcinoma and acts as a candidate tumor suppressor gene. Am J Pathol 2001; 159: 1635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hanafusa T, Yumoto Y, Nouso K et al. Reduced expression of insulin‐like growth factor binding protein‐3 and its promoter hypermethylation in human hepatocellular carcinoma. Cancer Lett 2002; 176: 149–58. [DOI] [PubMed] [Google Scholar]
- 29. Komatsu S, Okazaki Y, Tateno M et al. Methylation and downregulated expression of mac25/insulin‐like growth factor binding protein‐7 is associated with liver tumorigenesis in SV40T/t antigen transgenic mice, screened by restriction landmark genomic scanning for methylation (RLGS‐M). Biochem Biophys Res Commun 2000; 267: 109–17. [DOI] [PubMed] [Google Scholar]
- 30. Toyota M, Ho C, Ahuja N et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res 1999; 59: 2307–12. [PubMed] [Google Scholar]
- 31. Abdollahi A, Pisarcik D, Roberts D, Weinstein J, Cairns P, Hamilton TC. LOT1 (PLAGL1/ZAC1), the candidate tumor suppressor gene at chromosome 6q24‐25 is epigenetically regulated in cancer. J Biol Chem 2003; 278: 6041–9. [DOI] [PubMed] [Google Scholar]
- 32. Bilanges B, Varrault A, Basyuk E et al. Loss of expression of the candidate tumor suppressor gene ZAC in breast cancer cell lines and primary tumors. Oncogene 1999; 18: 3979–88. [DOI] [PubMed] [Google Scholar]
- 33. Xing RH, Rabbani SA. Transcriptional regulation of urokinase (uPA) gene expression in breast cancer cells: role of DNA methylation. Int J Cancer 1999; 81: 443–50. [DOI] [PubMed] [Google Scholar]
- 34. Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo . Faseb J 2003; 17: 1081–8. [DOI] [PubMed] [Google Scholar]
- 35. Kwong J, Lo KW, Shuk‐Nga Chow L et al. Epigenetic silencing of cellular retinol‐binding proteins in nasopharyngeal carcinoma. Neoplasia 2005; 7: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mazieres J, He B, You L et al. Wnt inhibitory factor‐1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res 2004; 64: 4717–20. [DOI] [PubMed] [Google Scholar]
- 37. Hagihara A, Miyamoto K, Furuta J et al. Identification of 27 5′ CpG islands aberrantly methylated and 13 genes silenced in human pancreatic cancers. Oncogene 2004; 23: 8705–10. [DOI] [PubMed] [Google Scholar]
- 38. Wang W, Huper G, Guo Y, Murphy SK, Olson JA Jr, Marks JR. Analysis of methylation‐sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene 2005; 24: 2705–14. [DOI] [PubMed] [Google Scholar]
- 39. Callahan CA, Ofstad T, Horng L et al. MIM/BEG4, a sonic hedgehog‐responsive gene that potentiates Gli‐dependent transcription. Genes Dev 2004; 18: 2724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee YG, Macoska JA, Korenchuk S, Pienta KJ. MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia 2002; 4: 291–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang S, Zhong C, Frenkel B, Reddi AH, Roy‐Burman P. Diverse biological effect and Smad signaling of bone morphogenetic protein 7 in prostate tumor cells. Cancer Res 2005; 65: 5769–77. [DOI] [PubMed] [Google Scholar]
- 42. Butt AJ, Firth SM, King MA, Baxter RC. Insulin‐like growth factor‐binding protein‐3 modulates expression of Bax and Bcl‐2 and potentiates p53‐independent radiation‐induced apoptosis in human breast cancer cells. J Biol Chem 2000; 275: 39 174–81. [DOI] [PubMed] [Google Scholar]


