Abstract
The cell cycle is strictly regulated by numerous mechanisms to ensure cell division. The transcriptional regulation of cell‐cycle‐related genes is poorly understood, with the exception of the E2F family that governs the cell cycle. Here, we show that a transcription factor, zinc finger protein 143 (ZNF143), positively regulates many cell‐cycle‐associated genes and is highly expressed in multiple solid tumors. RNA‐interference (RNAi)‐mediated knockdown of ZNF143 showed that expression of 152 genes was downregulated in human prostate cancer PC3 cells. Among these ZNF143 targets, 41 genes (27%) were associated with cell cycle and DNA replication including cell division cycle 6 homolog (CDC6), polo‐like kinase 1 (PLK1) and minichromosome maintenance complex component (MCM) DNA replication proteins. Furthermore, RNAi of ZNF143 induced apoptosis following G2/M cell cycle arrest. Cell growth of 10 lung cancer cell lines was significantly correlated with cellular expression of ZNF143. Our data suggest that ZNF143 might be a master regulator of the cell cycle. Our findings also indicate that ZNF143 is a member of the growing list of non‐oncogenes that are promising cancer drug targets. (Cancer Sci 2010; 101: 2538–2545)
Transcriptional regulation of gene expression requires the orchestrated recruitment of transcription factors by sequence‐specific DNA binding regulators. Staf was initially identified as the transcriptional activator of the RNA polymerase III‐dependent Xenopus tRNA gene.( 1 ) It has recently been shown that its human ortholog zinc finger protein 143 (ZNF143; formerly known as hStaf) is also involved in RNA‐polymerase‐II‐dependent gene transcription.( 2 , 3 ) The DNA binding domain of ZNF143 is located in the central part of the protein and consists of seven zinc finger domains.( 1 , 4 ) The ZNF143 DNA binding site is thought to be at least 18 bp long because the protein has seven zinc fingers; by analogy, the well‐known zinc finger transcription factor Sp family and members of the KLF family contain three zinc fingers and recognize DNA sequences approximately 6 bp long.( 5 ) Recently, it has been shown by a bioinformatics approach that ZNF143 binding sites are widely distributed in the CpG island‐type promoters of the human genome.( 6 ) Functional classification of ZNF143 target genes has revealed that many of the identified genes are important for cell growth: 27% of the genes are categorized as cell cycle/DNA replication/DNA repair proteins.
We have previously reported that expression of ZNF143 is induced by DNA‐damaging agents and is enhanced in cisplatin‐resistant cell lines.( 7 ) ZNF143 binds preferentially to cisplatin‐modified DNA. We also found that ZNF143 binding sites are frequently found in the promoter region of DNA repair genes.
Here, we investigated the ZNF143 target genes by RNA interference (RNAi). One hundred and fifty‐two genes were downregulated by ZNF143‐specific small interfering RNA (siRNA) transfection. Among them, 41 genes are categorized as concerned with cell cycle and DNA replication. ZNF143 was highly expressed in cancer cells when compared with non‐tumor regions. Downregulation of ZNF143 effectively induced G2/M cell cycle arrest and apoptosis, indicating that ZNF143 might be a promising molecular target for anti‐cancer drugs.
Materials and Methods
Cell culture and antibodies. Human prostate cancer cells PC3,( 7 ) human cervical cancer cells HeLa( 7 ) and human bladder cancer cells T24( 8 ) were cultured in minimum essential medium (MEM). Human colon cancer cells Caco‐2( 9 ) and DLD1,( 10 ) human glioblastoma CCF‐STTG1( 11 ) and human astrocytoma cells U343( 11 ) were cultured in Dulbecco’s modified Eagle’s minimal essential medium. Human lung cancer cells A549,( 12 ) B203L,( 12 ) PC9,( 12 ) A110L,( 12 ) QG56,( 12 ) SQ1,( 12 ) B1203L,( 12 ) PC10,( 12 ) 904L( 12 ) and A529L( 12 ) were cultured in RPMI 1640 medium. These media were purchased from Nissui Seiyaku (Tokyo, Japan) and contained 10% fetal bovine serum. Cell lines were maintained in a 5% CO2 atmosphere at 37°C. Antibodies against peroxiredoxin 4 (PRDX4) (sc‐23974), minichromosome maintenance complex component 2 (MCM2) (sc‐9839), MCM3 (sc‐9850), MCM4 (sc‐28317), MCM5 (sc‐22780) and MCM6 (sc‐9843) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against Flag (M2) and β‐actin (A5441) were purchased from Sigma Aldrich (St Louis, MO, USA). Antibodies against aurora B kinase (AURKB) (1788‐1) and nuclear factor (erythroid‐derived 2)‐like 2 (Nrf2) (2073‐1) were purchased from Epitomics (Burlingame, CA, USA). Antibodies against PLK1 (37‐7000) and CDC6 (05‐550) were purchased from Invitrogen (Carlsbad, CA, USA) and Upstate Biotech (Lake Placid, NY, USA), respectively. The polyclonal antibody against the high mobility group B2 (HMGB2) was described previously.( 13 ) The polyclonal antibody against ZNF143 was raised by multiple immunizations of a New Zealand white rabbit with synthetic peptides. The synthetic peptide sequences were MLLAQINRDSQGMTEFPGGGMEAQHVTLC and QLGEQPSLEEAIRIASRIQQGETPGLDD for ZNF143.
Plasmid construction. To prepare luciferase (luc) reporter plasmids, genomic DNA was amplified with the following primer pairs: CDC6 luc1, 5′‐AGATCTCGCCTCCCAGCGTGCTTTGCGG‐3′ and 5′‐AAGCTTCACCCAGCTCCGCTGCCTCAC‐3′; CDC6 luc2, 5′‐AGATCTACTACGCCTCCCAGCGTGCTTTGCGG‐3′ and 5′‐AAGCTTCACCCAGCTCCGCTGCCTCAC‐3′; PLK1 luc, 5′‐AGATCTCCGCATCCACGCCGGGTTTGG‐3′ and 5′‐AAGCTTGCGCTGCAGACCTCGATCCGAGC‐3′; AURKB luc, 5′‐AGATCTCACTGGGGGAATTTGGGGAAAC‐3′ and 5′‐AAGCTTGGGGTCCAAGGCACTGCTACTC‐3′; MCM2 luc, 5′‐AGATCTCGAACTCCTGAGCTTGTGATCC‐3′ and 5′‐AAGCTTCTCCGCCACTACAGCAACAACC‐3′; and HMGB2 luc, 5′‐AGATCTGGGCGGCGCACGGGAGACCC‐3′ and 5′‐AAGCTTCCCGCAGAGCGGCCGGACCC‐3′. Restriction enzyme sites are underlined. These PCR products were cloned and ligated into the BglII‐HindIII site of the pGL3‐basic vector (Promega, Madison, WI, USA). The pGL3‐P2 (promoter vector) containing an SV40 promoter upstream of the luciferase gene was purchased from Promega. Preparation of 3xFlag‐ZNF143 expression plasmid was described previously.( 7 )
Knockdown with siRNA. The following double‐stranded RNA 25‐bp oligonucleotides were commercially generated (Invitrogen): ZNF143 siRNA, siZNF #1, 5′‐GCTGGAAGATGGTA‐CCACAGCTTAT‐3′ (sense) and 5′‐ATAAGCTGTGGTACC‐ATCTTCCAGC‐3′ (antisense); and siZNF #2, 5′‐GGACGAC‐GTTGTTTCTACACAAGTA‐3′ (sense) and 5′‐TACTTGTGT‐AGAAACAACGTCGTCC‐3′ (antisense). Two hundred picomoles of siRNA were mixed with 10 μL Lipofectamine 2000 (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. After 20 min, 1 × 106 cancer cells were gently mixed and incubated for a further 20 min. Transfected cells were used for oligonucleotide microarray study, cell proliferation assays, flow cytometry and western blotting.
Oligonucleotide microarray study and microarray analysis. A microarray procedure was performed as described previously.( 14 ) In brief, total RNA extracts were collected from PC3 cells transfected with ZNF143 #2 siRNA or control siRNA in duplicate, as described above. Eight GeneChips (Affymetrix, Santa Clara, CA, USA) were used for analysis. The microarray analysis was performed using the BRB Array Tools software ver. 3.3.0 (http://linus.nci.nih.gov/BRB‐ArrayTools.html) developed by Dr Richard Simon and Amy Peng.
Cell proliferation assays. The cell proliferation assay was described previously.( 15 ) Briefly, siRNA‐transfected cancer cells were seeded into 12‐well plates at a density of 5 × 104 cells per well. Twenty‐four hours after transfection was set as time zero. The cells were harvested by trypsinization and counted every 24 h with a Coulter‐type cell size analyzer (CDA‐500; Sysmex Corp., Kobe, Japan).
Flow cytometry. Flow cytometry was described previously.( 15 ) Briefly, siRNA‐transfected PC3 cells (1 × 106) were seeded into 90‐mm plates. After 72 h, the cells were analyzed using an EpicsXL‐MCL flow cytometer (Beckman‐Coulter, Miami, FL, USA). For assessment of apoptosis, an Annexin V‐fluorescein isothiocyanate (FITC) Apoptosis Detection kit II (BD Biosciences, San Jose, CA, USA) was used.
Western blotting. Transfection of expression plasmid and preparation of nuclear proteins was described previously.( 7 ) ZNF143 expression plasmid‐ or siRNA‐transfected PC3 cells were collected after 48–72 h and nuclear protein or whole cell lysate was subjected to western blotting. Detection was performed using enhanced chemiluminescence (Amersham, Piscataway, NJ, USA). The protein expression levels were quantitated using a Multi Gauge Version 3.0 (Fujifilm, Tokyo, Japan).
Chromatin immunoprecipitation (ChIP) assays. The cloning of PC3 cells stably expressing 3xFlag‐tagged ZNF143 has been described previously.( 7 ) Purified DNA from these cells was used for PCR analysis with the indicated primer pairs (Table S1). The PCR products were separated by electrophoresis on 2% agarose gels and were stained with ethidium bromide.
Reporter assays. One × 105 PC3 cells per well were seeded into 12‐well plates. The following day, 1 μg luciferase reporter plasmid was transfected with 30 pmol ZNF143 siRNA or control siRNA using 5 μL Lipofectamine 2000 (Qiagen). After 48 h, luciferase activity was detected using a Picagene kit (Toyo Ink, Tokyo, Japan). Light intensity was measured using a luminometer (Luminescencer JNII RAB‐2300; Atto, Japan). The results shown are normalized for protein concentration measured using the Bradford method, and are representative of at least three independent experiments.
Human tissue samples. Samples were obtained from the surgical pathology archive of the Department of Pathology and Cell Biology of The University of Occupational and Environmental Health in Kitakyushu, Japan. The selected tissues consisted of one surgically resected case each of esophageal carcinoma, gastric adenocarcinoma, squamous cell carcinoma of the lung, urothelial carcinoma of the urinary bladder, testicular seminoma and cerebral astrocytoma. These cases were classified according to the World Health Organization Histological Typing of each tissue. The diagnosis was re‐evaluated and confirmed by at least three board‐certified surgical pathologists who had examined formalin‐fixed, paraffin‐embedded tissue sections stained with hematoxylin and eosin or other appropriate immunohistochemical stains.
Immunohistochemistry. The specimens were fixed in 20% formaldehyde and embedded in paraffin. Four micrometer‐thick tissue sections were deparaffinized, dehydrated with graded xylene and alcohol, and incubated in 3% hydrogen peroxide for 5 min at room temperature to eliminate endogenous peroxidase activity. Antigen retrieval for the anti‐ZNF143 antibody was performed with 0.1 M citrate buffer (pH 6.0) in an autoclave for 15 min. The sections were then incubated with anti‐ZNF143 antibody (dilution ratio 1:200). The Envision plus system (Dako, Carpinteria, CA, USA) was used for antibody‐bridge labeling, with hematoxylin counterstaining.
Statistical analysis. Pearson’s correlation was used for statistical analysis, and significance was set at the 5% level.
Results
ZNF143 expression is required for cancer cell growth. We estimated specificity of both ZNF143 antibody and ZNF143 siRNA. ZNF143‐specific antibody recognized both endogenous 90 kDa protein and exogenous 3xFlag‐ZNF143 protein (Fig. 1A). Endogenous ZNF143 protein was completely abolished with treatment of specific siRNA against ZNF143 (Fig. 1B). An in silico genome‐wide screen for ZNF143 binding sites suggested that many of the target genes are involved in cell growth.( 6 ) To confirm these results, we first investigated whether ZNF143 expression is required for cancer cell growth. At first, downregulation of ZNF143 expression effectively reduced cell growth in PC3 cells (Fig. 2A). Analysis of cell cycle profiles was performed on human prostate cancer PC3 cells after transfection with two different ZNF143‐specific siRNA. A marked increase in G2/M phase cells was observed when cells were transfected with ZNF143 siRNA (Fig. 2B,C). In addition, knockdown of ZNF143 expression induced apoptosis, as shown by an increased percentage of cells in the subG1 population (∼5%), whereas only 1% of cells treated with control siRNA was in the subG1 population (Fig. 2C). To extend these results, the cells were stained with an anti‐Annexin V‐FITC antibody. Treatment with control siRNA did not induce apoptosis, whereas knockdown of ZNF143 expression resulted in an increase in the percentage of annexin V positive cells (Fig. 2D).
Figure 1.
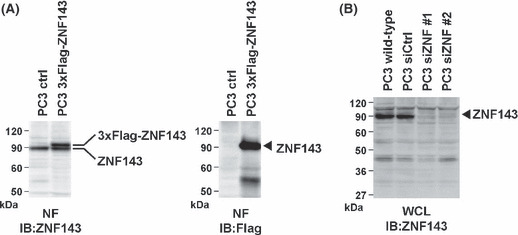
Specificity of anti‐zinc finger protein 143 (ZNF143) antibody and ZNF143 small interfering RNA (siRNA). (A) PC3 cells were transfected with or without 3xFlag‐ZNF143 and nuclear proteins (50 μg) were used for western blotting with anti‐ZNF143 and anti‐Flag antibodies. (B) PC3 cells were transfected with or without ZNF143 siRNA and whole cell lysate (100 μg) were used for western blotting with anti‐ZNF143 antibody. siZNF #1 and #2 indicate ZNF143 siRNA #1 and #2, respectively.
Figure 2.
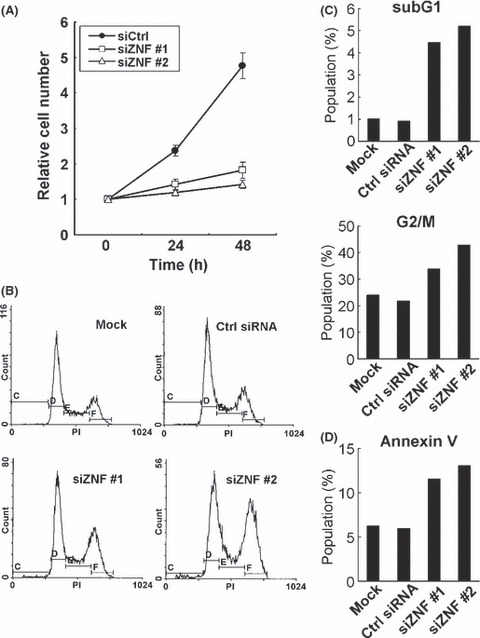
Knockdown of ZNF143 induces G2/M arrest. (A) PC3 cells were transfected with control siRNA (siCtrl), ZNF143 siRNA #1 (siZNF #1) or ZNF143 siRNA #2 (siZNF #2). Cells were counted after the indicated times, with time zero being 24 h after transfection. The results were normalized to cell numbers at time zero. The points represent the mean of at least three independent experiments; the bars show the SD. (B) PC3 cells were transfected with or without the indicated siRNA. Cells were harvested after 72 h and used for propidium iodide (PI) staining. DNA content in single cells was measured. siZNF #1 and #2 indicate ZNF143 siRNAs #1 and #2, respectively. (C) SubG1 and G2/M populations were calculated from the results of part (B). (D) siRNA‐transfected PC3 cells were stained with Annexin‐V‐fluorescein isothiocyanate and PI, and the percentage of apoptotic cells (annexin V positive and PI negative) was determined by flow cytometry. siZNF #1 and #2 indicate ZNF143 siRNA #1 and #2, respectively.
DNA microarray analysis with ZNF143‐specific siRNA. We then evaluated global expression profiles to determine the real ZNF143 target genes by cDNA microarray. One hundred and fifty‐two genes were downregulated <0.4‐fold after ZNF143‐specific siRNA transfection, whereas 84 genes were upregulated more than 2.5‐fold after downregulation of ZNF143 expression (Table S2). To categorize the 152 downregulated genes, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources Program (http://david.abcc.ncifcrf.gov/). Among 152 genes, 41 (27.0%) were categorized as concerned with the cell cycle/DNA replication (Table 1). Next, we searched for putative ZNF143 binding sites in the promoter region of these genes. All 41 genes had at least one ZNF143 binding site within 1 kb of the putative transcription start site (Table 1).
Table 1.
Cell cycle/DNA replication associated genes downregulated following zinc finger protein 143 (ZNF143) knockdown
| Gene ID | Symbol | Accession | Gene name | Fold change† |
|---|---|---|---|---|
| 23397 | NCAPH | NM_015341 | Barren homolog 1 (Drosophila) | 0.174 |
| 991 | CDC20 | NM_001255 | CDC20 cell division cycle 20 homolog (Saccharomyces cerevisiae) | 0.221 |
| 57405 | SPC25 | NM_020675 | Spindle pole body component 25 homolog (S. cerevisiae) | 0.234 |
| 4174 | MCM5 | NM_006739 | MCM5 minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) | 0.234 |
| 9212 | AURKB | NM_004217 | Aurora kinase B | 0.239 |
| 890 | CCNA2 | NM_001237 | Cyclin A2 | 0.248 |
| 990 | CDC6 | NM_001254 | CDC6 cell division cycle 6 homolog (S. cerevisiae) | 0.252 |
| 4173 | MCM4 | NM_005914 | MCM4 minichromosome maintenance deficient 4 (S. cerevisiae) | 0.256 |
| 4176 | MCM7 | NM_005916 | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 0.258 |
| 5347 | PLK1 | NM_005030 | Polo‐like kinase 1 (Drosophila) | 0.266 |
| 7465 | WEE1 | NM_003390 | WEE1 homolog (Schizosaccharomyces pombe) | 0.278 |
| 81620 | CDT1 | NM_030928 | DNA replication factor | 0.284 |
| 83461 | CDCA3 | NM_031299 | Cell division cycle associated 3 | 0.288 |
| 5888 | RAD51 | NM_002875 | RAD51 homolog (RECA homolog, Escherichia coli) (S. cerevisiae) | 0.289 |
| 4171 | MCM2 | NM_004526 | MCM2 minichromosome maintenance deficient 2, mitotin (S. cerevisiae) | 0.289 |
| 56992 | KIF15 | NM_020242 | Kinesin family member 15 | 0.290 |
| 3148 | HMGB2 | NM_002129 | High‐mobility group box 2 | 0.295 |
| 113130 | CDCA5 | NM_080668 | Cell division cycle associated 5 | 0.301 |
| 54443 | ANLN | NM_018685 | Anillin, actin binding protein (Scraps homolog, Drosophila) | 0.320 |
| 55388 | MCM10 | NM_018518 | MCM10 minichromosome maintenance deficient 10 (S. cerevisiae) | 0.332 |
| 4172 | MCM3 | NM_002388 | MCM3 minichromosome maintenance deficient 3 (S. cerevisiae) | 0.334 |
| 9918 | NCAPD2 | NM_014865 | Chromosome condensation‐related SMC‐associated protein 1 | 0.336 |
| 11004 | KIF2C | NM_006845 | Kinesin family member 2C | 0.348 |
| 1017 | CDK2 | NM_001798 | Cyclin‐dependent kinase 2 | 0.348 |
| 51512 | GTSE1 | NM_016426 | G‐2 and S‐phase expressed 1 | 0.353 |
| 9088 | PKMYT1 | NM_004203 | Protein kinase, membrane associated tyrosine/threonine 1 | 0.356 |
| 5984 | RFC4 | NM_002916 | Replication factor C (Activator 1) 4, 37KDA | 0.357 |
| 3835 | KIF22 | NM_007317 | Kinesin family member 22 | 0.358 |
| 10403 | NDC80 | NM_006101 | Kinetochore associated 2 | 0.359 |
| 699 | BUB1 | NM_004336 | BUB1 budding uninhibited by benzimidazoles 1 homolog (Yeast) | 0.360 |
| 9055 | PRC1 | NM_003981 | Protein regulator of cytokinesis 1 | 0.363 |
| 9700 | ESPL1 | NM_012291 | Extra spindle poles like 1 (S. cerevisiae) | 0.365 |
| 55143 | CDCA8 | NM_018101 | Cell division cycle associated 8 | 0.369 |
| 1104 | RCC1 | NM_001269 | Regulator of chromosome condensation 1 | 0.376 |
| 5982 | RFC2 | NM_002914 | Replication factor C (Activator 1) 2, 40KDA | 0.376 |
| 9787 | DLG7 | NM_014750 | Discs, large homolog 7 (Drosophila) | 0.391 |
| 10051 | SMC4 | NM_005496 | SMC4 structural maintenance of chromosomes 4‐like 1 (Yeast) | 0.391 |
| 9493 | KIF23 | NM_004856 | Kinesin family member 23 | 0.397 |
| 4751 | NEK2 | NM_002497 | NIMA (never in mitosis gene A)‐related kinase 2 | 0.399 |
| 899 | CCNF | NM_001761 | Cyclin F | 0.399 |
| 22974 | TPX2 | NM_012112 | TPX2, microtubule‐associated, homolog (Xenopus laevis) | 0.399 |
List of cell cycle/DNA‐replication‐associated genes with fold change marked <0.4. †Fold change indicates the average of expression data of ZNF143 siRNA/control siRNA.
Validation of the ZNF143 target genes. To validate the microarray expression data, we performed western blotting, ChIP assays and reporter assays. We obtained eight specific antibodies for proteins among the 41 cell cycle/DNA replication regulators with ZNF143 binding site(s). As shown in Figure 3, the expression of nine proteins was decreased by ZNF143 siRNA transfection. In particular, PLK1, CDC6 and HMGB2 were substantially decreased, whereas β‐actin and PRDX4, which do not have a ZNF143 binding site, were not affected by ZNF143 siRNA transfection. Next, we examined whether ZNF143 directly regulates the expression of these genes using ChIP and luciferase reporter assays. For ChIP assays, we used stable transfectants that expressed three flag tags at the N‐terminus of ZNF143.( 7 ) As shown in Figure 4A, ZNF143 bound to the promoters of the seven genes that contained a ZNF143 binding site. The promoter region of PRDX4, which does not have a ZNF143 binding site, was used as a negative control. We also investigated the luciferase reporter activities of five genes. The promoter activities of CDC6, PLK1, AURKB, MCM2 and HMGB2 were clearly decreased by transfection with ZNF143‐specific siRNA #2 (Fig. 4B). Myslinski et al. ( 6 ) reported that 58% of genes that had a ZNF143 binding site contained the submotif ACTACN in their 5′ regions, and that their promoter activities were higher than those of promoters that lacked the submotif. To confirm these results, we prepared two CDC6 promoters: CDC6 luc2 contained the ACTACN sequence and CDC6 luc1 did not. As shown in Figure 4C, our results were consistent with the findings of Myslinski et al., and ZNF143 siRNA reduced both CDC6 promoter activities, with or without the ACTACN motif, in the same ratio (Fig. 4B).
Figure 3.
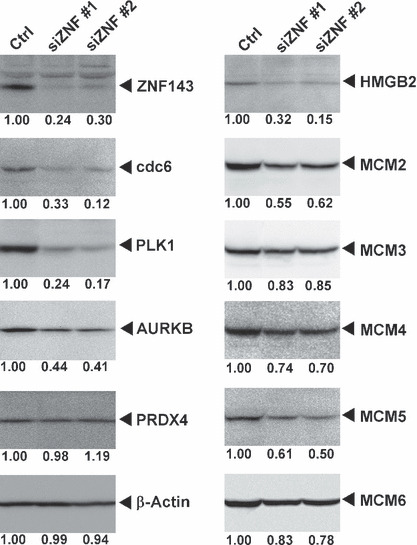
ZNF143 small interfering RNA (siRNA) reduced the expression of cell cycle/DNA replication‐related genes. siRNA‐transfected PC3 cells were used for western blotting. Quantitation of protein expression levels is shown under the panels. siZNF #1 and #2 indicate ZNF143 siRNA #1 and #2, respectively.
Figure 4.
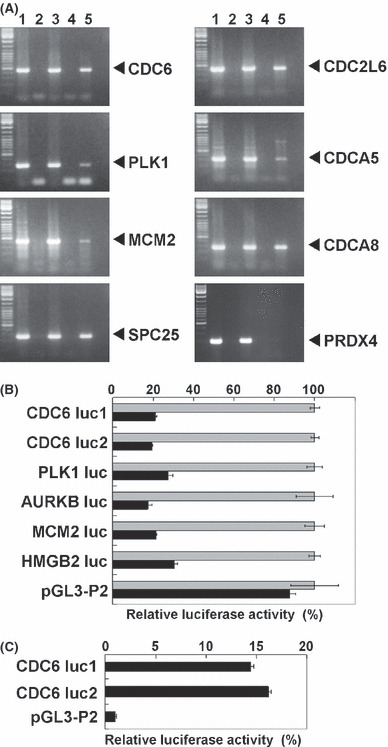
Luciferase reporter and chromatin immunoprecipitation assays. (A) Soluble chromatin was prepared from flag tagged zinc finger protein 143 (ZNF143) stable transfectants (lanes 3–5) or mock transfectants (lanes 1 and 2), and immunoprecipitated with anti‐mouse IgG (lane 4) or anti‐Flag (M2) antibodies (lanes 2 and 5). Extracted immunoprecipitated DNA (lanes 2, 4 and 5) and soluble chromatin (lanes 1 and 3) were amplified using specific primer pairs for the indicated promoter regions. The PCR products for CDC6, PLK1, MCM2, SPC25, CDC2L6, CDCA5, CDCA8 and PRDX4 were 463, 296, 423, 258, 411, 342, 260 and 153 bp, respectively. (B) PC3 cells were transfected with reporter plasmid and ZNF143 small interfering RNA (siRNA). Luciferase activities were normalized to each reporter activity with control siRNA. The gray and black bars indicate control siRNA and ZNF143 siRNA #2, respectively. Bars represent ± SD. (C) PC3 cells were transfected with reporter plasmid. The results were normalized to the protein concentration and are representative of at least three independent experiments. Both CDC6 luc1 and luc2 have a ZNF143 binding site, and CDC6 luc2 also has the ACTACN motif in the ZNF143 binding sequence. Luciferase activities were normalized to the empty vector pGL3‐P2. Bars represent ± SD.
Correlation between ZNF143 expression and cell growth in human lung cancer. ZNF143 regulates the gene expressions associated with the cell cycle and DNA replication. We investigated the cellular expression level of ZNF143 and doubling time with 10 lung cancer cell lines. ZNF143 expression was different in each cancer cell line as compared with Nrf2 and inversely correlated with doubling time (Fig. 5A,B). This result is consistent with repression of cell growth by downregulation of ZNF143 in lung cell cancer A549 and 904L (Fig. 5C). Repression of cell growth by downregulation of ZNF143 was also observed in HeLa, DLD1, Caco‐2, T24, CCF‐STTG1 and U343 (data not shown).
Figure 5.
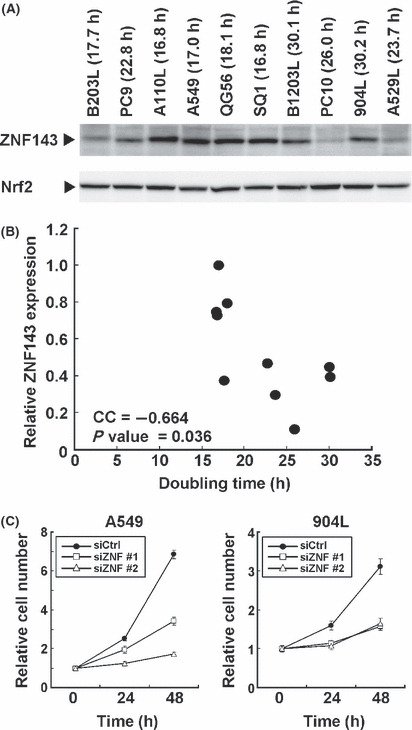
Correlation between zinc finger protein 143 (ZNF143) expression and cell growth. (A) Nuclear protein (50 μg) was used for western blotting with anti‐ZNF143 and anti‐Nrf2 antibodies. The doubling time for each cell lines is also shown. (B) Expression of ZNF143 and Nrf2 were quantitated using a Multi Gauge Version 3.0. using Figure 5A, and ZNF143 expression was normalized by Nrf2. The maximum expression levels of ZNF143 were set to 100. CC, coefficient of correlation. (C) A549 and 904L cells were transfected with control siRNA (siCtrl), ZNF143 siRNA #1 (siZNF #1) or ZNF143 siRNA #2 (siZNF #2). The cell proliferation assay is described in Figure 2A.
ZNF143 expression in human cancer specimens. To investigate further whether highly proliferative cancer cells are characterized by increased ZNF143 expression, tissue sections of surgical specimens from cancer patients were analyzed by immunohistochemistry (Fig. 6). Six solid tumors (esophageal carcinoma, gastric adenocarcinoma, squamous cell carcinoma of the lung, urothelial carcinoma of the urinary bladder, testicular seminoma and cerebral astrocytoma) were analyzed in comparison with adjacent non‐tumor samples. Nuclear‐localized ZNF143 was strongly expressed in cancer cells. Expression of ZNF143 was low or weak in non‐tumor tissue.
Figure 6.
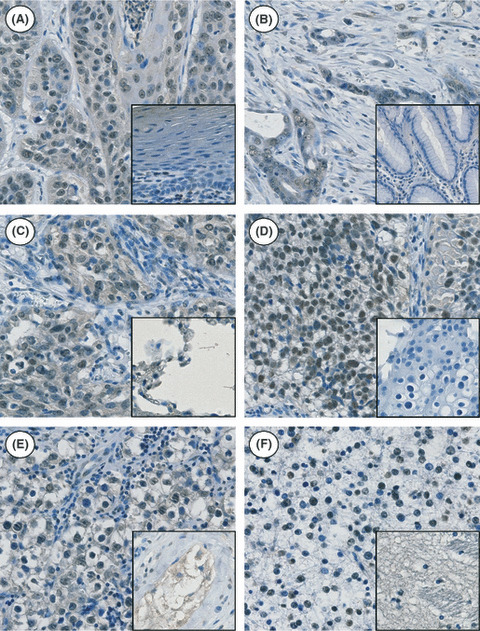
Immunohistochemical analysis of zinc finger protein 143 (ZNF143) expression in various human tumor tissues. Nuclear expression of the ZNF143 protein was seen in all cancer cells, while low or weak nuclear expression was seen in normal adjacent tissues (inset). The selected tissues consisted of surgically resected cases of esophageal carcinoma (A), gastric adenocarcinoma (B), squamous cell carcinoma of the lung (C), urothelial carcinoma of the urinary bladder (D), testicular seminoma (E) and cerebral astrocytoma (F).
Discussion
We have identified a number of genes that are induced by treatment with anti‐cancer agents and/or overexpressed in drug‐resistant cells.( 7 , 14 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 ) The drug sensitivity of cancer cells is governed by the complex pathways involved in the pathogenesis of cancer,( 25 , 26 ) and identification of the molecules governing cancer cell growth is emerging as a primary theme of molecular therapies. The transcription factor ZNF143 is stress‐inducible and overexpressed in cisplatin‐resistant cells.( 7 , 27 ) We found ZNF143 binding sites in the promoter region of many DNA repair genes and propose that ZNF143 regulates the expression of DNA repair genes. It has been shown recently that ZNF143 binding sites are widely distributed in the human genome and are located in the promoters of approximately 1000 human genes.( 6 ) This suggests that ZNF143 might regulate basic cellular functions at the transcriptional level.
Many studies have indicated that the cellular content of cell‐cycle‐related gene products is strictly regulated post‐translationally by the ubiquitin–proteasome system.( 28 , 29 ) On the other hand, little is known about the transcriptional regulation of cell‐cycle‐related genes. The E2F family of transcription factors is well known to be a master regulator of cell proliferation and is regulated by the retinoblastoma tumor suppressor gene.( 30 ) In this report, downregulation of ZNF143 strongly induced G2/M arrest. On the contrary, downregulation of E2F induced G1 cell cycle arrest, indicating that the role of ZNF143 may be quite distinct from that of E2F in the cell cycle. ZNF143 target genes have been identified by a combination of in silico and experimental approaches.( 6 ) Myslinski et al. ( 6 ) used eleven 18‐bp binding sites in a genome‐wide motif search and identified approximately 1000 genes as ZNF143 target genes. Functional classification of these genes indicated that the majority of the identified genes are important for cell growth. Consistently, our results clearly showed that ZNF143 is required for cancer cell growth (2, 5). RNAi‐mediated knockdown of ZNF143 showed that the expression of 152 genes was downregulated in human prostate cancer PC3 cells (Table S1). Among these, 41 genes are categorized as concerned with cell cycle/DNA replication (Table 1). The human genome database suggests that approximately 500–600 genes are assigned for cell cycle/DNA replication. Based on our data, approximately 7.5% (41 genes/500–600 genes) of cell cycle/DNA replication genes might be directly or indirectly regulated by ZNF143. On the other hand, ZNF143 regulates only approximately 0.5% (110 genes/23 000–27 000 genes) of other genes except for cell cycle/DNA replication. It is noteworthy that important cell‐cycle‐associated kinases such as PLK1, AURKB, WEE1, budding uninhibited by benzimidazoles (BUB1) and cyclin‐dependent kinase 2 (CDK2) are target genes for ZNF143. Only nine (SPC25, CDC6, PLK1, RAD51, MCM3, KIF2C, GTSE1, ESPL1, CDCA8) (22%) of 41 genes were present in the ZNF143 target gene list identified by the in silico genome screen.( 6 ) We also found putative ZNF143 binding sites in many DNA repair genes;( 7 ) however, only four (Rad51, BRCA1, FEN1, EXO1) genes were downregulated by ZNF143 siRNA. It has been shown that BUB family expression is required for cancer cell growth but not for normal cells, indicating that the BUB family is one of the genes involved in non‐oncogene addiction.( 31 , 32 ) It is an interesting finding that BUB1 and BUB1B( 33 ) are target genes of ZNF143. These indicate that ZNF143 specifically regulates genes associated with cell cycle/DNA replication and is one of the master regulators of DNA metabolism including cell cycle and cell growth.
It is well established that transcription factors do not make good targets for drug design, because transcription factors form multiple complexes with other cofactors. Our immunohistochemical studies have shown that ZNF143 is highly expressed in cancer cells (Fig. 6). RNAi‐mediated knockdown of ZNF143 induced apoptosis following G2/M cell cycle arrest (Fig. 2). Because cell‐cycle‐associated kinases have a critical role in cell cycle progression, these kinases comprise a promising set of targets for anti‐cancer drugs.( 34 , 35 ) Among the ZNF143 target cell‐cycle‐associated kinases, both PLK1 and AURKB are overexpressed in many types of cancer.( 36 , 37 , 38 , 39 ) Our data showed that RNAi‐mediated knockdown of ZNF143 effectively downregulated the expression of both PLK1 and AURKB (Fig. 3). The MCM proteins are required for pre‐replication complex formation, DNA replication initiation and DNA synthesis. It is well known that the MCM proteins are highly expressed in human cancers. Therefore, the MCM proteins are diagnostic markers and promising targets for anti‐cancer drug development.( 40 ) As shown in Figure 3, several MCM proteins are targets of ZNF143. Because ZNF143 regulates the expression of both DNA replication and cell cycle regulatory members, ZNF143 may be a promising target for cancer diagnostics and therapies.
We found Ets binding sites in the promoter region of the ZNF143 gene. The Ets family is one of the largest families of transcription factors and is known to be involved in diverse cellular functions. The Ets family is a proto‐oncogene and highly expressed in various cancers,( 41 ) indicating that ZNF143 expression might be regulated by the Ets family. Therefore, further studies to investigate the correlation between Ets and ZNF143 expression with clinicopathological data should help in understanding the role of ZNF143 in cancer.
Disclosure Statement
R. Yamazaki is an employee of Yakult Central Institute for Microbiological Research, Tokyo, Japan.
Supporting information
Table S1. Primer pairs used for Chromatin immunoprecipitation (ChIP) assays.
Table S2. The list of selected genes with fold change marked >2.5 or <0.4.
Supporting info item
Supporting info item
Acknowledgments
This study was supported by Grants‐in‐Aid for Scientific Research from the Ministry for Education, Culture, Sports, Science and Technology of Japan (17016075), University of Occupational and Environmental Health (UOEH) Japan, Grant for Advanced Research and the Vehicle Racing Commemorative Foundation. K. Kohno received a research grant from Yakult Co., Ltd., Tokyo, Japan. The authors thank Ms H. Isagai for her technical assistance.
References
- 1. Myslinski E, Krol A, Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J Biol Chem 1998; 273: 21998–2006. [DOI] [PubMed] [Google Scholar]
- 2. Schaub M, Myslinski E, Schuster C, Krol A, Carbon P. Staf, a promiscuous activator for enhanced transcription by RNA polymerase II and III. EMBO J 1997; 16: 173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rincon JC, Engler SK, Hargrove BW, Kunkel GR. Molecular cloning of a cDNA encoding human SPH‐binding factor, a conserved protein that binds to the enhancer‐like region of the U6 small nuclear RNA gene promoter. Nucleic Acids Res 1998; 26: 4846–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schaub M, Krol A, Carbon P. Structural organization of Staf‐DNA complexes. Nucleic Acids Res 2000; 28: 2114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kubota H, Yokota S, Yanagi H, Yura T. Transcriptional regulation of the mouse cytosolic chaperonin subunit gene Ccta/t‐complex polypeptide 1 by selenocysteine tRNA gene transcription activating factor family zinc finger proteins. J Biol Chem 2000; 275: 28641–8. [DOI] [PubMed] [Google Scholar]
- 6. Myslinski E, Gérard MA, Krol A, Carbon P. A genome scale location analysis of human Staf/ZNF143‐binding sites suggests a widespread role for human Staf/ZNF143 in mammalian promoters. J Biol Chem 2006; 281: 39953–62. [DOI] [PubMed] [Google Scholar]
- 7. Wakasugi T, Izumi H, Uchiumi T et al. ZNF143 interacts with p73 and is involved in cisplatin resistance through the transcriptional regulation of DNA repair genes. Oncogene 2007; 26: 5194–203. [DOI] [PubMed] [Google Scholar]
- 8. Takano H, Ise T, Nomoto M et al. Structural and functional analysis of the control region of the human DNA topoisomerase II alpha gene in drug‐resistant cells. Anticancer Drug Des 1999; 14: 87–92. [PubMed] [Google Scholar]
- 9. Torigoe T, Izumi H, Wakasugi T et al. DNA topoisomerase II poison TAS‐103 transactivates GC‐box‐dependent transcription via acetylation of Sp1. J Biol Chem 2005; 280: 1179–85. [DOI] [PubMed] [Google Scholar]
- 10. Minagawa N, Nakayama Y, Inoue Y et al. 4‐[3,5‐Bis(trimethylsilyl)benzamido] benzoic acid inhibits angiogenesis in colon cancer through reduced expression of vascular endothelial growth factor. Oncol Res 2004; 14: 407–14. [DOI] [PubMed] [Google Scholar]
- 11. Abe T, Mori T, Kohno K et al. Expression of 72 kDa type IV collagenase and invasion activity of human glioma cells. Clin Exp Metastasis 1994; 12: 296–304. [DOI] [PubMed] [Google Scholar]
- 12. Tanabe M, Izumi H, Ise T et al. Activating transcription factor 4 increases the cisplatin resistance of human cancer cell lines. Cancer Res 2003; 63: 8592–5. [PubMed] [Google Scholar]
- 13. Imamura T, Izumi H, Nagatani G et al. Interaction with p53 enhances binding of cisplatin‐modified DNA by high mobility group 1 protein. J Biol Chem 2001; 276: 7534–40. [DOI] [PubMed] [Google Scholar]
- 14. Igarashi T, Izumi H, Uchiumi T et al. Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene 2007; 26: 4749–60. [DOI] [PubMed] [Google Scholar]
- 15. Takahashi M, Shimajiri S, Izumi H et al. Y‐box binding protein‐1 is a novel molecular target for tumor vessels. Cancer Sci 2010; 101: 1367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kidani A, Izumi H, Yoshida Y et al. Thioredoxin2 enhances the damaged DNA binding activity of mtTFA through direct interaction. Int J Oncol 2009; 35: 1435–40. [DOI] [PubMed] [Google Scholar]
- 17. Yokomizo A, Ono M, Nanri H et al. Cellular levels of thioredoxin associated with drug sensitivity to cisplatin, mitomycin C, doxorubicin, and etoposide. Cancer Res 1995; 55: 4293–6. [PubMed] [Google Scholar]
- 18. Ohga T, Koike K, Ono M et al. Role of the human Y box‐binding protein YB‐1 in cellular sensitivity to the DNA‐damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res 1996; 56: 4224–8. [PubMed] [Google Scholar]
- 19. Ise T, Izumi H, Nagatani G et al. Structural characterization of the human interleukin‐13 receptor alpha1 gene promoter. Biochem Biophys Res Commun 1999; 265: 387–94. [DOI] [PubMed] [Google Scholar]
- 20. Kato K, Nomoto M, Izumi H et al. Structure and functional analysis of the human STAT3 gene promoter: alteration of chromatin structure as a possible mechanism for the upregulation in cisplatin‐resistant cells. Biochim Biophys Acta 2000; 1493: 91–100. [DOI] [PubMed] [Google Scholar]
- 21. Nagatani G, Nomoto M, Takano H et al. Transcriptional activation of the human HMG1 gene in cisplatin‐resistant human cancer cells. Cancer Res 2001; 61: 1592–7. [PubMed] [Google Scholar]
- 22. Murakami T, Shibuya I, Ise T et al. Elevated expression of vacuolar proton pump genes and cellular PH in cisplatin resistance. Int J Cancer 2001; 93: 869–74. [DOI] [PubMed] [Google Scholar]
- 23. Shiota M, Izumi H, Onitsuka T et al. Twist promotes tumor cell growth through YB‐1 expression. Cancer Res 2008; 68: 98–105. [DOI] [PubMed] [Google Scholar]
- 24. Miyamoto N, Izumi H, Noguchi T et al. Tip60 is regulated by circadian transcription factor clock and is involved in cisplatin resistance. J Biol Chem 2008; 283: 18218–26. [DOI] [PubMed] [Google Scholar]
- 25. Torigoe T, Izumi H, Ishiguchi H et al. Cisplatin resistance and transcription factors. Curr Med Chem Anticancer Agents 2005; 5: 15–27. [DOI] [PubMed] [Google Scholar]
- 26. Kohno K, Uchiumi T, Niina I et al. Transcription factors and drug resistance. Eur J Cancer 2005; 41: 2577–86. [DOI] [PubMed] [Google Scholar]
- 27. Ishiguchi H, Izumi H, Torigoe T et al. ZNF143 activates gene expression in response to DNA damage and binds to cisplatin‐modified DNA. Int J Cancer 2004; 111: 900–9. [DOI] [PubMed] [Google Scholar]
- 28. Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin‐proteasome system. Nat Rev Drug Discov 2006; 5: 596–613. [DOI] [PubMed] [Google Scholar]
- 29. Huang L, Chen CH. Proteasome regulators: activators and inhibitors. Curr Med Chem 2009; 16: 931–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer 2009; 9: 785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malumbres M, Barbacid M. Cell cycle kinases in cancer. Curr Opin Genet Dev 2007; 17: 60–5. [DOI] [PubMed] [Google Scholar]
- 32. Schlabach MR, Luo J, Solimini NL et al. Cancer proliferation gene discovery through functional genomics. Science 2008; 319: 620–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Myslinski E, Gérard MA, Krol A, Carbon P. Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res 2007; 35: 3453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov 2009; 8: 547–66. [DOI] [PubMed] [Google Scholar]
- 35. De Cárcer G, Pérez de Castro I, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem 2007; 14: 969–85. [DOI] [PubMed] [Google Scholar]
- 36. Strebhardt K, Ullrich A. Targeting polo‐like kinase 1 for cancer therapy. Nat Rev Cancer 2006; 6: 321–30. [DOI] [PubMed] [Google Scholar]
- 37. Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo‐like kinases (Plks) and cancer. Oncogene 2005; 24: 287–91. [DOI] [PubMed] [Google Scholar]
- 38. Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN Jr, Gandara DR. Aurora kinases as anticancer drug targets. Clin Cancer Res 2008; 14: 1639–48. [DOI] [PubMed] [Google Scholar]
- 39. Mountzios G, Terpos E, Dimopoulos MA. Aurora kinases as targets for cancer therapy. Cancer Treat Rev 2008; 34: 175–82. [DOI] [PubMed] [Google Scholar]
- 40. Lei M. The MCM complex: its role in DNA replication and implications for cancer therapy. Curr Cancer Drug Targets 2005; 5: 365–80. [DOI] [PubMed] [Google Scholar]
- 41. Tsantoulis PK, Gorgoulis VG. Involvement of E2F transcription factor family in cancer. Eur J Cancer 2005; 41: 2403–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer pairs used for Chromatin immunoprecipitation (ChIP) assays.
Table S2. The list of selected genes with fold change marked >2.5 or <0.4.
Supporting info item
Supporting info item


