Abstract
Methylenetetrahydrofolate reductase is a key enzyme in folate metabolism, which affects DNA synthesis and methylation and is possibly linked to colorectal carcinogenesis. Alcohol and acetaldehyde have an adverse effect on folate metabolism. This study investigated the relationship of functional MTHFR C677T and ALDH2 polymorphisms to colorectal adenomas with reference to alcohol consumption in a case‐control study of male officials in the Self‐Defense Forces (SDF) who received a preretirement health examination at two SDF hospitals. The study subjects were 452 cases of colorectal adenoma and 1050 controls with no polyp who underwent total colonoscopy. Genotypes were determined by the PCR‐RFLP method using genomic DNA extracted from the buffy coat. Statistical adjustment was made for age, hospital, rank in the SDF, body mass index, cigarette‐years and alcohol intake. Neither MTHFR C677T nor ALDH2 showed a measurable association with colorectal adenoma. While high alcohol consumption was associated with a moderately increased risk of colorectal adenoma, neither of the two polymorphisms showed a significant effect on the association between alcohol and colorectal adenoma. Individuals with the variant alleles ALDH2*2 and MTHFR 677T had a decreased risk of colorectal adenomas, showing adjusted odds ratios of 0.70 (95% confidence interval 0.49–1.00) for all adenomas and 0.57 (0.34–0.95) for large adenomas (≥ 5 mm), as compared to individuals with ALDH2*1/1 and MTHFR 677CC genotypes combined. The findings may be interpreted as suggesting that folate inhibits the growth of colorectal adenomas, but further confirmation is needed. (Cancer Sci 2005; 96: 513 –518)
Folate metabolism has drawn much attention in relation to colorectal carcinogenesis.( 1 , 2 ) Methylenetetrahydrofolate reductase (MTHFR) is a key enzyme regulating folate metabolism. MTHFR irreversibly converts 5,10‐methylenetetrahydrofolate to 5‐methyltetrahydrofolate.( 2 ) The substrate of MTHFR, 5,10‐methylenetetrahydrofolate, is required for conversion of deoxyuridylate to thymidylate. Insufficient thymidylate results in uracil misincorporation into DNA, leading to single‐strand and double‐strand breaks and increasing the incidence of DNA misrepair.( 3 , 4 ) On the other hand, 5‐methyltetrahydrofolate is a methyl donor in the remethylation of homocystein to methionine, which is required for DNA methylation. Imbalanced DNA methylation has been implicated in colorectal carcinogenesis.( 5 , 6 )
The MTHFR C677T polymorphism is a common functional polymorphism in exon 4, resulting in an alanine‐to‐valine substitution at codon 222.( 7 ) The homozygous variant (677TT) has no more than 30% of normal enzyme activity, and heterozygotes (677 CT) also seem to have lower activity of the enzyme.( 7 ) Several,( 8 , 9 , 10 , 11 , 12 ) but not all,( 13 , 14 , 15 ) epidemiological studies have shown a decreased risk of colorectal cancer associated with the MTHFR 677TT genotype, especially in individuals with high folate intake and/or low alcohol intake. On the other hand, a limited number of studies addressed the relationship between the MTHFR C677T polymorphism and colorectal adenoma, a well‐established precursor lesion of colorectal cancer.( 16 , 17 , 18 , 19 , 20 ) These studies generally showed no association between the MTHFR C677T polymorphism and the risk of colorectal adenoma, but reported variable associations between alcohol intake and colorectal adenoma, dependent on the MTHFR C677T polymorphism.( 16 , 17 , 18 , 19 , 20 )
Alcohol has fairly consistently been shown to be associated with increased risk of colorectal cancer and adenoma.( 21 ) Alcohol is known to exert adverse effects on folate metabolism by decreasing intestinal absorption and hepatic uptake, increasing renal excretion and cleaving folate.( 22 ) It is also known that acetaldehyde, the first metabolite of ethanol, has a carcinogenic effect in animals.( 23 ) In humans, most of the acetaldehyde is oxidized by the mitochondrial enzyme aldehyde dehydrogenase 2 (ALDH2) in the liver. The ALDH2 gene contains the variant ALDH2*2 allele, which results in an inactive enzyme by substitution of lysine for glutamine at codon 487.( 24 ) Few studies reported that the heterozygous variant of ALDH2*1/2 was associated with an increased risk of alcohol‐related cancers and of colon cancer.( 25 , 26 , 27 ) In the present study, we investigated the relationship between the MTHFR C677T and ALDH2 polymorphisms and colorectal adenomas with reference to the interaction of alcohol consumption in middle‐aged Japanese men. The gene–gene interaction was also a matter of interest.
Materials and Methods
Subjects
Study subjects were male officials in the Self‐Defense Forces (SDF) who received a preretirement health examination at the SDF Fukuoka Hospital (Kasuga, Japan) or Kumamoto Hospital (Kumamoto, Japan) during the period January 1997 to March 2001. The preretirement health examination is a nationwide program offering a comprehensive medical examination to those retiring from the SDF. Details of the preretirement health examination have been described elsewhere.( 28 , 29 ) In addition to blood samples for routine use in the health examination, a sample of 7 mL fasting venous blood was obtained for the purpose of medical research in general, not specifically for genetic study, with written informed consent. At the time of recruitment of the study subjects, there was no committee investigating ethical issues of epidemiological or genetic studies. However, the study was approved by the ethical committee of Kyushu University (Fukuoka, Japan) in the year 2002, and genotyping was carried out after individuals were made completely anonymous.
The present study included 452 cases of histologically confirmed colorectal adenoma and 1050 controls with no polyp who underwent total colonoscopy. In a consecutive series of 2459 men, five men refused to participate in the survey and a total of 319 men were excluded for not receiving colonoscopy (n = 77) or having a prior history of colectomy (n = 17), colorectal polypectomy (n = 226), malignant neoplasms (n = 33) or inflammatory bowel disease (n = 2). Some of the men had two or more reasons for exclusion. In the remaining 2135 men, colonoscopic findings were classified as colorectal cancer (n = 1), polyp (n = 938), non‐polyp benign lesion such as diverticula (n = 123) and normal (n = 1073). The cases studied comprised 461 men out of the 938 with colorectal polyps who were found to have adenoma without in situ or invasive carcinoma, and the controls were 1067 men of the 1196 with normal or non‐polyp benign lesions who underwent total colonoscopy. Finally, 26 men were excluded due to lack of DNA sample or unsuccessful genotyping of either polymorphism, leaving 452 cases and 1050 controls in the analysis.
Cases of adenoma with sizes of < 5, 5–9 and ≥ 10 mm (the largest size for multiple adenomas) numbered 264, 151 and 37, respectively. The analysis by subsite was confined to adenoma cases with total colonoscopy (n = 433). Numbers of cases having adenomas at the proximal colon alone, the distal colon and/or rectum alone, and both proximal and distal segments were 109, 218 and 106, respectively; rectal adenomas and distal colon adenomas were combined because cases with rectal adenomas alone were few (n = 40). Cases of large adenomas (≥ 5 mm) accounted for 37%, 32% and 63% of those having adenomas at the proximal segment only, distal segment only and both segments, respectively.
Lifestyle questionnaire
A self‐administered questionnaire was used to ascertain smoking habits, alcohol consumption and other lifestyle factors prior to colonoscopy. Smokers were defined as those who had ever smoked cigarettes daily for at least 1 year. Both current and past smokers were asked about the average number of cigarettes smoked per day and total years of smoking. Cumulative exposure to cigarette smoking was expressed as cigarette‐years, which were calculated by multiplying the average number of cigarettes per day by the total years of smoking. Alcohol drinkers were defined as those having drunk alcoholic beverages at least once a week for at least 1 year. Current drinkers reported the consumption of five types of alcoholic beverages (sake, shochu, beer, whisky/brandy and wine) on average in the past year, and their daily intake of ethanol was estimated. Cigarette smoking was classified into 0, 1–399, 400–799 and 800 cigarette‐years, and alcohol use was categorized into never, past and current use with consumption of < 30, 30–59 or 60 mL ethanol per day. Body mass index was calculated by dividing weight in kilograms by squared height in meters, and was categorized into four levels using quartiles in the distribution in the control group. The SDF rank was classified into low, middle and high ranks.
Genotyping
DNA was extracted from the buffy coat using a commercial kit (Qiagen, Hilden, Germany). Genotyping was carried out using the PCR‐RFLP method with electrophoresis on a 3% agarose gel (NuiSieve GTG) and visualization by ethidium bromide. The PCR was carried out in a reaction mixture of 10 µL containing 0.5 U Taq and 1 µL template DNA with a concentration of approximately 50–150 ng/µL. The MTHFR C677T genotype was determined, as described by Frosst et al.,( 7 ) using primers 5′‐TGAAG GAGAA GGTGT CTGCG GGA‐3′ (sense) and 5′‐AGGAC GGTGC GGTGA GAGTG‐3′ (antisense). The PCR product was digested with HinfI, which cleaves the 198‐bp PCR product into two fragments of 175 and 23 bp when the C677T mutation exists.
The ALDH2 polymorphism was determined using a method described elsewhere.( 25 ) The primers used were 5′‐CAAAT TACAG GGTCA ACTGC T‐3′ (sense) and 5′‐CCACA CTCAC AGTTT TCTCT T‐3′ (antisense). The restriction enzyme EarI digested the 135‐bp PCR product into two fragments of 112 and 23 bp in the case of the wild‐type allele (ALDH2*1).
Statistical analysis
Odds ratio (OR) and 95% confidence interval (CI) were obtained by logistic regression analysis; 95% CI was derived from the standard error for the logistic regression coefficient. Statistical adjustment was made for age (continuous variable), hospital, SDF rank, body mass index, cigarette‐years and alcohol intake using indicator variables for the above‐mentioned categories of the covariates. In evaluating the interaction with alcohol use, the highest two categories of alcohol intake (30–59 mL/day and 60 mL/day) were combined, and the remaining categories (lifelong non‐drinking, past alcohol use and < 30 mL/day) were also combined into one group because no measurable difference in the OR was observed within each of the two combined groups (see below). When the gene–gene interaction was examined, the rare variant homozygotes of ALDH2*2/2 were excluded, and the MTHFR 677CT and 677TT genotypes were combined in some analyses to avoid potential random variation due to small numbers. Statistical assessment of the interaction was carried out on the basis of the likelihood ratio test by adding cross‐product terms of indicator variables representing genotypes and alcohol drinking categories. Two‐sided P‐values less than 0.05 were regarded as statistically significant. All computations in these analyses were carried out using the Stata Statistical Software Release 8.0 (Stata Corporation, College Station, TX, USA).
Results
Among the controls, the frequencies of the CC, CT and TT genotypes of the MTHFR C677T polymorphism were 38%, 47% and 15%, respectively. The frequencies of ALDH2*1/1, ALDH2*1/2 and ALDH2*2/2 were 58%, 37% and 5%, respectively (Table 1). The distributions of the MTHFR C677T and ALDH2 genotypes were each in agreement with the Hardy–Weinberg equilibrium. While alcohol use did not vary with the MTHFR C677T polymorphism, alcohol consumption was progressively less frequent with increasing numbers of the ALDH2*2 allele (Fig. 1). High consumption of alcohol was associated with a moderate increase in the risk of colorectal adenoma. Adjusted OR (95% CI) of colorectal adenomas for lifelong non‐drinkers, past drinkers and current drinkers consuming < 30, 30–59 or > 60 mL alcohol per day were 1.00 (referent), 1.08 (0.52–2.23), 0.90 (0.60–1.34), 1.46 (1.00–2.14) and 1.50 (1.02–2.22), respectively, when the genetic polymorphisms were not taken into account.
Table 1.
Relationship between MTHFR C677T and ALDH2 polymorphisms and colorectal adenoma
| Genotype | Cases | Controls | Crude OR | |||
|---|---|---|---|---|---|---|
| n | % | n | % | Adjusted OR | (95% CI) † | |
| MTHFR C677T | ||||||
| CC | 182 | 40.3 | 399 | 38.0 | 1.00 | 1.00 (referent) |
| CT | 203 | 44.9 | 496 | 47.2 | 0.90 | 0.90 (0.71–1.16) |
| TT | 67 | 14.8 | 155 | 14.8 | 0.95 | 0.93 (0.66–1.32) |
| ALDH2 | ||||||
| 1/1 (wild type) | 299 | 66.2 | 605 | 57.6 | 1.00 | 1.00 (referent) |
| 1/2 | 137 | 30.3 | 390 | 37.1 | 0.71 | 0.81 (0.62–1.05) |
| 2/2 (variant) | 16 | 3.5 | 55 | 5.2 | 0.59 | 0.67 (0.35–1.27) |
Adjusted for age, hospital, rank, cigarette smoking, alcohol consumption (lifelong non‐use, former use, current use of < 30, 30–59, or ≥ 60 mL alcohol/day) and body mass index. CI, confidence interval; OR, odds ratio.
Figure 1.
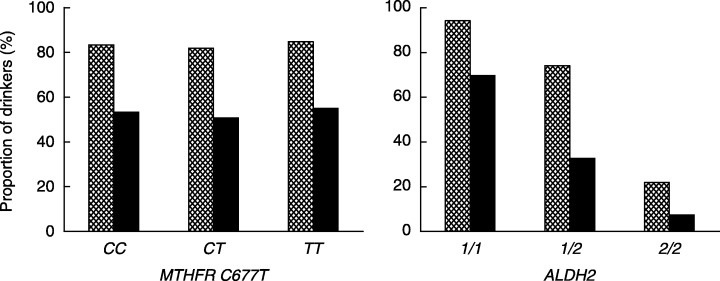
Proportions (%) of current alcohol drinkers (hatched bar) and those consuming ≥ 30 mL alcohol per day (black bar) according to MTHFR C677T and ALDH2 genotypes.
There was no measurable association between the MTHFR C677T polymorphism and colorectal adenoma (Table 1). Individuals with the ALDH2*2 allele showed non‐significant decreases in the OR of colorectal adenomas even after adjustment for alcohol consumption and other covariates.
When the relationship between the MTHFR C677T and ALDH2 polymorphisms and colorectal adenoma was analyzed in combination with alcohol consumption (Table 2), the interaction with alcohol consumption was almost null for either MTHFR C677T or ALDH2. High alcohol consumption (≥ 30 mL/day) was related to a moderate increase in the risk of colorectal adenoma regardless of these genetic polymorphisms. The adjusted OR for high versus low alcohol consumption were 1.44 (95% CI 1.04–2.01) among the ALDH2*1/1 homozygotes and 1.53 (95% CI 1.01–2.32) among the ALDH2*1/2 heterozygotes. A separate analysis was not carried out for the ALDH2*2/2 variant homozygotes because cases and controls with high alcohol consumption were extremely few.
Table 2.
Relationship between the MTHFR C677T and ALDH2 polymorphisms and colorectal adenoma in combination with alcohol consumption
| Genotype | < 30 mL alcohol/day | ≥ 30 mL alcohol/day | Interaction | ||
|---|---|---|---|---|---|
| n † | OR (95% CI) ‡ | n † | OR (95% CI) ‡ | ||
| MTHFR C677T | |||||
| CC | 62/186 | 1.00 (referent) | 120/213 | 1.58 (1.09–2.29) | P = 0.99 |
| CT | 75/244 | 0.90 (0.61–1.34) | 128/252 | 1.43 (0.99–2.06) | |
| TT | 23/70 | 0.95 (0.54–1.67) | 44/85 | 1.44 (0.90–2.32) | |
| ALDH2 | |||||
| 1/1 (wild type) | 67/185 | 1.00 (referent) | 232/420 | 1.41 (1.01–1.96) | P = 0.90 |
| 1/2 | 79/264 | 0.80 (0.55–1.18) | 58/126 | 1.18 (0.77–1.82) | |
| 2/2 (variant) | 14/51 | 0.70 (0.36–1.37) | 2/4 | 1.50 (0.26–8.61) | |
Numbers of cases/controls.
‡ Adjusted for age, hospital, rank, cigarette smoking and body mass index. CI, confidence interval; OR, odds ratio.
The association of the combined genotype of MTHFR C677T and ALDH2 with colorectal adenomas was examined after exclusion of those with the ALDH2*2/2 genotype (Table 3). Although the interaction between the two polymorphisms was not statistically significant, individuals with the MTHFR 677T allele seemed to have a lowered risk of colorectal adenoma when they were ALDH2*1/2 heterozygotes. Compared with those individuals having the ALDH2*1/1 and MTHFR 677CC genotypes, individuals with the ALDH2*1/2 genotype and MTHFR 677T allele (677CT and 677TT combined) had a statistically significant decrease in the risk of adenomas. There was no interaction between the combined genotype and alcohol consumption (data not shown). With allowance for the combined genotypes of the MTHFR C677T and ALDH2 polymorphism, adjusted OR for high (≥ 30 mL/day) versus low (< 30 mL/day) consumption of alcohol was 1.43 (95% CI 1.10–1.85).
Table 3.
Relationship between the combined genotypes of MTHFR C677T and ALDH2 and colorectal adenoma
| MTHFR C677T | ALDH2*1/1 | ALDH2*1/2 | Interaction | ||
|---|---|---|---|---|---|
| n † | OR (95% CI) ‡ | n † | OR (95% CI) ‡ | ||
| CC | 122/242 | 1.00 (referent) | 58/132 | 1.02 (0.69–1.52) | P = 0.41 |
| CT | 132/272 | 0.96 (0.71–1.31) | 60/200 | 0.69 (0.47–1.02) | |
| TT | 45/91 | 0.97 (0.63–1.49) | 19/58 | 0.72 (0.40–1.29) | |
| CC | 122/242 | 1.00 (referent) | 58/132 | 1.02 (0.69–1.52) | P = 0.18 |
| CT + TT | 177/363 | 0.96 (0.72–1.29) | 79/258 | 0.70 (0.49–1.00) § | |
Numbers of cases/controls.
Adjusted for age, hospital, rank, cigarette smoking, alcohol consumption (< 30 or ≥ 30 mL/day) and body mass index.
P = 0.048. CI, confidence interval; OR, odds ratio.
A decreased risk of colorectal adenoma associated with the MTHFR 677T allele among ALDH2*1/2 heterozygotes was more evident for large adenomas than for small adenomas, and slightly so for distal adenomas than for proximal adenomas (Table 4). The decrease in the OR of large adenomas was statistically significant. Again, the combined genotype of MTHFR C677T and ALDH2 did not modify the association with alcohol consumption for either small or large adenomas and for either proximal or distal adenomas (data not shown). The adjusted OR (95% CI) for high versus low alcohol consumption with further adjustment for the combined genotype were: small adenomas, 1.44 (1.05–1.96); large adenomas, 1.45 (1.00–2.10); proximal adenomas, 1.31 (0.84–2.06); and distal adenomas, 1.72 (1.22–2.44).
Table 4.
Relationship between the combined genotypes of MTHFR C677T and ALDH2 and colorectal adenoma according to size and site of adenoma
| MTHFR C677T | ALDH2*1/1 | ALDH2*1/2 | Interaction | ||
|---|---|---|---|---|---|
| n † | OR (95% CI) ‡ | n † | OR (95% CI) ‡ | ||
| Small adenomas | |||||
| CC | 65/242 | 1.00 (referent) | 35/132 | 1.12 (0.69–1.81) | P = 0.25 |
| CT + TT | 104/363 | 1.04 (0.73–1.48) | 50/258 | 0.81 (0.53–1.25) | |
| Large adenomas | |||||
| CC | 57/242 | 1.00 (referent) | 23/132 | 0.91 (0.52–1.58) | P = 0.36 |
| CT + TT | 73/363 | 0.87 (0.59–1.29) | 29/258 | 0.57 (0.34–0.95) | |
| Proximal colon adenomas | |||||
| CC | 28/242 | 1.00 (referent) | 12/132 | 0.92 (0.44–1.93) | P = 0.98 |
| CT + TT | 42/363 | 1.04 (0.62–1.73) | 25/258 | 0.97 (0.53–1.77) | |
| Distal colon and rectal adenomas | |||||
| CC | 58/242 | 1.00 (referent) | 30/132 | 1.13 (0.68–1.89) | P = 0.30 |
| CT + TT | 83/363 | 0.96 (0.65–1.40) | 38/258 | 0.77 (0.48–1.23) | |
Numbers of cases/controls.
Adjusted for age, hospital, rank, cigarette smoking, alcohol consumption (< 30 or ≥ 30 mL/day) and body mass index. CI, confidence interval; OR, odds ratio.
Discussion
The present study showed no measurable association between either the MTHFR C677T or ALDH2 polymorphisms and colorectal adenoma. Neither of the polymorphisms showed an effect on the association between alcohol consumption and colorectal adenomas.
Previous studies have generally found no measurable association between the MTHFR C677T polymorphism and colorectal adenoma.( 16 , 17 , 18 , 19 , 20 ) Results from these studies, however, are disparate regarding the interaction between the MTHFR C677T polymorphism and alcohol consumption on the risk of colorectal adenoma. Of the five studies,( 16 , 17 , 18 , 19 , 20 ) two showed an increased risk of colorectal adenoma associated with high alcohol intake among those with the 677TT genotype, while no clear association with alcohol was seen among those with the 677CC or CT genotypes.( 18 , 20 ) An earlier study of officials in the SDF also observed an increased risk among the 677TT homozygotes with high alcohol consumption.( 19 ) On the contrary, the Minnesota case‐control study reported a significant positive association with alcohol intake in those with the 677CC genotype (trend P = 0.005) and an inverse association in those with the 677TT genotype (trend P = 0.10), showing a statistically significant interaction.( 17 ) In the Nurses’ Health Study in the United States,( 16 ) a significant increase in risk was observed among the 677TT homozygotes with low alcohol consumption. Some of the inconsistent findings as to the interaction between alcohol intake and the MTHFR C677T polymorphism may be due to chance. Alternatively, inconsistency among the studies may be related to different folate levels in the study populations. The MTHFR C677T polymorphism could exert opposite effects in colorectal carcinogenesis depending on the folate pool because thymidylate synthesis and DNA methylation proceed at the expense of each other.
The present study did not examine the association between risk of colorectal adenoma and the MTHFR A1298C polymorphism, another functional polymorphism in the MTHFR gene.( 2 ) This polymorphism seems less relevant to the risk of colorectal cancer or adenomas,( 11 , 12 , 20 ) although a decreased risk of colon cancer was reported inconsistently for the 1298CC genotype in a subgroup analysis.( 13 ) The A1298C polymorphism was virtually unrelated to colorectal cancer in a large case‐control study in Japan.( 12 )
The ALDH2*1/2 heterozygosity was shown to be associated with an increased risk of alcohol‐related cancers and of colon cancer among Japanese alcoholics.( 25 ) A case‐control study in Japan also showed a slightly greater increase in the risk of colon cancer, but not rectal cancer, associated with high alcohol consumption among ALDH2 heterozygtes compared with ALDH2 wild‐type homozygotes.( 26 ) These results, taken together with the carcinogenic property of acetaldehyde, suggest that the ALDH2 variant allele might confer a greater risk of colorectal adenoma associated with alcohol consumption. The present study, however, provided no evidence for an interaction between the ALDH2 polymorphism and alcohol use on the risk of colorectal adenoma, whereas alcohol use itself was associated with a moderately increased risk of colorectal adenoma. The lack of interaction between alcohol and the ALDH2 polymorphism indicates that acetaldehyde metabolism in the liver may not be linked with the development of colorectal adenoma or cancer. Bacterial production of acetaldehyde in the colon is an alternative mechanism by which alcohol may enhance colorectal carcinogenesis.( 30 ) In rats, ethanol consumption results in a substantial increase in the concentration of acetaldehyde and decreases folate levels in the colonic mucosa.( 31 ) Furthermore, incubation of human colonic contents with alcohol results in significant production of acetaldehyde.( 32 ) It is notable that an increased risk of colorectal adenoma and cancer associated with alcohol consumption have been observed fairly consistently in different populations, including Caucasians( 21 ) in whom the frequency of the ALDH2*2 allele is virtually zero.( 24 )
Interestingly, the combination of variant alleles (ALDH2*2 and MTHFR 677T) was associated with a decreased risk of colorectal adenoma, especially of large adenomas. The findings suggest that individuals with the MTHFR 677T allele may have a lower risk of colorectal adenoma under certain conditions. It is possible that individuals with the ALDH2*2 allele may have had favorable dietary patterns other than lower consumption of alcohol, especially in terms of folate intake. The seemingly greater decrease in the risk of large adenomas associated with the coexistence of the ALDH2*2 and MTHFR C677T alleles may be ascribed to chance because cases were much fewer in the subgroup analysis by size. Nonetheless, this finding is compatible with a decreased risk of colorectal cancer associated with the MTHFR 677TT genotype under high‐folate and low‐alcohol conditions, and may be interpreted as suggesting that folate inhibits the growth of colorectal adenoma.
Different molecular alterations have been implicated in carcinogenesis of the proximal and distal sites of the colorectum.( 33 , 34 ) The MTHFR 677TT genotype was more strongly( 10 ) or exclusively( 14 ) associated with decreased risk of proximal colon cancer. In the present study, however, there was no clear difference in the association with either the MTHFR C677T or ALDH2 polymorphism between proximal and distal adenomas, although an increased risk of adenomas associated with high alcohol consumption seemed to be slightly greater for distal adenomas.
The present study had methodological advantages in that colonoscopy was done almost unselectively in a defined population and the absence of polyp lesions was confirmed in the control subjects by total colonoscopy. The study subjects were not representative of Japanese men in the general population, but selection was unlikely to exist with regard to the genetic polymorphisms under study. The allele frequency of MTHFR 677T (38% in the controls) is quite similar to those reported in random or non‐random samples of Japanese populations elsewhere.( 12 , 35 , 36 ) The frequency of the ALDH2*2 allele (24% in the controls) did not differ greatly from the somewhat variable frequencies observed in Japan; for example, the variant allele accounted for 17% in a random sample of 324 adult residents( 35 ) and 28% among 241 non‐cancer outpatients at a hospital.( 36 )
The lack of information regarding folate intake was a weakness of the present study. Although the questionnaire included questions on consumption frequency of selected food items, the method of estimating folate intake has not been established in the Self‐Defense Forces Health Study. Such information would be of value in clarifying the role of the MTHFR C677T polymorphism in the occurrence of colorectal adenoma. Whereas the statistical power was fairly high in addressing the overall association with each polymorphism, smaller numbers in the analysis of interaction necessarily resulted in a substantial decrease in the power. For example, the power of detecting a 1.5‐fold increase in risk for the ALDH2*1/2 versus ALDH2*1/1 genotype at the 5% significance level (two‐sided) was roughly estimated to be 93%, but it was no more than 44% when such an increase in risk was sought in individuals with ALDH2*1/2 and high alcohol use compared with those with ALDH2*1/1 and low alcohol use.
In summary, a case‐control study of Japanese men showed no measurable association of either the MTHFR C677T or ALDH2 polymorphisms with colorectal adenoma. While alcohol intake was associated with a moderate increase in the risk of colorectal adenoma, neither of the two polymorphisms modified the relationship between alcohol consumption and colorectal adenoma. The combination of ALDH2*2 and MTHFR 677T alleles was associated with a decreased risk of colorectal adenoma, especially of large adenomas, but the findings need further confirmation.
Acknowledgments
This work was supported by Grants‐in‐Aid for Scientific Research (B) (15390204) from the Japan Society for the Promotion of Science, and for Scientific Research on Priority Areas (12218226) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. The authors acknowledge supportive work by ward nurses at the SDF Fukuoka and Kumamoto Hospitals and the assistance of Ms Masumi Koga, Ms Kumiko Arie and Ms Ryoko Tanaka.
References
- 1. Lucock M. Folic acid: nutritional biochemistry, molecular biology and role in disease processes. Mol Genet Metab 2000; 71: 121–38. [DOI] [PubMed] [Google Scholar]
- 2. Sharp L, Little J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: a HuGE review. Am J Epidemiol 2004; 159: 423–43. [DOI] [PubMed] [Google Scholar]
- 3. Blount BC, Mack MM, Wehr CM et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 1997; 94: 3290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duthie SJ. Folic acid deficiency and cancer: mechanisms of DNA instability. Br Med Bull 1999; 55: 578–92. [DOI] [PubMed] [Google Scholar]
- 5. Toyota M, Ahuja N, Ohe‐Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999; 96: 8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pufulete M, Al‐Ghnaniem R, Leather AJ et al. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 2003; 124: 1240–8. [DOI] [PubMed] [Google Scholar]
- 7. Frosst P, Blom HJ, Milos R et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 1995; 10: 111–13. [DOI] [PubMed] [Google Scholar]
- 8. Chen J, Giovannucci E, Kelsey K et al. A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res 1996; 56: 4862–4. [PubMed] [Google Scholar]
- 9. Ma J, Stampfer MJ, Giovannucci E et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res 1997; 57: 1098–102. [PubMed] [Google Scholar]
- 10. Slattery ML, Potter JD, Samowitz W, Schaffer D, Leppert M. Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol Biomarkers Prev 1999; 8: 513–18. [PubMed] [Google Scholar]
- 11. Le Marchand L, Donlon T, Hankin JH, Kolonel LN, Wilkens LR, Seifried A. B‐vitamin intake, metabolic genes, and colorectal cancer risk (United States). Cancer Causes Control 2002; 13: 239–48. [DOI] [PubMed] [Google Scholar]
- 12. Yin G, Kono S, Toyomura K et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and colorectal cancer: The Fukuoka Colorectal Cancer Study. Cancer Sci 2004; 95: 908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keku T, Millikan R, Worley K et al. 5,10‐Methylenetetrahydrofolate reductase codon 677 and 1298 polymorphisms and colon cancer in African Americans and whites. Cancer Epidemiol Biomarkers Prev 2002; 11: 1611–20. [PubMed] [Google Scholar]
- 14. Toffoli G, Gafa R, Russo A et al. Methylenetetrahydrofolate reductase 677 C→T polymorphism and risk of proximal colon cancer in north Italy. Clin Cancer Res 2003; 9: 743–8. [PubMed] [Google Scholar]
- 15. Kim DH, Ahn YO, Lee BH, Tsuji E, Kiyohara C, Kono S. Methylenetetrahydrofolate reductase polymorphism, alcohol intake, and risks of colon and rectal cancers in Korea. Cancer Lett 2004; 216: 199–205. [DOI] [PubMed] [Google Scholar]
- 16. Chen J, Giovannucci E, Hankinson SE et al. A prospective study of methylenetetrahydrofolate reductase and methionine synthase gene polymorphisms, and risk of colorectal adenoma. Carcinogenesis 1998; 19: 2129–32. [DOI] [PubMed] [Google Scholar]
- 17. Ulrich CM, Kampman E, Bigler J et al. Colorectal adenomas and the C677T MTHFR polymorphism: evidence for gene–environment interaction? Cancer Epidemiol Biomarkers Prev 1999; 8: 659–68. [PubMed] [Google Scholar]
- 18. Levine AJ, Siegmund KD, Ervin CM et al. The methylenetetrahydrofolate reductase 677C>T polymorphism and distal colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev 2000; 9: 657–63. [PubMed] [Google Scholar]
- 19. Marugame T, Tsuji E, Inoue H et al. Methylenetetrahydrofolate reductase polymorphism and risk of colorectal adenomas. Cancer Lett 2000; 151: 181–6. [DOI] [PubMed] [Google Scholar]
- 20. Giovannucci E, Chen J, Smith‐Warner SA et al. Methylenetetrahydrofolate reductase, alcohol dehydrogenase, diet, and risk of colorectal adenomas. Cancer Epidemiol Biomarkers Prev 2003; 12: 970–9. [PubMed] [Google Scholar]
- 21. World Cancer Research Fund and American Institute for Cancer Research. Food, Nutrtion and the Prevention of Cancer: A Global Perspective. Washington, DC: American Institute for Cancer Research, 1997. [DOI] [PubMed] [Google Scholar]
- 22. Halsted CH, Villanueva JA, Devlin AM, Chandler CJ. Metabolic interactions of alcohol and folate. J Nutr 2002; 132: 2367S–72S. [DOI] [PubMed] [Google Scholar]
- 23. Feron VJ, Til HP, De Vrijer F, Woutersen RA, Cassee FR, Van Bladeren PJ. Aldehydes: occurrence, carcinogenic potential, mechanism of action and risk assessment. Mutat Res 1991; 259: 363–85. [DOI] [PubMed] [Google Scholar]
- 24. Brennan P, Lewis S, Hashibe M et al. Pooled analysis of alcohol dehydrogenase genotypes and head and neck cancer: a HuGE review. Am J Epidemiol 2004; 159: 1–16. [DOI] [PubMed] [Google Scholar]
- 25. Yokoyama A, Muramatsu T, Ohmori T et al. Alcohol‐related cancers and aldehyde dehydrogenase‐2 in Japanese alcoholics. Carcinogenesis 1998; 19: 1383–7. [DOI] [PubMed] [Google Scholar]
- 26. Murata M, Tagawa M, Watanabe S, Kimura H, Takeshita T, Morimoto K. Genotype difference of aldehyde dehydrogenase 2 gene in alcohol drinkers influences the incidence of Japanese colorectal cancer patients. Jpn J Cancer Res 1999; 90: 711–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuo K, Hamajima N, Shinoda M et al. Gene‐environment interaction between an aldehyde dehydrogenase‐2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis 2001; 22: 913–16. [DOI] [PubMed] [Google Scholar]
- 28. Kono S, Handa K, Hayabuchi H et al. Obesity, weight gain and risk of colon adenomas in Japanese men. Jpn J Cancer Res 1999; 90: 805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Toyomura K, Yamaguchi K, Kawamoto H et al. Relation of cigarette smoking and alcohol use to colorectal adenomas by subsite: the self‐defense forces health study. Cancer Sci 2004; 95: 72–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Visapaa JP, Tillonen J, Salaspuro M. Microbes and mucosa in the regulation of intracolonic acetaldehyde concentration during ethanol challenge. Alcohol Alcohol 2002; 37: 322–6. [DOI] [PubMed] [Google Scholar]
- 31. Homann N, Tillonen J, Salaspuro M. Microbially produced acetaldehyde from ethanol may increase the risk of colon cancer via folate deficiency. Int J Cancer 2000; 86: 169–73. [DOI] [PubMed] [Google Scholar]
- 32. Jokelainen K, Roine RP, Vaananen H, Farkkila M, Salaspuro M. In vitro acetaldehyde formation by human colonic bacteria. Gut 1994; 35: 1271–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993; 260: 816–19. [DOI] [PubMed] [Google Scholar]
- 34. Breivik J, Lothe RA, Meling GI, Rognum TO, Borresen‐Dale AL, Gaudernack G. Different genetic pathways to proximal and distal colorectal cancer influenced by sex‐related factors. Int J Cancer 1997; 74: 664–9. [DOI] [PubMed] [Google Scholar]
- 35. Yoshimura K, Hanaoka T, Ohnami S et al. Allele frequencies of single nucleotide polymorphisms (SNPs) in 40 candidate genes for gene–environment studies on cancer: data from population‐based Japanese random samples. J Hum Genet 2003; 48: 654–8. [DOI] [PubMed] [Google Scholar]
- 36. Hamajima N, Saito T, Matsuo K et al. Genotype frequencies of 50 polymorphisms for 241 Japanese non‐cancer patients. J Epidemiol 2002; 12: 229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]


