Abstract
Immune‐associated molecules play important roles in cancer development and progression. The aims of this study were to determine the diagnostic utility of uric acid (UA) and soluble MHC class I chain‐related molecules A (sMICA) and B (sMICB) in pancreatic ductal adenocarcinoma (PDAC) compared with those of cancer antigen 19‐9 (CA19‐9), the most commonly available tumor marker for PDAC. We evaluated serum levels of UA, sMICA and sMICB along the carcinogenic process of PDAC obtained from 148 individuals composed of normal (n = 70), chronic pancreatitis (n = 23) and PDAC (n = 55), and compared them with those of CA19‐9. We also evaluated the correlations of these biomarkers with tumor size, resectability or TNM stage, and tested logistic regression to ascertain the potential usability of these markers for the detection of PDAC. We also investigated the correlations among these biomarkers. Serum UA, sMICA and sMICB differed significantly according to groups (Kruskal–Wallis, P < 0.05), and were closely correlated with the development of PDAC. Serum sMICA were correlated with distant metastasis and sMICB were correlated with unresectability. Sensitivity and specificity of sMICA and sMICB were higher than CA19‐9, and a multi‐maker panel using all tested markers (UA, sMICA, sMICB and CA19‐9) demonstrated the best potential for detecting PDAC (94.2% sensitivity at 93.3% specificity). The three tested markers also showed added diagnostic potentials to overcome the limitation of CA19‐9 by differentiation of PDAC from non‐cancerous conditions when CA19‐9 is inappropriate. In conclusion, serum UA, sMICA and sMICB might be useful screening or differential diagnostic biomarkers for PDAC to complement CA19‐9. (Cancer Sci 2011; 102: 1673–1679)
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies characterized by poor responsiveness to conventional anti‐cancer therapy.( 1 , 2 ) Because more than 80% of patients are surgically unresectable at the time of diagnosis,( 3 ) development of an early diagnosis tool is very important. However, current conventional diagnostic methods for PDAC are somewhat complex, expensive and often invasive. Furthermore, diagnosis can be delayed because symptoms of PDAC are non‐specific unless it induces jaundice. Thus, it is clinically important to develop comfortable, inexpensive, non‐invasive and easily accessible screening methods such as blood biomarkers.
The serum glycosylation marker cancer antigen 19‐9 (CA19‐9) is the most commonly available biomarker for PDAC. However, the American Society of Clinical Oncology (ASCO) does not recommend this marker for diagnostic purposes of PDAC because of its inadequate sensitivity and/or specificity for detection of PDAC with or without symptoms.( 4 , 5 , 6 ) Cancer antigen 19‐9 (CA19‐9) can be elevated in patients with biliary obstruction in the absence of a tumor such as benign inflammatory head masses in chronic pancreatitis, and in other benign conditions such as digestive pathologies, renal cysts and rheumatological disorders.( 7 , 8 ) Therefore, it might be disappointing to distinguish benign inflammatory head masses in chronic pancreatitis from overt PDAC. Hence, it is required that new effective serum tumor markers for the early detection of PDAC are investigated to provide a chance for cure and for differentiation of overt malignancy from benign conditions when serum CA19‐9 does not correspond to a disease entity.
Because every change in body status such as development of a malignancy leads to an immune response, serum immune‐associated molecules can reflect the initiation or progression of cancer. Uric acid (UA) is a chemical produced by xanthine oxidase when DNA and RNA are degraded. A variety of medical diseases such as gout, cardiovascular disease and diabetes show abnormal concentrations of UA.( 9 , 10 ) Uric acid is also known as an important endogenous damage‐associated molecular pattern (DAMP) released from injured cells.( 11 ) As a DAMP, it can stimulate dendritic cell (DC) maturation and enhance the innate natural killer (NK) cell‐mediated cytotoxicity and the adaptive cytotoxic T‐lymphocyte (CTL) response in vivo; thus, serum levels of UA can reflect anti‐tumor activity of the immune system.( 12 ) Therefore, decreased serum levels of UA might reflect impairment of anti‐tumor immunity, and the presence of initiation or progression of cancer.
Major histocompatibility complex (MHC) class I‐related molecules A (MICA) and B (MICB) are stress‐inducible ligands of immunoreceptor, NK group 2, member D (NKG2D) expressed on cytotoxic NK and T‐cells, resulting in enhancing their cytotoxic activity.( 13 ) MICA/B can be released into the blood stream as soluble forms (sMICA/B) from the surfaces of cancer cells by proteolytic shedding,( 14 , 15 ) and act as negative regulators of anti‐tumor immunity by downregulating expression of the NKG2D receptor. Thus, they might be meaningful diagnostic serum biomarkers for malignancy.( 14 , 15 , 16 , 17 , 18 ) According to a previous study, the ability of sMICA or sMICB to discriminate between cancer and non‐cancer cases might differ depending on the tumor type.( 16 ) Therefore, individual studies for sMICA or sMICB are required according to tumor types to obtain a more accurate understanding of the exact roles of sMICA/B in the development or progression of cancer.
Several recent studies have demonstrated the potential use of sMICA as a diagnostic and prognostic marker for PDAC.( 17 ) However, there has been very limited available data about sMICB as a diagnostic tumor marker for PDAC, and those studies have shown conflicting results.( 17 , 18 ) Also, UA has rarely been evaluated as a diagnostic marker for PDAC. In the present study, we evaluated the clinical implication of three immune‐associated molecules, namely UA, sMICA and sMICB, as potential diagnostic serum biomarkers for PDAC with comparison with those of serum CA19‐9, the most commonly available biomarker for PDAC.
Materials and Methods
Patients and blood samples. The statistical significance level (α) was set at 0.05 and statistical power was set at 0.8. With this setting, we calculated the minimal sample size of each group to be 23 individuals. Based on this calculation, blood samples were collected from 70 healthy controls (40 men and 30 women; median age, 53 years), 23 patients with chronic pancreatitis (12 men and 11 women; median age, 52 years) and 55 patients with PDAC (33 men and 22 women; median age, 58 years) who visited the Yonsei University Health System. For the healthy controls, age‐ and sex‐matched individuals visiting for a medical checkup and showing a normal pancreas in imaging and biochemical tests without any disease and risk factors for PDAC were enrolled. For the chronic pancreatitis group, patients showing typical radiological or histopathological findings of chronic pancreatitis without other disease were enrolled. All PDAC patients were diagnosed histopathologically through biopsy or surgical specimen. In all groups, individuals suffering from chronic illnesses such as gout, cardiovascular disease or diabetes, and taking medications that might affect serum UA levels were excluded. Patients with other cancers and/or other pancreatic malignancies were also excluded.
In the cancer groups, all patients underwent radiological procedures for staging including abdominal helical computed tomography, magnetic resonance imaging and whole body positron emission tomography scans. The TNM stage classification was analyzed according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual (6th edition).( 19 ) Blood samples were obtained prior to the start of any treatment modalities. This research was approved by the Institutional Review Board of Yonsei University Health System and all participants provided written informed consent.
Measurement of serum levels of CA19‐9 and UA. Approximately 10 mL of whole blood was allowed to clot at room temperature for 30 min and was centrifuged at 1500g for 15 min. Serum fraction was used to measure the CA19‐9 and UA levels using a Vitros‐3600 automatic analyzer (Ortho Clinical Diagnostic, New York, NY, USA) and Hitachi 7600 automatic analyzer (Hitachi, Tokyo, Japan), respectively.
Measurement of serum levels of MICA and MICB by enzyme‐linked immunosorbent assay (ELISA). Levels of sMICA and sMICB were measured using a previously described sandwich‐ELISA.( 17 ) In brief, to determine sMICA levels, the monoclonal antibodies (mAb) AMO1 and BAMO3 were used at concentrations of 5 and 1 μg/mL, respectively, with recombinant sMICA*04 as a standard. To determine sMICB levels, mAb BAMO1 and BMO2 were used at concentrations of 2 and 1 μg/mL, respectively, with recombinant MICB*02 as a standard. Absorbance was measured at 450 nm.
Measurement of serum levels of immune‐related cytokines using a chemiluminescent immunoassay. To evaluate the serum levels of interleukin (IL)‐2, interferon (IFN)‐γ and IL‐10, we used a commercially available MILLIPLEX MAP Human Cytokine/Chemokine Kit (Millipore, Billerica, MA, USA) for sera according to the recommended protocol. This allowed for the simultaneous quantification of three immune‐related cytokines.
Statistical analysis. Serum levels of UA, sMICA, sMICB and CA19‐9 were expressed as means and 25–75% standard deviations (SD). The Kruskal–Wallis one‐way analysis of variance on ranks was used to evaluate the differences of expression levels of markers among the three disease groups. Other non‐parametric values were also compared by the Kruskal–Wallis test among the three groups. Nominal data was compared by Chi‐squared test. For non‐parametric comparisons of two variables, the Mann–Whitney U‐test was used. Spearman’s correlation was performed to assess the correlations between serum values and non‐continuous variables and Pearson’s correlation analysis was performed to assess the correlations between serum values and continuous variables. The receiver operating characteristic (ROC) curves were plotted to determine the best cut‐off point and sensitivity/specificity for screening of PDAC. To ascertain the potential usability of multiple markers composed of UA, sMICA, sMICB and CA19‐9 for screening of PDAC, we used logistic regression, which estimates the probability of having PDAC among the tested individuals.( 20 ) Each marker was included as a linear term and evaluated from one to four panel size. For comparison among each panel, the cut‐off point ensured a target specificity of around 90%. For all statistical analyses, the social sciences software package (version 13.0, SPSS, Chicago, IL, USA) was used and a P‐value <0.05 was considered statistically significant.
Results
Serum levels of UA, sMICA, sMICB and CA19‐9 along the carcinogenic process of PDAC (normal–chronic pancreatitis–PDAC sequence). Statistical analysis (Kruskal–Wallis test) showed that serum UA was significantly decreased as the carcinogenic process proceeded (normal: mean, 5.24 mg/dL; chronic pancreatitis: mean, 4.63 mg/dL; PDAC: mean, 3.59 mg/dL; P < 0.001, Table 1). Serum sMICA were significantly higher in patients with PDAC (mean, 179.41 pg/mL) than patients with chronic pancreatitis (mean, 44.10 pg/mL; P < 0.001) and those who were normal (mean, 7.51 pg/mL; P < 0.001). However, there was no significant difference between the normal and chronic pancreatitis groups (P > 0.05) (Table 1). Similar to sMICA, serum sMICB were also significantly higher in patients with PDAC (mean, 39.68 pg/mL) than in patients with chronic pancreatitis (mean, 7.61 pg/mL; P < 0.001) and those who were normal (mean, 1.02 pg/mL; P < 0.001), whereas there was no significant difference between the normal and chronic pancreatitis groups (P > 0.05) (Table 1). Overall, serum sMICB were lower than sMICA and rarely detected in both the normal and chronic pancreatitis groups, while sMICA were modestly detected in the non‐cancer groups. Serum CA19‐9 also tended to increase significantly along the carcinogenic process of PDAC (P < 0.001) (Table 1).
Table 1.
Serum levels of UA, sMICA, sMICB and cancer antigen 19‐9 (CA19‐9) according to disease groups and pathological parameters of PDAC
| UA (mg/dL) (mean ± SD) | sMICA (pg/mL) (mean ± SD) | sMICB (pg/mL) (mean ± SD) | CA19‐9 (U/mL) (mean ± SD) | |
|---|---|---|---|---|
| Disease group (P‐value) | 0.001 | 0.001 | 0.001 | 0.001 |
| Normal (n = 70) | 5.24 ± 1.35 | 7.51 ± 6.91 | 1.02 ± 3.18 | 8.50 ± 7.21 |
| Chronic pancreatitis (n = 23) | 4.63 ± 1.34 | 44.10 ± 56.42 | 7.61 ± 12.23 | 144.15 ± 336.87 |
| PDAC (n = 55) | 3.59 ± 1.37 | 179.41 ± 146.70 | 39.68 ± 36.48 | 4077.81 ± 6858.11 |
| Cancer pathological parameters† | ||||
| Size (P‐value) | 0.901 | 0.119 | 0.043 | 0.068‡ |
| <2 cm (n = 5) | 3.56 ± 0.92 | 117.00 ± 66.02 | 11.0 ± 0.10 | 277.20 ± 475.37 |
| >2 cm and <5 cm (n = 40) | 3.63 ± 1.52 | 171.28 ± 110.60 | 31.64 ± 17.92 | 3728.90 ± 6889.85 |
| >5 cm (n = 10) | 3.46 ± 1.38 | 239.84 ± 238.87 | 94.35 ± 76.71 | 7173.40 ± 7591.70 |
| Unresectability (P‐value)§ | 0.659 | 0.617 | 0.026 | 0.037 |
| Resectable (n = 11) | 3.78 ± 1.26 | 138.45 ± 54.23 | 10.05 ± 1.32 | 231.01 ± 422.31 |
| Unresectable (n = 44) | 3.56 ± 1.53 | 183.97 ± 140.13 | 45.06 ± 37.38 | 5391.78 ± 7691.23 |
| Nodal metastasis (P‐value) | 0.944 | 0.600 | 0.710 | 0.020 |
| Negative (n = 15) | 3.58 ± 1.22 | 161.93 ± 127.72 | 38.20 ± 21.31 | 2345.33 ± 5971.70 |
| Positive (n = 40) | 3.59 ± 1.51 | 188.58 ± 149.74 | 40.78 ± 42.75 | 4837.23 ± 7180.39 |
| Distant metastasis (P‐value) | 0.374 | 0.022 | 0.116 | 0.001 |
| Negative (n = 30) | 3.80 ± 1.44 | 136.76 ± 69.20 | 25.51 ± 16.00 | 1353.41 ± 3429.42 |
| Positive (n = 25) | 3.88 ± 2.12 | 237.45 ± 189.88 | 56.20 ± 47.86 | 8028.75 ± 8586.38 |
†In these analyses, only PDAC patients were included. ‡This variable tends to be higher according to increased size, although it is not statistically significant. §A tumor was defined as resectable when it did not invade the celiac axis or the superior mesenteric, although it extended beyond the pancreas (T1‐3, N0‐1, M0, stage I–II), and was defined as unresectable when it invaded the celiac axis, the superior mesenteric artery or distant organs (T4, N0‐1, M0‐1, stage III–IV). PDAC, pancreatic ductal adenocarcinoma; SD, standard deviation; SMICA, soluble MHC class I chain‐related molecules A; SMICB, soluble MHC class I chain‐related molecules B; UA, uric acid. Statistical significance was assessed by the Kruskal–Wallis test or the Mann–Whitney U‐test. P < 0.05 (two‐tailed) was considered to be statistically significant. Values in bold font are statistically significant.
Correlations between serum levels of UA, sMICA, sMICB or CA19‐9 and clinicopathological characteristics. Serum UA, sMICA, sMICB and CA19‐9 were not significantly affected by age (P > 0.05) and gender (P > 0.05), except for the correlation of serum UA with gender; it was higher in men than women (Spearman’s correlation, γs = −0.322, P = 0.001) (Table 2). Relationships between serum UA, sMICA, sMICB or CA19‐9 and pathological parameters of PDAC such as tumor size, unresectability and TNM stage were also evaluated to reveal the roles of these values in the progression of PDAC (1, 2). Serum sMICA were strongly correlated with distant metastasis (Spearman’s correlation, γs = 0.323, P = 0.021) (1, 2), whereas serum sMICB were closely correlated with unresectability (γs = 0.627, P = 0.022) and size (γs = 0.562, P = 0.045) (1, 2). However, contrary to our expectation, serum UA was not correlated with all progression parameters of PDAC (1, 2). Serum CA19‐9 showed close correlation with several progression parameters of PDAC; size: γs = 0.381, P = 0.007; node metastasis: γs = 0.337, P = 0.018; resectability: γs = 0.300, P = 0.036; and distant metastasis: γs = 0.491, P = 0.001 (1, 2).
Table 2.
Correlations between serum levels of UA, sMICA, sMICB or cancer antigen 19‐9 (CA19‐9) and clinicopathological characteristics of PDAC
| UA | sMICA | sMICB | CA19‐9 | |
|---|---|---|---|---|
| γs (P‐value) | γs (P‐value) | γs (P‐value) | γs (P‐value) | |
| Gender | −0.322(0.001) | −0.090 (0.351) | −0.183 (0.111) | −0.078 (0.408) |
| Age | −0.053 (0.575) | 0.128 (0.190) | 0.073 (0.530) | 0.155 (0.098) |
| Size† | −0.042 (0.776) | 0.192 (0.201) | 0.562 (0.045) | 0.381 (0.007) |
| Unresectability‡ | −0.066 (0.656) | 0.074 (0.604) | 0.627 (0.022) | 0.300 (0.036) |
| Node metastasis | −0.010 (0.945) | 0.073 (0.610) | −0.134 (0.663) | 0.337 (0.018) |
| Distant metastasis | −0.130 (0.380) | 0.323 (0.021) | 0.454 (0.119) | 0.491 (0.001) |
†Analyzed by being divided into three groups according to size as follows: <2 cm; >2 cm and <5 cm; >5 cm. ‡Analyzed by being divided into resectable and unresectable; resectable was defined as tumor extending beyond the pancreas but without involvement of the celiac axis or the superior mesenteric artery (T1‐3, N0‐1, M0, stage I–II), and unresectable was defined as tumor invading the celiac axis or the superior mesenteric artery and extending into distant organs (T4, N0‐1, M0‐1, stage III–IV). γs, Spearman correlation coefficient; PDAC, pancreatic ductal adenocarcinoma; sMICA, soluble MHC class I chain‐related molecules A; sMICB, soluble MHC class I chain‐related molecules B; UA, uric acid. Values in bold font are statistically significant.
Correlations among serum levels of UA, sMICA, sMICB and CA19‐9. We evaluated the correlations among tested potential markers in all patients (n = 148) including whole disease states of malignant transformation of PDAC (Table 3A) and in overt PDAC (n = 55) including just the PDAC state (Table 3B). Along the carcinogenic process of PDAC, serum UA showed a negative correlation with sMICA (Pearson’s correlation, γp = −0.441, P = 0.001), sMICB (γp = −0.360, P = 0.002) and CA19‐9 (γp = −0.221, P = 0.020). Serum sMICA were also positively correlated with sMICB (γp = 0.919, P = 0.001) and CA19‐9 (γp = 0.473, P = 0.001), and sMICB were also positively correlated with CA19‐9 (γp = 0.508, P = 0.001) (Table 3A). In the PDAC state, the correlations among tested potential markers shown were similar patterns to those when all samples were enrolled in the test excluding serum CA19‐9, which was not correlated with UA and sMICB (Table 3B).
Table 3.
Pearson’s correlation among serum levels of UA, sMICA and sMICB in all subjects with pancreatic carcinogenesis (A) and overt pancreatic ductal adenocarcinoma (B)
| UA | sMICA | sMICB | CA19‐9 | |
|---|---|---|---|---|
| γp (P‐value) | γp (P‐value) | γp (P‐value) | γp (P‐value) | |
| (A) | ||||
| UA | – | −0.441 (0.001) | −0.360 (0.002) | −0.221 (0.020) |
| sMICA | −0.441 (0.001) | – | 0.919 (0.001) | 0.473 (0.001) |
| sMICB | −0.360 (0.002) | 0.919 (0.001) | – | 0.508 (0.001) |
| CA19‐9 | −0.221 (0.020) | 0.473 (0.001) | 0.508 (0.001) | – |
| (B) | ||||
| UA | – | −0.328 (0.023) | −0.751 (0.008) | −0.069 (0.648) |
| sMICA | −0.328 (0.023) | – | 0.857 (0.001) | 0.317 (0.027) |
| sMICB | −0.751 (0.008) | 0.857 (0.001) | – | 0.347 (0.245) |
| CA19‐9 | −0.069 (0.648) | 0.317 (0.027) | 0.347 (0.245) | – |
γp, Pearson correlation coefficient; CA19‐9, cancer antigen 19‐9; sMICA, soluble MHC class I chain‐related molecules A; sMICB, soluble MHC class I chain‐related molecules B; UA, uric acid. Values in bold font are statistically significant.
Correlation between cytokines reflecting the functional cytotoxic activity of NK or T‐cells and serum levels of UA, sMICA and sMICB along the carcinogenic process of PDAC. Because UA is considered as an immune stimulator, lower UA levels might reflect the immunosuppressive status of patients susceptible to cancer development. Similarly, increased serum levels of sMICA and sMICB might reflect the immunosuppressive status susceptible to cancer because sMICA and sMICB can inhibit cellular cytotoxicity of NK or T‐cells directly or indirectly.( 14 , 15 , 18 ) To validate this, we evaluated the circulating levels of immune‐related cytokines reflecting NK or T‐cell activities. Interleukin‐2 and IFN‐γ were evaluated as pro‐inflammatory cytokines with antitumor activity, and IL‐10 was evaluated as an anti‐inflammatory cytokine with protumorigenic activity along the carcinogenic process of PDAC. Table 4(A) shows that serum levels of IL‐10 downregulating expression of T‐helper 1 (Th1) cytokines were significantly increased in the PDAC group compared with those of the normal and chronic pancreatitis groups (Kruskal–Wallis, P < 0.05), whereas IL‐2 and IFN‐γ having a Th1 cytokine‐stimulating function were significantly reduced along the carcinogenic process of PDAC (P < 0.05). These results suggest that antitumor immunity was suppressed along the carcinogenic process of PDAC. Serum UA was negatively correlated with serum IL‐10 whereas sMICA were positively correlated with IL‐10 and negatively correlated with IFN‐γ, and sMICB was negatively correlated with IL‐2 (Table 4B).
Table 4.
Serum levels of cytokines involved in antitumor immunity (IL‐2, IFN‐γ, IL‐10) as functional immunological parameters along the carcinogenic process of PDAC (A), and correlations between immune‐related cytokines and UA, sMICA or sMICB in carcinogenesis of PDAC (B)
| Cytokines (mean ± SD, ng/mL) | Normal (n = 70) | Chronic pancreatitis (n = 23) | PDAC (n = 55) | Kruskal–Wallis (P‐value) |
|---|---|---|---|---|
| (A) | ||||
| IL‐2 | 4.71 ± 21.80 | 0.76 ± 0.74 | 0.03 ± 0.12 | 0.001 |
| IFN‐γ | 10.23 ± 14.49 | 2.92 ± 4.79 | 2.14 ± 4.03 | 0.001 |
| IL‐10 | 3.02 ± 12.23 | 1.52 ± 3.51 | 14.42 ± 42.50 | 0.001 |
| IL‐2 γp (P‐value) | IFN‐γ γp (P‐value) | IL‐10 γp (P‐value) | |
|---|---|---|---|
| (B) | |||
| UA | 0.058 (0.540) | 0.092 (0.333) | −0.256 (0.006) |
| sMICA | −0.082 (0.375) | −0.222 (0.015) | 0.189 (0.039) |
| sMICB | −0.382 (0.001) | −0.172 (0.132) | 0.143 (0.212) |
γp, Pearson’s correlation coefficient; IFN‐γ, interferon‐γ; IL, interleukin; PDAC, pancreatic ductal adenocarcinoma; SD, standard deviation; sMICA, soluble MHC class I chain‐related molecules A; sMICB, soluble MHC class I chain‐related molecules B; UA, uric acid. Values in bold font are statistically significant.
Logistic regression for validation of the potential usability of a multi‐marker panel for detection of PDAC. To validate the potential usability of serum UA, sMICA and sMICB for screening of PDAC, we used linear logistic regression. Table 5 lists the unbiased results of sensitivities from one to four panel size at a fixed specificity of around 90% by logistic regression. As a single marker, both sMICA (76.5% sensitivity at 91.2% specificity) and sMICB (76.5% sensitivity at 90.6% specificity) demonstrated superior sensitivity to detect PDAC compared with CA19‐9 (73.5% sensitivity at 90.9% specificity), although UA did not (39.6% sensitivity at 88.0% specificity). The ROC curves also revealed a higher power of sMICA and sMICB to detect PDAC compared with CA19‐9 (Fig. 1). When markers were combined as a multi‐marker panel, almost all combinations achieved superior diagnostic potentials compared with the use of CA19‐9 alone. Although the use of UA alone was inferior to CA19‐9, it became similar or superior to CA19‐9 when combined with sMICA or sMICB. The multi‐maker panel using all tested markers (UA, sMICA, sMICB and CA19‐9) demonstrated the best potential to distinguish PDAC from non‐cancerous conditions (94.2% sensitivity at 93.3% specificity).
Table 5.
Sensitivity and specificity from validation of an algorithm based on combining biomarkers using linear logistic regression
| Variables | Sensitivity, % | Specificity, % |
|---|---|---|
| Single‐marker panel | ||
| sMICA | 76.5 | 91.2 |
| sMICB | 76.9 | 90.6 |
| UA | 39.6 | 88.0 |
| CA19‐9 | 73.5 | 90.9 |
| Two‐marker panel | ||
| sMICA, sMICB | 84.6 | 92.2 |
| sMICA, UA | 81.3 | 90.8 |
| sMICB, UA | 72.7 | 90.2 |
| sMICA, CA19‐9 | 87.8 | 89.4 |
| sMICB, CA19‐9 | 84.6 | 91.9 |
| CA19‐9, UA | 65.2 | 89.1 |
| Three‐marker panel | ||
| sMICA, sMICB, UA | 90.9 | 93.4 |
| sMICA, sMICB, CA19‐9 | 92.3 | 93.5 |
| sMICA, UA, CA19‐9 | 84.8 | 92.2 |
| sMICB, UA, CA19‐9 | 81.8 | 91.7 |
| Four‐marker panel | ||
| sMICA, sMICB, UA, CA19‐9 | 94.2 | 93.3 |
CA19‐9, cancer antigen 19‐9; sMICA, soluble MHC class I chain‐related molecules A; sMICB, soluble MHC class I chain‐related molecules B; UA, uric acid.
Figure 1.
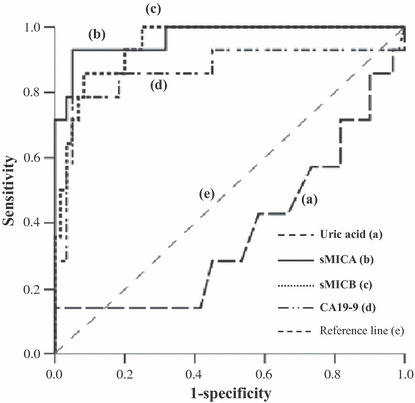
Receiver operator characteristics (ROC) curves generated with serum uric acid (a), soluble MHC class I chain‐related molecules A (sMICA) (b), sMICB (c) and cancer antigen 19‐9 (CA19‐9) (d) for the detection of pancreatic ductal adenocarcinoma (PDAC). The figure shows that serum sMICA and sMICB showed higher sensitivity and specificity for the diagnosis of PDAC compared with CA19‐9, although uric acid did not.
Differential diagnostic potentials of serum UA, sMICA and sMICB between benign and malignant conditions when serum CA19‐9 is inappropriate. Although serum CA19‐9 is a commonly available biomarker for PDAC, this value is disappointing for differentiating between benign and malignant conditions when it does not correspond to a disease entity. Hence, we compared the serum levels of UA, sMICA and sMICB between individuals with low CA19‐9 (≤30 U/mL) and high CA19‐9 (>30 U/mL) in both the non‐cancer and cancer groups, and evaluated the sensitivity and specificity of these markers to differentiate between benign and malignant conditions in cases where CA19‐9 was inappropriate. Serum levels of UA, sMICA and sMICB were not different between individuals with low CA19‐9 and high CA19‐9 in both the non‐cancer (P > 0.05) and cancer groups (P > 0.05) (Table 6A). Also, all three markers provided acceptable sensitivity and specificity to differentiate benign conditions from PDAC in both groups with low CA19‐9 and high CA19‐9 (Table 6B).
Table 6.
(A) Serum levels of UA, sMICA and sMICB between individuals with low cancer antigen 19‐9 (CA19‐9) (≤30 U/mL) and those with high CA19‐9 (>30 U/mL) in the non‐cancer and cancer groups, respectively. (B) sensitivity and specificity of UA, sMICA and sMICB to differentiate between the presence and absence of overt pancreatic ductal adenocarcinoma in patients with high CA19‐9 (>30 U/mL) or patients with low CA19‐9 (≤30 U/mL), respectively
| Non‐cancer patients | P‐value* | Cancer patients | P‐value* | |||
|---|---|---|---|---|---|---|
| Low CA 19‐9 (mean ± SD) | High CA 19‐9 (mean ± SD) | Low CA 19‐9 (mean ± SD) | High CA 19‐9 (mean ± SD) | |||
| (A) | ||||||
| CA19‐9 (U/mL) | 5.87 ± 5.09 | 322.85 ± 402.11 | 0.001 | 6223.98 ± 7987.79 | 7.11 ± 0.16 | 0.001 |
| Uric acid (ng/mL) | 5.14 ± 1.21 | 4.88 ± 1.59 | 0.588 | 3.79 ± 1.79 | 3.74 ± 1.41 | 0.711 |
| sMICA (pg/mL) | 9.17 ± 18.23 | 19.21 ± 26.51 | 0.081 | 205.60 ± 154.12 | 128.51 ± 91.24 | 0.068 |
| sMICB (pg/mL) | 1.87 ± 5.72 | 1.89 ± 3.86 | 0.122 | 39.55 ± 37.40 | 71.33 ± 75.19 | 0.451 |
| Cut‐off value | High CA19‐9 patients | Low CA19‐9 patients | |||
|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | ||
| (B) | |||||
| Uric acid (ng/mL) | 4.5 | 73.7 | 68.4 | 75.0 | 71.7 |
| sMICA (pg/mL) | 80 | 76.3 | 66.7 | 72.9 | 81.7 |
| sMICB (pg/mL) | 15 | 75.0 | 66.7 | 100.0 | 85.0 |
*Statistical significance was assessed by the Mann–Whitney U‐test. SD, standard deviation; sMICA, soluble MHC class I chain‐related molecules A; sMICB, soluble MHC class I chain‐related molecules B; UA, uric acid.
Discussion
Serum UA has been considered to have a protective effect against the development of cancer as an anti‐oxidant( 21 ) or as an immune‐stimulating DAMP.( 11 ) Because the immunosuppressive state might accelerate development of PDAC, serum UA might be negatively correlated along the carcinogenic process of PDAC. However, few epidemiological investigations have addressed this association, especially in PDAC, and previous findings are inconsistent.( 22 , 23 , 24 , 25 ) Strasak and colleagues showed a positive correlation between elevated serum levels of UA and a poor prognosis of cancer.( 22 ) However, this study was limited to older women and mainly focused on the cancer outcome rather than incidence. In contrast, Lickl and colleagues demonstrated serum UA levels were significantly lower in cancer patients (n = 501) than healthy people (n = 15 150) for both genders.( 23 ) Our observations were similar to those of Lickl et al. We demonstrated that lower UA levels were closely correlated with the immunosuppressive state (Table 4) and development of PDAC (Table 1). Also, serum UA was negatively correlated with sMICA and sMICB, which are disruptors of NK cells; this might additionally verify the immune impairment state in PDAC compared with non‐cancerous conditions. Although we cannot explain the reason for this discrepancy, we guess it might originate from the many factors such as drugs, food or disease that can influence serum UA; however, previous studies( 22 , 23 , 24 , 25 ) might fail to exclude all of the interfering factors completely. We tried to exclude as many of the interfering factors as possible in the present study.
Because anti‐tumor immunity is generally suppressed according to tumor size, serum UA might be correlated with tumor size. However, we could not observe a correlation between serum UA and tumor size (1, 2). In fact, serum UA can be unrelated to tumor size in the advanced stage because it can be affected by the malignant process itself, that is, it can be elevated according to the increased turnover rate of nucleic acid in rapidly proliferating advanced‐stage cancer.( 26 ) Nevertheless, it might be clinically useful as a diagnostic biomarker for PDAC because it can differentiate PDAC from non‐cancerous conditions when CA19‐9 is inappropriate (Table 6).
Several studies have demonstrated sMICA and sMICB can disrupt the cytotoxicity of NK or T cells,( 14 , 15 , 18 ) thereby the resultant elevated levels of sMICA and sMICB can indicate the presence or progression of cancer.( 14 , 15 , 16 , 17 , 18 ) In the current study, we demonstrated the correlations between serum sMIC levels and the immunosuppressive state of patients through the expression patterns of immune‐related cytokines (IL‐2, IFN‐γ and IL‐10). Interleukin‐2 and IFN‐γ are potent Th1‐related cytokines that act on NK and T cells towards anti‐tumor activity,( 27 , 28 ) whereas IL‐10 downregulates expression of Th1 cytokines and suppresses the function of NK and T cells.( 29 ) The results revealed that serum IL‐2 and IFN‐γ were significantly reduced (Kruskal–Wallis, P < 0.05), whereas serum IL‐10 was significantly increased (Kruskal–Wallis, P < 0.05) along the PDAC carcinogenesis (Table 4A). Serum UA was negatively correlated with IL‐10, whereas serum sMICA was strongly correlated with IL‐10 and negatively correlated with IFN‐γ. Serum sMICB were negatively correlated with IL‐2 (Table 4B). These results might indicate that the immunosuppressive status of patients reflects the carcinogenesis or tumor progression of PDAC, and it was negatively correlated with serum UA and positively correlated with serum sMICA and sMICB.
We observed serum sMICA were closely correlated with distant metastasis of PDAC (Table 2), corresponding to previous studies.( 17 , 18 ) However, it was not correlated with the parameters of local tumor extension such as tumor size, resectability and node metastasis (Table 2). In contrast, we could not observe correlations between serum sMICB and distant metastasis different from two previous studies although previous results for sMICB as a biomarker of PDAC were conflicting.( 18 , 19 ) Instead, we could observe that it was correlated with unresectability and tumor size reflecting local tumor extension (1, 2).
As no data are available regarding a potential association of either MIC molecule with certain pathophysiological conditions or tumor entities as yet, we cannot explain exactly the reason why correlations with clinicopathological characteristics were different between sMICA and sMICB, although similar physiological roles are expected because they are close relatives in the MIC family. However, several studies suggest that the physiological roles of both MIC are different from each other; for example, only MICB is efficiently targeted by herpesvirus microRNA,( 30 ) or upon stress MICB displays a more substantial upregulation than MICA.( 31 ) Additionally, a different pathophysiological mechanism has been proposed to underlie between the two MIC for the surface expression and release of MICA versus MICB.( 32 ) Thus, serum sMICA and sMICB might be involved in somewhat different pathways to disturb antitumor immunity and induce the malignant transformation or progression of PDAC. In fact, we observed that overall serum sMICB was lower than sMICA, and it was rarely detected in both normal and chronic pancreatitis groups, while sMICA was modestly detected in the non‐cancer groups. Also, sMICA was closely correlated with systemic spread of tumor, while sMICB was correlated with local tumor extension.
Although serum UA, sMICA and sMICB could not exactly reflect the progression of PDAC better than serum CA19‐9, they might be clinically relevant in several aspects. Differentiation between overt PDAC and chronic pancreatitis, especially superimposed with benign inflammatory head mass, is clinically important to prevent unnecessary extensive surgery and to decide proper treatment. However, this task is sometimes difficult and serum CA19‐9 has proved to be disappointing for this differentiation because it can be inappropriately elevated in several benign conditions including mass‐forming chronic pancreatitis. In the current study, we showed that three tested potential biomarkers (UA, sMICA and sMICB) have additional diagnostic potentials to differentiate between overt PDAC and benign conditions when serum CA19‐9 is inappropriate (Table 6). Thus, the detection power might be increased when these markers are used in combination with CA19‐9. Additionally, our data showed that serum sMICB was correlated with unresectability. The ability to predict unresectability is clinically very important because treatment strategy and prognosis are remarkably different between resectable and unresectable cancers, and complete resection alone can offer a chance of curing PDAC patients.
Moreover, sMICA and sMICB were shown to have higher sensitivity and specificity than CA19‐9 to detect PDAC as a single marker. As multi‐marker panels, the usefulness of serum UA, sMICA and sMICB were also validated by logistic regression for multivariate analysis; logistic regression revealed that the best power to detect PDAC was achieved when all markers were used, including CA19‐9 (94.2% sensitivity at 93.3% specificity) (Table 5).
Taken together, we validated the clinical significance of immune‐associated molecules, serum UA, sMICA and sMICB, as biomarkers for PDAC by showing diagnostic potentials to overcome the limitation of CA19‐9 when, for example, it is needed to differentiate between overt PDAC and mass‐forming chronic pancreatitis or between resectable and unresectable cancer, and by revealing higher sensitivity and specificity of sMICA and sMICB than CA19‐9 to detect PDAC. In conclusion, serum UA, sMICA and sMICB might be useful serum biomarkers for PDAC, especially when CA19‐9 is inappropriate for a disease entity. However, the present study used a relatively small sample size. Therefore, in the future more large‐scaled studies are needed to confirm our findings.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This work was supported by the National Research Foundation (NRF) in South Korea through National Core Research Center for Nanomedical Technology (R15‐2004‐024‐00000‐0).
References
- 1. Lillemoe KD, Yeo CJ, Cameron JL. Pancreatic cancer: state of‐ the‐art care. CA Cancer J Clin 2000; 50: 241–68. [DOI] [PubMed] [Google Scholar]
- 2. Mulcahy MF, Wahl AO, Small W Jr. The current status of combined radiotherapy and chemotherapy for locally advanced or resected pancreas cancer. J Natl Compr Canc Netw 2005; 3: 637–42. [DOI] [PubMed] [Google Scholar]
- 3. Camacho D, Reichenbach D, Duerr GD, Venema TL, Sweeney JF, Fisher WE. Value of laparoscopy in the staging of pancreatic cancer. J Ophthalmol 2005; 6: 552–61. [PubMed] [Google Scholar]
- 4. Locker GY, Hamilton S, Harris J et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol 2006; 24: 5313–27. [DOI] [PubMed] [Google Scholar]
- 5. Chang CY, Huang SP, Chiu HM, Lee YC, Chen MF, Lin JT. Low efficacy of serum levels of CA 19‐9 in prediction of malignant diseases in asymptomatic population in Taiwan. Hepatogastroenterology 2006; 53: 1–4. [PubMed] [Google Scholar]
- 6. Clavé P, Boadas J, González‐Carro P et al. Accuracy of imaging techniques and tumor markers in the diagnosis of pancreatic cancer. Gastroenterol Hepatol 1999; 22: 335–41. [PubMed] [Google Scholar]
- 7. Akdoğan M, Saşmaz N, Kayhan B, Biyikoğlu I, Dişibeyaz S, Sahin B. Extraordinarily elevated CA19‐9 in benign conditions: a case report and review of the literature. Tumori 2001; 87: 337–9. [DOI] [PubMed] [Google Scholar]
- 8. Kayhan B, Kayhan B, Akdogan M. Can IL‐2R alpha be a valuable marker along with CA 19‐9 in the diagnosis of chronic pancreatitis and pancreatic cancer. Int J Biol Markers 2004; 19: 196–02. [DOI] [PubMed] [Google Scholar]
- 9. Heinig M, Johnson RJ. Role of uric acid in hypertension, renal disease, and metabolic syndrome. Cleve Clin J Med 2006; 73: 1059–64. [DOI] [PubMed] [Google Scholar]
- 10. Dehghan A, Van Hoek M, Sijbrands EJ, Hofman A, Witteman JC. High serum uric acid as a novel risk factor for type 2 diabetes. Diabetes Care 2008; 31: 361–2. [DOI] [PubMed] [Google Scholar]
- 11. Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature 2003; 425: 516–21. [DOI] [PubMed] [Google Scholar]
- 12. Ullrich E, Ménard C, Flament C et al. Dendritic cells and innate defense against tumor cells. Cytokine Growth Factor Rev 2008; 19: 79–92. [DOI] [PubMed] [Google Scholar]
- 13. Bauer S, Groh V, Wu J et al. Activation of NK cells and T cells by NKG2D, a receptor for stress inducible MICA. Science 1999; 285: 727–9. [DOI] [PubMed] [Google Scholar]
- 14. Salih HR, Rammensee HG, Steinle A. Cutting edge: down‐regulation of MICA on human tumors by proteolytic shedding. J Immunol 2002; 169: 4098–102. [DOI] [PubMed] [Google Scholar]
- 15. Groh V, Wu J, Yee C, Spies T. Tumour‐derived soluble MIC ligands impair expression of NKG2D and T‐cell activation. Nature 2002; 419: 734–8. [DOI] [PubMed] [Google Scholar]
- 16. Holdenrieder S, Stieber P, Peterfi A, Nagel D, Steinle A, Salih HR. Soluble MICA in malignant diseases. Int J Cancer 2006; 118: 684–7. [DOI] [PubMed] [Google Scholar]
- 17. Märten A, von Lilienfeld‐Toal M, Büchler MW, Schmidt J. Soluble MIC is elevated in the serum of patients with pancreatic carcinoma diminishing gammadelta T cell cytotoxicity. Int J Cancer 2006; 119: 2359–65. [DOI] [PubMed] [Google Scholar]
- 18. Holdenrieder S, Stieber P, Peterfi A, Nagel D, Steinle A, Salih HR. Soluble MICB in malignant diseases: analysis of diagnostic significance and correlation with soluble MICA. Cancer Immunol Immunother 2006; 55: 1584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Greene FL, Page DL, Fleming ID et al. , eds. AJCC Cancer Staging Manual. Ann Arbor, MI: Springer, 2002. [Google Scholar]
- 20. McIntosh MW, Pepe MS. Combining several screening tests: optimality of the risk score. Biometrics 2002; 58: 657–64. [DOI] [PubMed] [Google Scholar]
- 21. Ames BN, Cathcart R, Schweirs E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant‐ and radical‐caused aging and cancer: a hypothesis. Proc Natl Acad Sci USA 1981; 78: 6858–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strasak AM, Rapp K, Hilbe W et al. The role of serum uric acid as an antioxidant protecting against cancer: prospective study in more than 28 000 older Austrian women. Ann Oncol 2007; 18: 1893–7. [DOI] [PubMed] [Google Scholar]
- 23. Lickl E, Alth G, Ebermann R, Beck RH, Tuma K. Serum urate level and cancer. Med Hypotheses 1989; 28: 193–5. [DOI] [PubMed] [Google Scholar]
- 24. Hiatt RA, Fireman BH. Serum uric acid unrelated to cancer incidence in humans. Cancer Res 1988; 48: 2916–18. [PubMed] [Google Scholar]
- 25. Yossepowitch O, Pinchuk I, Gur U, Neumann A, Lichtenberg D, Baniel J. Advanced but not localized prostate cancer is associated with increased oxidative stress. J Urol 2007; 178: 1238–43. [DOI] [PubMed] [Google Scholar]
- 26. Ultmann JE. Hyperuricemia in disseminated neoplastic disease other than lypmphomas and leukemias. Cancer 1962; 15: 122–9. [DOI] [PubMed] [Google Scholar]
- 27. Thornton AM, Donovan EE, Piccirillo CA, Shevach EM. Cutting edge: IL‐2 is critically required for the in vitro activation of CD4+CD25+ T cell suppressor function. J Immunol 2004; 172: 6519–23. [DOI] [PubMed] [Google Scholar]
- 28. Lange F, Rateitschak K, Fitzner B, Pöhland R, Wolkenhauer O, Jaster R. Studies on mechanisms of interferon‐gamma action in pancreatic cancer using a data‐driven and model‐based approach. Mol Cancer 2011; 10: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin‐10 and related cytokines and receptors. Annu Rev Immunol 2004; 22: 929–79. [DOI] [PubMed] [Google Scholar]
- 30. Nachmani D, Stern‐Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress‐induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009; 5: 376–85. [DOI] [PubMed] [Google Scholar]
- 31. Spies T. Regulation of NKG2D ligands: a purposeful but delicate affair. Nat Immunol 2008; 9: 1013–5. [DOI] [PubMed] [Google Scholar]
- 32. Salih HR, Goehlsdorf D, Steinle A. Release of MICB molecules by tumor cells: mechanism and soluble MICB in sera of cancer patients. Hum Immunol 2006; 67: 188–95. [DOI] [PubMed] [Google Scholar]


