Abstract
Genetic alterations in cancer can provide information for predicting a tumor's sensitivity to chemotherapeutic drugs. But although such information is certainly useful, the relatively low frequency of mutations seen in many cancers limits the utility of pharmacogenomics in large numbers of cancer patients, necessitating consideration of other approaches. Epigenetic changes such as DNA methylation are a hallmark of human cancers. Methylation of genes involved in DNA repair and maintaining genome integrity (e.g. MGMT, hMLH1, WRN, and FANCF), and cell‐cycle checkpoint genes (e.g. CHFR and 14‐3‐3 σ, CDK10, and p73), all reportedly influence the sensitivity to chemotherapeutic drugs, suggesting that DNA methylation could serve as a molecular marker for predicting the responsiveness of tumors to chemotherapy. However, the comprehensive study of pharmacoepigenomics awaits the advent of genome‐wide analysis of DNA methylation using microarrays and next‐generation sequencers. (Cancer Sci 2009; 100: 787–791)
Abbreviations:
- 5‐aza‐dC
5‐aza‐2′‐deoxycitidine
- 5‐FU
5‐fluorouracil
- BCNU
1,3‐bis(2‐chloroethyl)‐1‐nitrosourea
- BCR‐ABL
breakpoint cluster region‐ABL
- CDK10
cyclin‐dependent kinase 10
- CHFR
checkpoint with ring finger
- CIMP
CpG island methylator phenotype
- DNMT
DNA methyltransferase
- EGFR
epidermal growth factor receptor
- FA
Fanconi anemia
- FANCF
Fanconi anemia protein F
- hMLH
human mutL homolog 1
- MAPK
mitogenactivated protein kinase
- MGMT
O 6‐methylguanine‐DNA‐methyltransferase
- RASSF
Ras association domain family
- SFRP
secreted frizzled‐related protein
- shRNA
short hairpin RNA
- siRNA
short interfering RNA
- WRN
Werner syndrome protein
Cancer arises through the accumulation of multiple genetic changes, including point mutations, gene amplifications and gene deletions, which ultimately lead to activation of oncogenes and inactivation of tumor‐suppressor genes.( 1 ) Moreover, it was recently proposed that cancer cells are ‘addicted’ to oncogenes for maintenance of the malignant phenotype.( 2 ) The most convincing evidence for the concept of oncogene addiction comes from the increasing number of studies showing the therapeutic efficacy of antibodies and drugs that selectively target specific oncogenes in human cancers. For example, mutation of EGFR indicates sensitivity to gefitinib,( 3 , 4 , 5 ) the presence of BCR‐ABL translocation or mutation of c‐kit indicates sensitivity to imatinib,( 6 ) and amplification or overexpression of human EGFR‐related 2 (Her‐2)/ErbB2 indicates sensitivity to herceptin.( 7 ) Thus genetic alterations in cancer can provide important information that enables one to predict the sensitivity of a given tumor to particular chemotherapeutic drugs.
Information about gene expression also can be used to predict the response to chemotherapy. Profiles of gene expression in cancer cell lines revealed an association between the expression of certain genes and the cells’ sensitivity to chemotherapeutic drugs.( 8 , 9 ) What's more, gene expression signatures have been used clinically to predict the likely responsiveness of tumors to chemotherapy.( 10 ) But although information about genetic changes certainly contributes to our ability to predict sensitivity to chemotherapeutic drugs, the relatively low frequency of mutations seen in many cancers limits the utility of pharmacogenomics in large numbers of cancer patients, necessitating consideration of other approaches. In this review, we focus on the implications of epigenetic alterations such as DNA methylation in predicting the efficacy of chemotherapeutic drugs in the treatment of cancer.
Role of DNA methylation in carcinogenesis
DNA methylation of the 5′‐CpG islands of genes plays an important role in gene regulation. Under normal physiological conditions, DNA methylation is involved in regulating genome imprinting, X‐chromosome inactivation, and inactivation of repetitive sequences. Three DNA methyltransferases (DNMT1, DNMT3A, DNMT3B) catalyze methylation of the promoter regions of a variety of genes, including genes involved in cell‐cycle checkpoints, apoptosis, DNA repair, cell adhesion, and signal transduction.( 11 , 12 , 13 ) Simultaneous methylation of multiple genes occurs in colorectal cancers that show the CIMP,( 14 ) and the majority of sporadic colorectal cancers that show microsatellite instability are associated with CIMP, which leads to inactivation of the mismatch repair gene hMLH1 and thus disruption of mismatch repair.( 15 ) DNA methylation also plays a role in altering signaling pathways in cancer. For example, epigenetic inactivation of SFRP1, SFRP2, SFRP5, DKK1, DKK2, and DKK3, six negative regulators of WNT signaling, contributes to the full activation of T cell Factor (TCF) β‐catenin activity in colorectal cancers (Fig. 1a,b),( 16 , 17 ) whereas epigenetic inactivation of RASSF1 and RASSF2, negative regulators of the Ras signaling pathway, contributes to full activation of oncogenic Ras signaling (Fig. 2).( 18 , 19 ) Although the molecular mechanisms underlying DNA methylation remain unclear, recent studies suggest that inflammation and pathogens are likely involved.( 20 , 21 )
Figure 1.
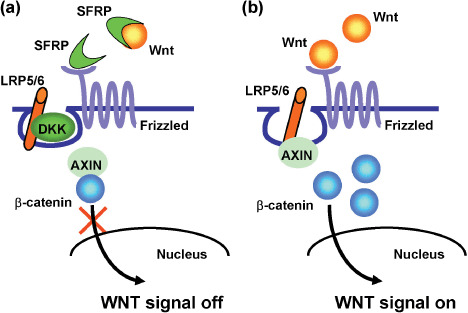
Epigenetic inactivation of negative regulators of WNT signaling. (a) In normal cells, SFRP and DKK are associated with key WNT signaling molecules such as WNT ligands and LRP5/6, which prevent translocation of β‐catenin to the nucleus. (b) In cancer cells, epigenetic inactivation of SFRP and DKK enables β‐catenin to translocate to the nucleus, which leads to activation of WNT signaling. SFRP, secreted frizzled‐related protein; DKK, Dickkopf; LRP, lipoprotein receptor‐related protein.
Figure 2.
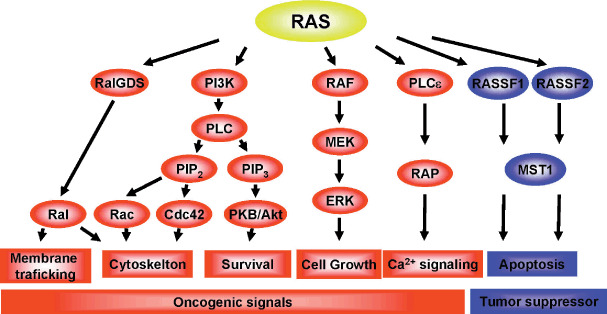
Positive and negative regulators of Ras signaling. The oncogenic and anti‐oncogenic functions of Ras are mediated by positive and negative effectors. Among the negative effectors of Ras, epigenetic inactivation of RASSF1 and RASSF2 is frequently observed in human tumors. Akt, v‐akt murine thymoma viral oncogene homolog; Cdc, cell‐division cycle; Erk, extracellular signal‐regulated kinase; MEK, Mitogen‐activated protein kinase; MST, mammalian STE20‐like protein kinase 1; PI3K, Phosphoinositide 3‐kinase; PIP, phosphatidylinositol phosphate; PKB, protein kinase B; RAP, ras‐related protein; RASSF, Ras association domain family.
Epigenetic inactivation of DNA repair and altered sensitivity to chemotherapeutic drugs
Genomic instability is an important phenotype that allows cancer cells to generate oncogenic translocations, inactivate tumor‐suppressor genes, and amplify oncogenes and drug‐resistance genes. Genomic instability is caused by impairment or inactivation of DNA repair systems, which could represent a molecular target of cancer therapy. Evidence suggests, for example, that epigenetic inactivation of DNA repair underlies tumor responsiveness to DNA‐damaging agents. The first reported epigenetic alteration associated with sensitivity to a chemotherapeutic drug was the association between methylation of the MGMT gene and sensitivity to alkylating agents.( 22 ) MGMT is a DNA repair enzyme that removes mutagenic adducts from O 6‐guanine in DNA,( 23 ) and its epigenetic silencing has been reported in a wide variety of tumors.( 24 ) This silencing of MGMT is associated with G : C to A : T transition mutations in K‐ras and p53, a mutator phenotype distinct from mismatch repair deficiency.( 25 , 26 ) Alkylating agents are one of the most widely used classes of chemotherapeutic drugs and frequently act by modifying the O 6 position of guanine. Consequently, their toxicity, and thus their efficacy, is diminished in tumors expressing MGMT.( 27 ) For example, Esteller et al. reported that MGMT gene methylation correlates with response of gliomas to BCNU (Fig. 3).( 22 ) Moreover, several clinical trials have shown MGMT gene methylation to be an independent predictor of outcome in glioblastoma patients treated with methylating agents.( 28 , 29 )
Figure 3.
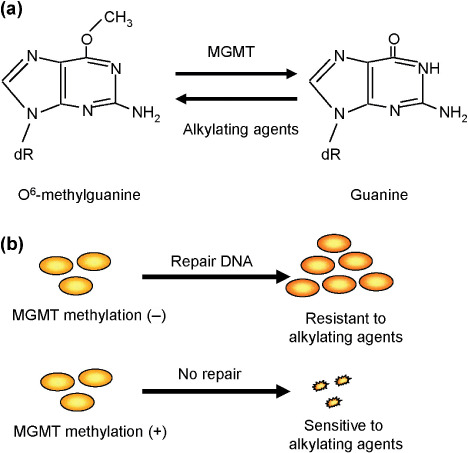
Epigenetic inactivation of O 6‐methylguanine‐DNA‐methyltransferase (MGMT) and sensitivity to alkylating agents. (a) MGMT repairs O 6‐methylguanine. (b) Cancers with MGMT methylation are sensitive to alkylating agents due to the absence of O 6‐methylguanine repair activity.
Approximately 15% of colorectal cancers show microsatellite instability due to methylation of the mismatch repair gene hMLH1.( 14 ) Clinically, colorectal cancers with hMLH1 methylation are less aggressive, but they do not respond to 5‐FU.( 30 ) Thymidylate synthase catalyzes the conversion of dUMP to dTMP, which is necessary for DNA synthesis, and inhibition of this enzyme is the major mechanism underlying the anticancer effects of 5‐FU. Ricciardiello et al. reported that colorectal cancers with hMLH1 methylation express high levels thymidylate synthase.( 31 ) Moreover, colorectal cancer cell lines displaying microsatellite instability are resistant to 5‐FU due to methylation of hMLH1, but they become susceptible to treatment upon exposure to 5‐aza‐dC.( 32 ) Thus methylation of hMLH1 appears to be a predictive molecular marker of the sensitivity of colorectal cancers to 5‐FU.
RecQ‐like helicases also reportedly play a role in the maintenance of genetic stability, and disruption of their activity results in chromosome breakage syndromes such as Bloom syndrome, Rothmund–Thomson syndrome, and Werner syndrome, the last of which is an inherited disorder characterized by the premature onset of aging and susceptibility to various types of cancer. Recently, Agrelo et al. reported that the WRN gene is frequently silenced by DNA methylation in colorectal cancers( 33 ) and that colorectal cancer cell lines showing WRN methylation are sensitive to the topoisomerase inhibitor camptothecin and to the interstrand crosslinker mitomycin C. Clinically, moreover, colorectal cancers exhibiting WRN methylation respond well to the topoisomerase inhibitor irinotecan. Hypermethylation of WRN in colorectal tumors could thus be a useful predictor of a robust clinical response to a topoisomerase inhibitor.
Fanconi anemia is an autosomal recessive chromosomal instability syndrome that causes FA patients to be prone to various types of malignancies. Taniguchi et al. reported that epigenetic inactivation of one of the FA complementation group genes, FANCF, is associated with resistance to cisplatin.( 34 ) Defects in the FA–Breast Cancer (BRCA) pathway are associated with genomic instability and increased sensitivity to DNA‐damaging agents such as mitomycin C and cisplatin, and there is a significant correlation between FANCF methylation and sensitivity to cisplatin in ovarian cancer cell lines, so that restoration of FNCAF expression using 5‐aza‐dC induces resistance to cisplatin. Methylation of FANCF has been found in 20% of primary ovarian cancers not previously exposed to cisplatin,( 34 ) but the correlation between chemosensitivity and FANCF methylation in primary tumors remains to be determined. Methylation of FANCF was also found in 30% of cervical cancers, 15% of head and neck squamous cell cancers, and 14% of non‐small cell lung cancers.( 35 , 36 ) Further study will be necessary to determine whether methylation of FANCF is a predictive marker of sensitivity to DNA‐damaging agents.
Cell‐cycle checkpoint defects and sensitivity to chemotherapeutic drugs
Impairment of cell‐cycle checkpoints is associated with sensitivity to chemotherapeutic agents. For example, overexpression of mitotic arrest difficient 2 (MAD2) sensitizes cancer cells to both cisplatin and vincristine,( 37 , 38 ) whereas overexpression of Aurora A induces chemoresistance.( 39 ) In addition, we recently found that two microtubule inhibitors, paclitaxel and docetaxel, induce apoptosis among gastric cancer cells showing CHFR methylation and that adenoviral introduction of CHFR into methylated cancer cell lines restores the checkpoint and reduces the incidence of apoptosis (Fig. 4).( 40 ) This correlation between CHFR methylation and sensitivity to microtubule inhibitors appears to be specific, as there was no correlation between CHFR methylation and sensitivity to other chemotherapeutic agents (e.g. VP16) or to UV. This suggests that CHFR methylation could serve as a clinically useful predictive marker of the sensitivity of tumors to microtubule inhibitors. Consistent with that idea, Koga et al. found that six of seven (86%) patients with methylated CHFR tumors showed some regression or no progression of their disease when treated with a microtubule inhibitor, whereas four of five (80%) patients with an unmethylated CHFR tumor showed progressive deterioration.( 41 ) A correlation between CHFR methylation and sensitivity to microtubule inhibitors also was noted in oral squamous cell carcinoma.( 42 )
Figure 4.
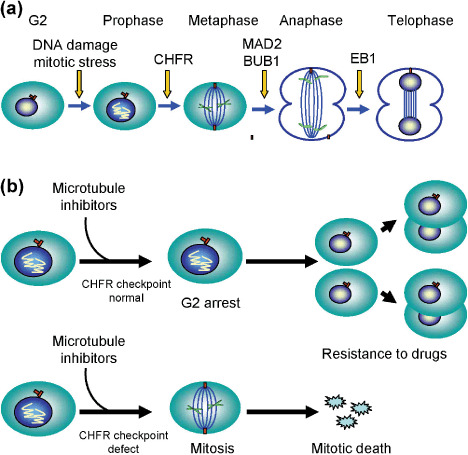
Epigenetic inactivation of a mitotic checkpoint gene, CHFR, and sensitivity to microtubule inhibitors. (a) Genes involved in the mitotic checkpoint. (b) CHFR and sensitivity to microtubule inhibitors. Cancer cells that show an intact CHFR checkpoint arrest at G2–M phase after treatment with microtubule inhibitors, which allows the cells to grow. These tumors are resistant to the drugs (top). By contrast, cancer cells that show methylation of CHFR do not arrest after treatment with microtubule inhibitors. These tumors are sensitive to the drugs (bottom). BUB, budding uninhibited by benzimidazoles; CHFR, checkpoint with ring finger; EB, end‐binding protein; MAD, mitotic arrest deficient.
The fact that CHFR is frequently inactivated by genetic or epigenetic alteration in human cancers suggests that this cancer‐specific checkpoint defect also could be a useful therapeutic target.( 40 , 43 ) Bearing that in mind, we recently established a system to knock down CHFR expression using shRNA.( 42 ) We found that CHFR expression was significantly suppressed in cancer cells transfected with shRNA, and the resultant impairment of the prophase checkpoint led to an increased mitotic index in cells treated with microtubule inhibitors, which in turn led to an increased incidence of apoptosis. This effect was specific to microtubule inhibitors, as no effect was seen when a DNA‐damaging agent (cisplatin or VP16) was used. In addition, an earlier finding that E3 ubiquitin ligases can be targeted using small molecules( 44 ) suggests drugs that inhibit CHFR's ubiquitin ligase activity also could be used to enhance the sensitivity of cancer cells to microtubule inhibitors.
Disruption of the G2–M checkpoint also appears to contribute to the sensitivity of chemotherapeutic drugs. Among the genes involved in the G2–M checkpoint, 14‐3‐3 σ, a transcriptional target of p53,( 45 ) is frequently silenced by DNA methylation in breast and gastric cancers,( 46 , 47 ) and it has been suggested that 14‐3‐3 σ is a critical regulator of G2–M that also has tumor‐suppressor activity. Knocking out 14‐3‐3 σ in cancer cells leads to mitotic catastrophe and cell death following DNA damage resulting form the absence of G2–M arrest.( 48 , 49 ) Consistent with those data, the G2–M checkpoint is impaired in gastric cancer cell lines that show methylation of 14‐3‐3 σ,( 47 ) and restoration of 14‐3‐3 σ expression using 5‐aza‐dC restores G2–M arrest induced by DNA damage. In addition, functional proteomic analysis revealed 14‐3‐3 σ to be a key molecule that contributes to resistance to mitoxantrone and adriamycin in breast cancer cells.( 50 )
Using high‐throughput siRNA screening, Iorns et al. identified CDK10 as an important determinant of resistance to endocrine therapy in breast cancer, and thus a major factor limiting successful treatment of the disease.( 51 ) They also found that knocking down CDK10 increases V‐ets erythroblastosis virus E26 oncogene homolog 2 (ETS2)‐driven transcription of C‐Raf, resulting in activation of the MAPK pathway and loss of tumor cell reliance on estrogen signaling, and that breast cancer patients with estrogen receptor‐α‐positive tumors expressing low levels of CDK10 relapse early on tamoxifen, which suggests that downregulation of CDK10 contributes to resistance to endocrine therapy. In that regard, DNA methylation of CDK10 was found in 18% of breast cancers, suggesting that methylation of CDK10 could be a predictive molecular marker of breast cancer sensitivity to tamoxifen.
In general, the studies cited above were carried out using a candidate gene approach, but recent progress in genome‐wide methylation analysis could enable performance of unbiased methylation analyses. For example, genome‐wide gene expression profiles in NCI‐60 cell lines are often used to assess the association between gene expression and sensitivity to chemotherapeutic drugs. By comparing the DNA methylation profiles for 32 genes with drug sensitivity in NCI‐60 cell lines, Shen et al. were able to identify a correlation between p73 methylation and sensitivity to alkylating agents.( 52 ) p73 is a member of the p53 family and, like other p53 family members, it is involved in cell‐cycle checkpoint function, apoptosis, DNA repair, and cellular differentiation.( 53 ) The findings of Shen et al. suggest that methylation of p73 could be a predictive marker of sensitivity to alkylating agents.( 52 ) Consistent with that idea, overexpression of p73 has been observed in cancers of the bladder, lung, and ovary, and is often associated with resistance to treatment with DNA‐damaging agents.( 54 , 55 , 56 ) In addition, knocking down p73 using siRNA reduced cellular viability after treatment with BCNU and cisplatin. The molecular mechanism by which silencing of p73 sensitizes cancer cells to alkylating agents remains unknown, however.
Epigenetic alteration of signaling pathways and resistance to therapy
Patients with K‐ras mutations reportedly do not respond to treatment with monoclonal anti‐EGFR antibodies such as cetuximab or panitumumab.( 57 , 58 ) Situated downstream of EGFR, K‐ras is a key component of the RAS–MAPK pathway and is involved in mediating cell proliferation. Its mutation may enable cells to circumvent the anti‐EGFR activity of cetuximab and panitumumab. That colorectal cancers with K‐ras mutations tend to show methylation of multiple CpG islands suggests that resistance to anti‐EGFR therapy in patients with K‐ras mutations may be associated with CIMP,( 59 ) and that DNA methylation of genes affected by CIMP may also contribute to the resistance to cetuximab or panitumumab. In fact, RASSF2, a negative effector of RAS, is silenced by DNA methylation in CIMP‐positive colorectal cancers.( 18 , 60 )
Future directions in cancer epigenomics: Genome‐wide approaches
Although DNA methylation of certain genes appears to influence sensitivity to chemotherapeutic drugs, the majority of studies carried out to date were done using cell line models or only a small number of subjects (Table 1). Large‐scale analyses will be necessary to confirm the utility of epigenetic information for prediction of responses to chemotherapeutic drugs. Comprehensive studies of pharmacoepigenomics in cancer await advances in genome‐wide DNA methylation analyses using microarrays and next‐generation sequencers.
Table 1.
Stages toward the clinical application of DNA methylation markers for prediction of sensitivity to chemotherapeutic drugs
| Gene | Function | Cancer type | Stages in clinical application | References |
|---|---|---|---|---|
| MGMT | DNA repair | Glioma | Several clinical studies to define sensitivity to alkylating agents have been reported. | 22, 28, 29 |
| hMLH1 | Mismatch repair | Colorectal cancer | Association with sensitivity to 5‐fluorouracil has been reported. | 30 |
| Independent experiments remain to be performed. | ||||
| WRN | DNA helicase | Colorectal cancer | Association with sensitivity to cisplatin has been reported. | 33 |
| Independent experiments remain be performed. | ||||
| FANCF | DNA repair | Ovarian cancer | Association with sensitivity to cisplatin has been reported. | 34 |
| Independent experiments remain to be performed. | ||||
| CHFR | Mitotic checkpoint | Colorectal cancer | Several clinical trials to define sensitivity to microtubule inhibitors have been reported. | 41, 42 |
| 14‐3‐3σ | G2–M checkpoint | Colorectal cancer | Association with sensitivity to DNA damaging agents has been shown in cell lines. | 47 |
| Clinical studies remain to be performed. | ||||
| CDK10 | G2–M checkpoint | Breast cancer | Association with sensitivity to tamoxifen has been reported. | 51 |
| Independent experiments remain to be performed. | ||||
| p73 | DNA damage checkpoint | Renal cancer | Association with sensitivity to cisplatin has been reported for cell lines. | 52 |
| Clinical studies remain to be performed. |
CHFR, checkpoint with ring finger; FANCF, Fanconi anemia protein F; hMLH1, human mutL homolog 1; MGMT, O 6‐methylguanine‐DNA‐methyltransferase; WRN, Werner syndrome protein.
Acknowledgments
We thank Dr William F. Goldman for editing the manuscript. This study was supported by Grants‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (M. Toyota, K. Imai, T. Tokino, and Y. Shinomura), a Grant‐in‐Aid for the Third‐term Cancer Control Strategy, and Grant‐in‐Aid for Cancer Research from the Ministry of Health, Labor, and Welfare, Japan (M. Toyota).
References
- 1. Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–70. [DOI] [PubMed] [Google Scholar]
- 2. Weinstein IB, Joe A. Oncogene addiction. Cancer Res 2008; 68: 3077–80. [DOI] [PubMed] [Google Scholar]
- 3. Lynch TJ, Bell DW, Sordella R et al . Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–39. [DOI] [PubMed] [Google Scholar]
- 4. Mitsudomi T, Kosaka T, Endoh H et al . Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non‐small‐cell lung cancer with postoperative recurrence. J Clin Oncol 2005; 23: 2513–20. [DOI] [PubMed] [Google Scholar]
- 5. Paez JG, Janne PA, Lee JC et al . EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–500. [DOI] [PubMed] [Google Scholar]
- 6. Pardanani A, Tefferi A. Imatinib targets other than bcr/abl and their clinical relevance in myeloid disorders. Blood 2004; 104: 1931–9. [DOI] [PubMed] [Google Scholar]
- 7. Hayes DF, Thor AD. c‐erbB‐2 in breast cancer: development of a clinically useful marker. Semin Oncol 2002; 29: 231–45. [DOI] [PubMed] [Google Scholar]
- 8. Dan S, Tsunoda T, Kitahara O et al . An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res 2002; 62: 1139–47. [PubMed] [Google Scholar]
- 9. Potti A, Dressman HK, Bild A et al . Genomic signatures to guide the use of chemotherapeutics. Nat Med 2006; 12: 1294–300. [DOI] [PubMed] [Google Scholar]
- 10. Minna JD, Girard L, Xie Y. Tumor mRNA expression profiles predict responses to chemotherapy. J Clin Oncol 2007; 25: 4329–36. [DOI] [PubMed] [Google Scholar]
- 11. Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128: 683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis 2007; 28: 2434–42. [DOI] [PubMed] [Google Scholar]
- 13. Ushijima T, Okochi‐Takada E. Aberrant methylations in cancer cells: where do they come from? Cancer Sci 2005; 96: 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toyota M, Ahuja N, Ohe‐Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999; 96: 8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin Cancer Biol 1999; 9: 349–57. [DOI] [PubMed] [Google Scholar]
- 16. Sato H, Suzuki H, Toyota M et al . Frequent epigenetic inactivation of DICKKOPF family genes in human gastrointestinal tumors. Carcinogenesis 2007; 28: 2459–66. [DOI] [PubMed] [Google Scholar]
- 17. Suzuki H, Watkins DN, Jair KW et al . Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004; 36: 417–22. [DOI] [PubMed] [Google Scholar]
- 18. Akino K, Toyota M, Suzuki H et al . The Ras effector RASSF2 is a novel tumor‐suppressor gene in human colorectal cancer. Gastroenterology 2005; 129: 156–69. [DOI] [PubMed] [Google Scholar]
- 19. Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21. 3 Nat Genet 2000; 25: 315–19. [DOI] [PubMed] [Google Scholar]
- 20. Fukayama M, Hino R, Uozaki H. Epstein–Barr virus and gastric carcinoma: virus–host interactions leading to carcinoma. Cancer Sci 2008; 99: 1726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maekita T, Nakazawa K, Mihara M et al . High levels of aberrant DNA methylation in Helicobacter pylori‐infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res 2006; 12: 989–95. [DOI] [PubMed] [Google Scholar]
- 22. Esteller M, Garcia‐Foncillas J, Andion E et al . Inactivation of the DNA‐repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000; 343: 1350–4. [DOI] [PubMed] [Google Scholar]
- 23. Pegg AE. Mammalian O 6‐alkylguanine‐DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res 1990; 50: 6119–29. [PubMed] [Google Scholar]
- 24. Esteller M, Herman JG. Generating mutations but providing chemosensitivity. the role of O 6‐methylguanine DNA methyltransferase in human cancer. Oncogene 2004; 23: 1–8. [DOI] [PubMed] [Google Scholar]
- 25. Esteller M, Risques RA, Toyota M et al . Promoter hypermethylation of the DNA repair gene O 6‐methylguanine‐DNA methyltransferase is associated with the presence of G : C to A : T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 2001; 61: 4689–92. [PubMed] [Google Scholar]
- 26. Esteller M, Toyota M, Sanchez‐Cespedes M et al . Inactivation of the DNA repair gene O 6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K‐ras in colorectal tumorigenesis. Cancer Res 2000; 60: 2368–71. [PubMed] [Google Scholar]
- 27. Ludlum DB. DNA alkylation by the haloethylnitrosoureas: nature of modifications produced and their enzymatic repair or removal. Mutat Res 1990; 233: 117–26. [DOI] [PubMed] [Google Scholar]
- 28. Brell M, Tortosa A, Verger E et al . Prognostic significance of O 6‐methylguanine‐DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res 2005; 11: 5167–74. [DOI] [PubMed] [Google Scholar]
- 29. Hegi ME, Diserens AC, Godard S et al . Clinical trial substantiates the predictive value of O‐6‐methylguanine‐DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res 2004; 10: 1871–4. [DOI] [PubMed] [Google Scholar]
- 30. Carethers JM, Chauhan DP, Fink D et al . Mismatch repair proficiency and in vitro response to 5‐fluorouracil. Gastroenterology 1999; 117: 123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ricciardiello L, Ceccarelli C, Angiolini G et al . High thymidylate synthase expression in colorectal cancer with microsatellite instability: implications for chemotherapeutic strategies. Clin Cancer Res 2005; 11: 4234–40. [DOI] [PubMed] [Google Scholar]
- 32. Arnold CN, Goel A, Boland CR. Role of hMLH1 promoter hypermethylation in drug resistance to 5‐fluorouracil in colorectal cancer cell lines. Int J Cancer 2003; 106: 66–73. [DOI] [PubMed] [Google Scholar]
- 33. Agrelo R, Cheng WH, Setien F et al . Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci USA 2006; 103: 8822–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taniguchi T, Tischkowitz M, Ameziane N et al . Disruption of the Fanconi anemia‐BRCA pathway in cisplatin‐sensitive ovarian tumors. Nat Med 2003; 9: 568–74. [DOI] [PubMed] [Google Scholar]
- 35. Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene 2004; 23: 1000–4. [DOI] [PubMed] [Google Scholar]
- 36. Narayan G, Arias‐Pulido H, Nandula SV et al . Promoter hypermethylation of FANCF: disruption of Fanconi Anemia–BRCA pathway in cervical cancer. Cancer Res 2004; 64: 2994–7. [DOI] [PubMed] [Google Scholar]
- 37. Cheung HW, Jin DY, Ling MT et al . Mitotic arrest deficient 2 expression induces chemosensitization to a DNA‐damaging agent, cisplatin, in nasopharyngeal carcinoma cells. Cancer Res 2005; 65: 1450–8. [DOI] [PubMed] [Google Scholar]
- 38. Wang X, Jin DY, Wong HL, Feng H, Wong YC, Tsao SW. MAD2‐induced sensitization to vincristine is associated with mitotic arrest and Raf/Bcl‐2 phosphorylation in nasopharyngeal carcinoma cells. Oncogene 2003; 22: 109–16. [DOI] [PubMed] [Google Scholar]
- 39. Anand S, Penrhyn‐Lowe S, Venkitaraman AR. AURORA‐A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell 2003; 3: 51–62. [DOI] [PubMed] [Google Scholar]
- 40. Satoh A, Toyota M, Itoh F et al . Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res 2003; 63: 8606–13. [PubMed] [Google Scholar]
- 41. Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K. The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol 2006; 41: 133–9. [DOI] [PubMed] [Google Scholar]
- 42. Ogi K, Toyota M, Mita H et al . Small interfering RNA‐induced CHFR silencing sensitizes oral squamous cell cancer cells to microtubule inhibitors. Cancer Biol Ther 2005; 4: 773–80. [DOI] [PubMed] [Google Scholar]
- 43. Toyota M, Sasaki Y, Satoh A et al . Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci USA 2003; 100: 7818–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Ther 2003; 2: 623–9. [PubMed] [Google Scholar]
- 45. Hermeking H, Lengauer C, Polyak K et al . 14‐3‐3 sigma is a p53‐regulated inhibitor of G2/M progression. Mol Cell 1997; 1: 3–11. [DOI] [PubMed] [Google Scholar]
- 46. Ferguson AT, Evron E, Umbricht CB et al . High frequency of hypermethylation at the 14‐3‐3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA 2000; 97: 6049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14‐3‐3 sigma gene is associated with 5′‐CpG island hypermethylation in human cancers. Cancer Res 2000; 60: 4353–7. [PubMed] [Google Scholar]
- 48. Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14‐3‐3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature 1999; 401: 616–20. [DOI] [PubMed] [Google Scholar]
- 49. Chan TA, Hwang PM, Hermeking H, Kinzler KW, Vogelstein B. Cooperative effects of genes controlling the G2/M checkpoint. Genes Dev 2000; 14: 1584–8. [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y, Liu H, Han B, Zhang JT. Identification of 14‐3‐3sigma as a contributor to drug resistance in human breast cancer cells using functional proteomic analysis. Cancer Res 2006; 66: 3248–55. [DOI] [PubMed] [Google Scholar]
- 51. Iorns E, Turner NC, Elliott R et al . Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell 2008; 13: 91–104. [DOI] [PubMed] [Google Scholar]
- 52. Shen L, Toyota M, Kondo Y et al . Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA 2007; 104: 18 654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ozaki T, Nakagawara A. p73, a sophisticated p53 family member in the cancer world. Cancer Sci 2005; 96: 729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nyman U, Sobczak‐Pluta A, Vlachos P, Perlmann T, Zhivotovsky B, Joseph B. Full‐length p73α represses drug‐induced apoptosis in small cell lung carcinoma cells. J Biol Chem 2005; 280: 34 159–69. [DOI] [PubMed] [Google Scholar]
- 55. Vikhanskaya F, Marchini S, Marabese M, Galliera E, Broggini M. P73a overexpression is associated with resistance to treatment with DNA‐damaging agents in a human ovarian cancer cell line. Cancer Res 2001; 61: 935–8. [PubMed] [Google Scholar]
- 56. Yokomizo A, Mai M, Tindall DJ et al . Overexpression of the wild type p73 gene in human bladder cancer. Oncogene 1999; 18: 1629–33. [DOI] [PubMed] [Google Scholar]
- 57. Freeman DJ, Juan T, Reiner M et al . Association of K‐ras mutational status and clinical outcomes in patients with metastatic colorectal cancer receiving panitumumab alone. Clin Colorectal Cancer 2008; 7: 184–90. [DOI] [PubMed] [Google Scholar]
- 58. Khambata‐Ford S, Garrett CR, Meropol NJ et al . Expression of epiregulin and amphiregulin and K‐ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007; 25: 3230–7. [DOI] [PubMed] [Google Scholar]
- 59. Toyota M, Ohe‐Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA 2000; 97: 710–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kakar S, Deng G, Cun L, Sahai V, Kim YS. CpG island methylation is frequently present in tubulovillous and villous adenomas and correlates with size, site, and villous component. Hum Pathol 2008; 39: 30–6. [DOI] [PubMed] [Google Scholar]


