Abstract
Identification of reliable markers of chemo‐ and radiosensitivity and the key molecules that enhance the susceptibility of squamous esophageal cancer cells to anticancer treatments would be highly desirable. To test whether regenerating gene (REG) I expression enhances chemo‐ and radiosensitivity in esophageal squamous cell carcinoma cells, we used MTT (3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide) assays to compare the chemo‐ and radiosensitivities of untransfected TE‐5 and TE‐9 cells with those of cells stably transfected with REG Iα and Iβ. We then used flow cytometry to determine whether REG I expression alters cell cycle progression. No REG I mRNA or protein were detected in untransfected TE‐5 and TE‐9 cells. Transfection with REG Iα and Iβ led to strong expression of both REG I mRNA and protein in TE‐5 and TE‐9 cells, which in turn led to significant increases in both chemo‐ and radiosensitivity. Cell cycle progression was unaffected by REG I expression. REG I thus appears to enhance the chemo‐ and radiosensitivity of squamous esophageal cancer cells, which suggests that it may be a useful target for improved and more individualized treatments for patients with esophageal squamous cell carcinoma. (Cancer Sci 2008; 99: 2491–2495)
Thoracic squamous cell esophageal carcinoma is known for its rapid clinical progression and poor prognosis.( 1 , 2 ) Esophagectomy with extensive lymph node dissection is performed as a standard treatment for thoracic esophageal cancer; however, this procedure induces severe surgical stress and depression of host immunological defenses, and is associated with various surgical complications.( 3 , 4 ) On the other hand, there recently have been remarkable advances in definitive chemoradiotherapy (CRT) for thoracic esophageal cancer that offer patients a choice between surgery and definitive CRT as their first‐line and primary treatments.( 5 ) Esophageal squamous cell carcinomas generally show some degree of radiosensitivity, but individual tumors can exhibit widely differing susceptibility to radiotherapy; although CRT may be effective in some patients, others may show either no response or experience adverse effects. For those individuals, valuable time has been wasted, and the opportunity to obtain a potentially curative surgery may be lost.( 6 ) Thus, identification of reliable markers of chemoradiosensitivity and the key molecules that enhance chemoradiosensitivity in esophageal cancer cells would be highly desirable and has long been sought.
The human regenerating gene (REG) family belongs to the lectin superfamily and encodes five small, secreted proteins. REG I was originally isolated as an endogenous growth factor from pancreatic islet β cells.( 7 , 8 , 9 ) Since then, there have been many reports suggesting that REG plays important roles in tissue regeneration, cell proliferation, differentiation, mitogenesis, and carcinogenesis in various gastric and enteric tissues.( 10 , 11 , 12 , 13 , 14 , 15 , 16 ) Among the various functions of REG I, we focused on the relationship between REG I and chemo‐ and radiosensitivity because our earlier clinical study suggested that REG Iα expression in squamous cell esophageal carcinoma cells was a reliable marker of chemoradiosensitivity in advanced esophageal squamous cell cancer patients.( 17 , 18 ) That study was the first clinical report to show a correlation between survival and/or chemoradiosensitivity and REG I expression in thoracic squamous cell esophageal cancer. To confirm those clinical results and to test whether REG I expression enhances chemo‐ and radiosensitivity in esophageal squamous cell carcinoma cells, we have now carried out the present in vitro study.
Materials and Methods
Cell lines and culture. We obtained the TE‐5 and TE‐9 esophageal squamous cell carcinoma lines from the Cell Resource Center for Biochemical Research Institute of Development, Aging, and Cancer at Tohoku University, Japan( 19 ) All cells were cultured in RPMI‐1640 (Sigma‐Aldrich, St Louis, MO, USA) supplemented with 10% heat‐inactivated fetal bovine serum (Gibco, Grand Island, NY, USA) and antibiotics (penicillin G/streptomycin/amphotericin B; Gibco) in a humidified incubator at 37°C under an atmosphere of 5% CO2/95% air.
Isolation of stable transfectants expressing REG I protein. cDNA fragments encoding human REG Iα or REG Iβ (nucleotides 15–597 of M18963 and nucleotides 58–619 of D16816, respectively) were inserted into the XhoI/XbaI site of the pCI‐neo mammalian expression vector (Promega, Madison, WI, USA). The expression vectors or control vector (without insert DNA) were then introduced into TE‐5 and TE‐9 cells by electroporation, after which the cells were cultured in RPMI‐1640 supplemented with 10% fetal calf serum and 500 µg/mL Geneticin (Invitrogen, Grand Island, NY, USA) for 2 weeks. Stable transfectants expressing REG I protein were identified by immunoblot analysis of the culture medium prior to their isolation.( 20 , 21 , 22 )
Immunoblot analysis. Cells were cultured in 10‐cm dishes for 24 h, after which serum‐free RPMI‐1640 medium was added, and the cells were cultured for an additional 48 h. The supernatant was then collected, and the protein concentration was determined. Samples of extract containing 20 µg of protein were then fractionated by sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to polyvinylidene difluoride membranes (Immobilon; Millipore, Bedford, MA, USA), after which the membranes were incubated first with anti‐human REG I antibody (diluted 1:500 in TBS) for 1 h and then with peroxidase‐conjugated secondary anti‐mouse IgG (diluted 1:1000 in TBS) for 1 h. Immunodetection was accomplished using an ECL Western blotting detection reagents and analysis system (GE Healthcare, Waukesha, WI, USA). The membranes were subsequently exposed to X‐ray film.
Reverse transcriptase–polymerase chain reaction. Total RNA was isolated from each cell type using ISOGEN (Nippon Gene Co., Tokyo, Japan). cDNA was then reverse‐transcribed from 1‐µg samples of total RNA using a SuperScriptIII reverse transcriptase kit (Invitrogen) according to the manufacturer's instructions. Polymerase chain reaction (PCR) was then carried out using the primers shown in Table 1 and Platinum Taq DNA polymerase (Invitrogen). The amplification protocol entailed incubation at 94°C for 2 min, followed by 35 cycles (for REG Iα and REG Iβ), 55°C for 30 s, and 30 cycles (for glyceraldhyde‐3‐phosphate dehydrogenase [GAPDH]) at 94°C for 30 s or 72°C for 60 s. The amplified products were subjected to 1.5% agarose gel electrophoresis and visualized by staining with ethidium bromide.
Table 1.
Primer sequences used for real‐time reverse transcriptase–polymerase chain reaction
| Primer sequence (5′–3′) | Size | |
|---|---|---|
| REG Iα | 5′‐AACATGAATTCGGGCAACC‐3′ 5′‐AGGAGAACTTGTCTTCACAA‐3′ | 480 |
| REG Iβ | 5′‐GCAAGAGATTCACTGCCGCTAA‐3′ 5′‐GCAGGACCAGTTCTAGACATCC‐3′ | 397 |
| GAPDH | 5′‐CGGAGTCAACGGATTTGGTCGTAT‐3′ 5′‐AGCCTTCTCCATGGTGGTGAAGAC‐3′ | 306 |
Radiotherapy and chemotherapy for cultured cells. Cells were exposed to 0, 5, or 10 Gy radiation using a SOFTEX M‐100WE operating at 100 KV and 5 mA, which delivered the dose at 0.01 Gy/min. Chemotherapy involved the application of 5‐fluorouracil (5‐FU; Kyowa Hakko Kogyo Co., Tokyo, Japan) and cisplatin (CDDP; Nihonkayaku Co., Tokyo, Japan) to the cultures to final concentrations of 0.1, 1.0, 5.0, or 10 µM.
Cell proliferation assay. Cell proliferation was assessed using a Cell Counting Kit‐8 (Dojindo, Kumamoto, Japan). Cells were plated in 96‐well plates to a density of 1 × 103 cells/well and incubated for 0, 1, 2, 3, or 4 days. After addition of 10 µL of Cell Counting Kit‐8 reagent, the cells were incubated for 2 h, after which the optical densities of the plates were read at 450 nm using a Model 550 Microplate Reader (Bio‐Rad Laboratories).
Cell viability was assessed using colorimetric MTT (3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide) assays (Sigma‐Aldrich). Cells were plated in 96‐well plates to a density of 1 × 103 cells/well and incubated for 24 h at 37°C. They were then irradiated at 0, 5, or 10 Gy, and treated with one of the indicated concentrations of either CDDP or 5‐FU. For concurrent therapy, the cells were plated as described above, and CDDP (0–10 µM) was added to the cultures; then 4 h later the cells were irradiated (5 Gy). Following incubation for an additional 72 h, 10 µL of 5.5 mg/mL MTT was added to the cultures, which were then incubated for 4 h at 37°C. Thereafter, 90 µL of extraction solution (40% N,N‐dimethylformamide, 2% CH3COOH, 20% SDS, and 0.03 N HCl) was added, and the mixture was incubated for 2 h at room temperature. Finally, cell viability was determined by measuring the optical density at 570 nm using a microplate reader.
Flow cytometry. Cells (1 × 103) were cultured in 6‐cm dishes for 2 days at 37°C, trypsinized, rinsed in phosphate‐buffered saline (PBS), and fixed in 70% ethanol. Prior to analysis, the cells were centrifuged to remove the ethanol and resuspended in 500 µL of 50 µg/mL propidium iodide, 20 µg/mL RNase A, 0.1% (w/v) sodium citrate, and 0.3% (w/v) Nonident P‐40. After incubation for 1 h at 37°C, the cells were examined using a flow cytometer (Cytomics FC500; Beckman Coulter, Miami, FL, USA). The data obtained were analyzed using CXP software version 2 (Beckman).
Statistical analysis. Data were expressed as mean values. Significant differences between groups were assessed using one‐way analysis of variance (anova) with the Dunnett multiple comparison test and χ2‐test (Stat View J‐5.0; Abacus Concepts, Berkeley, CA, USA). Values of P < 0.01 were considered significant.
Results
Chemo‐ and radiosensitivity of cells stably transfected with REG I. We stably transfected TE‐5 and TE‐9 cells with REG Iα and REG Iβ DNA, after which the expression of REG I mRNA and protein was assessed. TE‐5 and TE‐9 transfectants (TE‐5 REG Iα/TE‐5 REG Iβ and TE‐9 REG Iα/TE‐9 REG Iβ cells) showed stronger expression of REG I than control cells, or cells transfected with the neomycin‐resistance gene alone (mock‐transfected) (Fig. 1). When we measured the chemo‐ and radiosensitivities of the transfectants in MTT assays, we found that there were no differences between the growth rates of cells stably transfected with REG I and the control cells (TE‐5 mock and TE‐9 mock) (Fig. 2a). On the other hand, both TE‐5 REG Iα/TE‐5 REG Iβ and TE‐9 REG Iα/TE‐9 REG I β cells showed a significant increase in radiosensitivity (5 and 10 Gy) and chemosensitivity (10 µM CDDP), as compared with TE‐5 mock and TE‐9 mock cells (Fig. 2b,c). And in concurrent experiments, TE‐5 REG Iα/TE‐5 REG Iβ and TE‐9 REG Iα/TE‐9 REG I β cells showed significantly higher chemoradiosensitivity than control cells (Fig. 2d). Thus exogenous expression of REG I by esophageal cancer cells enhanced their chemo‐ and radiosensitivity.
Figure 1.
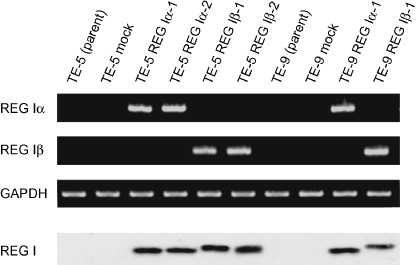
TE‐5 and TE‐9 cells stably transfected with regenerating gene (REG) Iα and REG Iβ DNA expressed REG I mRNA and protein.
Figure 2.

Proliferation of esophageal cancer cells stably transfected with regenerating gene (REG) I was measured as a function of WST‐8 cleavage (a). Cell viability was assessed using MTT (3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide) assays. Cells were treated with radiation at dose of 5 Gy or 10 Gy (b), with 0.1, 1.0, 5.0, or 10 µM cisplatin (c), or with a combination of 5 Gy radiation and 0.1, 1.0, 5.0, or 10 µM cisplatin (d). REG I transfectants were significantly more susceptible to chemo‐, radio‐ and chemoradiotherapy than control cells. All results are expressed as the means ± SD of 10 samples; *P < 0.01.
Cell cycle progression in TE cells stably transfected with REG I. Next, we used flow cytometry to test whether cell cycle progression was altered by REG I transfection. We found that all of the cell lines had very similar cell cycle distribution profiles, indicating that REG I expression does not affect cell cycle progression (Table 2).
Table 2.
Cell‐cycle distribution in control cells and regenerating gene (REG) I stable transfectants
| Cell lines | G1/G0 (%) | S (%) | G2/M (%) |
|---|---|---|---|
| TE‐5 (parent) | 34 | 23 | 43 |
| TE‐5 mock | 38 | 25 | 37 |
| TE‐5 REG Iα‐1 | 40 | 36 | 34 |
| TE‐5 REG Iα‐2 | 40 | 25 | 35 |
| TE‐5 REG Iβ‐1 | 42 | 24 | 34 |
| TE‐5 REG Iβ‐2 | 38 | 27 | 35 |
| TE‐9 (parent) | 39 | 28 | 33 |
| TE‐9 mock | 44 | 25 | 31 |
| TE‐9 REG Iα‐1 | 42 | 27 | 31 |
| TE‐9 REG Iβ‐1 | 40 | 29 | 31 |
There were no significant differences.
Discussion
In the present study, we showed that REG I transfection enhanced both chemo‐ and radiosensitivity in squamous cell esophageal cancer.
Cellular responses to DNA damage constitute an important field in cancer biology. Most therapeutic modalities currently used to treat malignancies, including radiation therapy and many chemotherapeutic agents, target DNA. When normal cells are damaged by radiation or chemotherapeutic agents, cell‐cycle checkpoints sense DNA damage and activate pathways that lead to DNA repair.( 23 , 24 ) Cancer cells that are highly proliferative are generally considered to be more susceptible to DNA damage because checkpoint dysfunction leads to enhanced cell cycle progression, which makes it more likely that there will be insufficient time to repair DNA damage and circumvent apoptosis.( 25 ) Thus the loss of cell‐cycle checkpoint responses likely results in increased cellular susceptibility to chemoradiotherapy, particularly if the affected checkpoint controls the G2 transition.
The molecular basis of both radiosensitivity and chemosensitivity is a complex product of both cellular and tissue responses. Among the affected molecules, p53 is reported to have a major impact on the cellular responses to ionizing radiation and cytotoxic drugs, but there are inconsistencies in the reported mechanisms by which p53 affects cell survival.( 26 ) When we compared the expression of p53 mRNA and protein in REG‐I transfected and mock‐transfected TE‐5 cells, we found no difference in the p53 status (data not shown). Moreover, Barnas et al. reported that TE‐9 cells express truncated forms of p53 as a result of frameshift mutations and that these truncated forms did not exhibit p53‐DNA binding activity.( 27 ) We therefore suggest that REG I enhances chemo‐ and radiosensitivity in a manner that is independent of p53. Numerous other molecules are known to be involved in the cellular responses to anticancer therapy, including those encoded by cyclin D, p21, p16, ras, raf‐1, bcl‐2, Ki‐67, EGFR, RUNX3, TRAIL, and PARP, among others, but the molecular mechanisms involved remain unclear.( 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 ) Overall, there is much that is still not understood about the complex roles played by many proteins during the various responses to treatment.
The results of this in vitro study demonstrate that REG I could enhance the sensitivity of squamous cell esophageal carcinoma cells to anticancer treatments. By contrast, Mitani et al. reported that overexpression of REG IV inhibited 5‐FU‐induced apoptosis.( 39 ) While the detailed mechanisms are unclear, it appears that the REG family can have opposing effects on chemo‐ and/or radiosensitivity. For instance, Takasawa et al. demonstrated that REG activates cyclin D1 signaling, which correlates with cell cycle progression and radiosensitivity,( 40 ) whereas Sekikawa et al. reported that the REG I mediates the anti‐apoptotic effects of STAT3 signaling by enhancing Akt activation, Bad phosphorylation, and Bcl‐xL expression.( 41 ) Considering that both the cyclin D1 and Akt/Bad/Bcl‐xL pathways can be activated by REG, the sensitivity of cancer cells to particular chemotherapeutic agents may reflect the relative impacts of these and other signaling pathways.
In conclusion, the present results demonstrate that REG I enhances the susceptibility of cancer cells to anticancer treatments. REG I may thus be a useful target for anticancer therapy, enabling us to design better, more individualized treatments for patients with advanced esophageal squamous cell carcinoma.
Acknowledgments
This work was supported, in part, by Grants‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sience, Sports, and Technology of Japan, and Charitable Trust Maeta Toyokichi Memorial Fund.
References
- 1. Enzinger PC, Mayer RJ. Esophageal cancer. N Eng J Med 2003; 349: 2241–51. [DOI] [PubMed] [Google Scholar]
- 2. Ando N, Ozawa S, Kitagawa Y, Shinozawa Y, Kitajima M. Improvement in the results of surgical treatment of advanced squamous esophageal carcinoma during 15 consecutive years. Ann Surg 2000; 232: 225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tashiro T, Yamamori H, Takagi K et al . Changes in immune function following surgery for esophageal carcinoma. Nutrition 1999; 15: 760–6. [DOI] [PubMed] [Google Scholar]
- 4. Fujita H, Kakegawa T, Yamana H et al . Mortality and morbidity rates, postoperative course, quality of life and prognosis after radical lymphadenectomy for esophageal cancer. Ann Surg 1995; 222: 654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hironaka S, Ohtsu A, Boku N et al . Nonrandomized comparison between definitive chemoradiotherapy and radical surgery in patients with T(2–3) N(any) M(0) squamous cell carcinoma of the esophagus. Int J Radiat Oncol 2003; 57: 425–33. [DOI] [PubMed] [Google Scholar]
- 6. Ishikura S, Nihei K, Outhu A, Boku N et al . Long‐term toxicity after definitive chemoradiotherapy for squamous cell carcinoma of the thoracic esophagus. J Clin Oncol 2003; 21: 2697–702. [DOI] [PubMed] [Google Scholar]
- 7. Terazono K, Yamamoto H, Takasawa S et al . A novel gene activated in regenerating islets. J Biol Chem 1988; 263: 2111–14. [PubMed] [Google Scholar]
- 8. Sanchez D, Figarella C, Marchand‐Pinatel S, Bruneau N, Guy‐Crotte O. Preferential expression of Reg Iβ gene in human adult pancreas. Biochem Biophys Res Commun 2001; 284: 729–37. [DOI] [PubMed] [Google Scholar]
- 9. Hervieu V, Christa L, Gouysse G et al . HIP/PAP, a number of the reg family, is expressed in glucagon‐producing enteropancreatic endocrine cells and tumors. Hum Pathol 2006; 37: 1066–75. [DOI] [PubMed] [Google Scholar]
- 10. Fukui H, Kinoshita Y, Maekawa T et al . Regenerating gene protein may mediate gastric mucosal proliferation induced by hypergastrinemia in rats. Gastroenterology 1998; 115: 1483–93. [DOI] [PubMed] [Google Scholar]
- 11. Bishinupuri KS, Luo Q, Murmu N et al . Reg IV activates the epidermal growth factor receptor/Akt/AP‐1 signaling pathway in colon adenocarcinomas. Gastroenterology 2006; 130: 137–49. [DOI] [PubMed] [Google Scholar]
- 12. Yonemura Y, Sakakura S, Yamamoto H et al . REG gene expression is associated with the infiltrating growth of gastric carcinoma. Cancer 2003; 98: 1394–400. [DOI] [PubMed] [Google Scholar]
- 13. Shinozaki S, Nakamura T, Iimura M et al . Upregulation of Reg Iα and GW112 in the epithelium of inflamed colonic mucosa. Gut 2001; 48: 623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ose T, Kadowaki Y, Fukuhara H et al . Reg I‐knockout mice reveal its role in regulation of cell growth that is required in generation and maintenance of the villous structure of small intestine. Oncogene 2007; 26: 349–59. [DOI] [PubMed] [Google Scholar]
- 15. Harada K, Zen Y, Kanemori Y et al . Human REG I gene is up‐regulated in intrahepatic cholangiocarcinoma and its precursor lesions. Hepatology 2001; 33: 1036–42. [DOI] [PubMed] [Google Scholar]
- 16. Cavard C, Terris B, Grimber G et al . Overexpression of regenerating islet‐derived 1 alpha and 3 alpha genes in human primary liver tumors with β‐catenin mutations. Oncogene 2006; 25: 599–608. [DOI] [PubMed] [Google Scholar]
- 17. Motoyama S, Sugiyama T, Ueno Y et al . REG I expression predicts long‐term survival among locally advanced thoracic squamous cell esophageal cancer patient treated with neoadjuvant chemoradiotherapy followed by esophagectomy. Ann Surg Oncol 2006; 13: 1724–31. [DOI] [PubMed] [Google Scholar]
- 18. Hayashi K, Motoyama S, Sugiyama T et al . REG Iα is a Reliable marker of chemoradiosensitivity in squamous cell esophageal cancer patients. Ann Surg Oncol 2008; 15: 1224–31. [DOI] [PubMed] [Google Scholar]
- 19. Nishihira T, Hashimoto Y, Katayama M et al . Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol 1993; 119: 441–9. [DOI] [PubMed] [Google Scholar]
- 20. Takasawa S, Akiyama T, Nata K et al . Cycslic ADP‐ribose and inositol 1,4,5‐trisphosphate as alternate second messengers for intracellular Ca2+ mobilization in normal and diabetic β‐cells. J Biol Chem 1998; 273: 2497–500. [DOI] [PubMed] [Google Scholar]
- 21. Kobayashi S, Akiyama T, Nata K et al . Identification of a receptor for Reg (regenerating gene) protein, a pancreatic β‐cell regenerating factor. J Biol Chem 2000; 275: 10723–6. [DOI] [PubMed] [Google Scholar]
- 22. Okamoto H, Takasawa S, Tohgo A et al . Synthesis and hydrolysis of cyclic ADP‐ribose by human leukocyte antigen CD38: inhibition of hydrolysis by ATP and physiological significance. Meth Enzymol 1997; 280: 306–18. [DOI] [PubMed] [Google Scholar]
- 23. Kastan MB, Bartek J. Cell‐cycle checkpoints and cancer. Nature 2004; 432: 316–23. [DOI] [PubMed] [Google Scholar]
- 24. Sheridan MT, O'Dwyer T, Seymour CB, Mothersill CE. Potential indicators of radiosensitivity in squamous cell carcinoma of the head and neck. Radiat Oncol Invest 1997; 5: 180–6. [DOI] [PubMed] [Google Scholar]
- 25. Xia F, Powell SN. The molecular basis of radiosensitivity and chemosensitivity in the treatment of breast cancer. Semin Radiat Oncol 2002; 12: 296–304. [DOI] [PubMed] [Google Scholar]
- 26. Okumura H, Natsugoe S, Matsumoto M et al . The predictive value of p53, p53R2, and p21 for the effect of chemoradiation therapy on oesophageal squamous cell carcinoma. Br J Surg 2005; 92: 284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barnas C, Martel‐Planche G, Furukawa Y et al . Inactivation of the p53 protein in cell lines derived from human esophageal cancers. Int J Cancer 1997; 71: 79–87. [DOI] [PubMed] [Google Scholar]
- 28. Trent S, Yang C, Li C, Lynch M, Schmidt EV. Heat shock protein B8, a cyclin‐dependent kinase‐independent cyclin D1 target gene, contributes to its effects on radiation sensitivity. Cancer Res 2007; 67: 10774–81. [DOI] [PubMed] [Google Scholar]
- 29. Hsaio M, Tse V, Carmel J et al . Functional expression of human p21 (WAFI/CIPI) gene in rat glioma cells suppresses tumor growth in vivo and induces radiosensitivity. Biochem Biophys Res Commun 1997; 233: 329–35. [DOI] [PubMed] [Google Scholar]
- 30. Matsumura Y, Yamagishi N, Miyakoshi J, Imamura S, Takebe H. Increase in radiation sensitivity of human malignant melanoma cells by expression of wild‐type p16 gene. Cancer Lett 1997; 115: 91–6. [DOI] [PubMed] [Google Scholar]
- 31. Sklar MD. The ras oncogenes increase the intrinsic resistance of NIH 3T3 cells to ionizing radiation. Science 1988; 239: 645–7. [DOI] [PubMed] [Google Scholar]
- 32. Kasid U, Pfeifer A, Weichselbaum RR, Dritschilo A, Mark GE. The raf oncogene is associated with a radiation‐resistant human laryngeal cancer. Science 1987; 237: 1039–41. [DOI] [PubMed] [Google Scholar]
- 33. Kumar P, Coltas IK, Kumar B, Chepeha DB, Bradford CR, Polverini PJ. Bcl‐2 protects endothelial cells against gamma‐radiation via a Raf‐MEK‐ERK‐survivin signaling pathway that is independent of cytochrome c release. Cancer Res 2007; 67: 1193–202. [DOI] [PubMed] [Google Scholar]
- 34. Takeuchi H, Ozawa S, Ando N et al . Cell‐cycle regulators and the Ki‐67 labeling index can predict the response to chemoradiotherapy and the survival of patients with locally advanced squamous cell carcinoma of the esophagus. Ann Surg Oncol 2003; 10: 792–800. [DOI] [PubMed] [Google Scholar]
- 35. Wang KL, Wu TT, Choi IS et al . Expression of epidermal growth factor receptor in esophageal and esophagogastric junction adenocarcinomas. Cancer 2007; 109: 658–67. [DOI] [PubMed] [Google Scholar]
- 36. Sakakura C, Miyagawa K, Fukuda KI et al . Frequent silencing of RUNX3 in esophageal cell carcinomas is associated with radioresistance and poor prognosis. Oncogene 2007; 26: 5927–38. [DOI] [PubMed] [Google Scholar]
- 37. Verbrugge I, Vries ED, Tait SWG et al . Ionizing radiation modulates the TRAIL death‐inducing signaling complex, allowing bypass of the mitochondrial apoptosis pathway. Oncogene 2008; 27: 574–84. [DOI] [PubMed] [Google Scholar]
- 38. Albert JM, Cao C, Kim KW et al . Inhibition of poly (ADP‐ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin Cancer Res 2007; 13: 3033–42. [DOI] [PubMed] [Google Scholar]
- 39. Mitani Y, Matsumura S, Yoshida K et al . Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5‐fluorouracil‐based chemotherapy. Oncogene 2007; 26: 4383–93. [DOI] [PubMed] [Google Scholar]
- 40. Takasawa S, Ikeda T, Akiyama T et al . Cyclin D1 activation through ATF‐2 in Reg‐induced pancreatic β‐cell regeneration. FEBS Lett 2006; 580: 585–91. [DOI] [PubMed] [Google Scholar]
- 41. Sekikawa A, Fukui H, Fujii S et al . REG Iα protein mediates an anti‐apoptotic effect of STAT3 signaling in gastric cancer cells. Carcinogenesis 2007; 29: 76–83. [DOI] [PubMed] [Google Scholar]


