Abstract
BCSC‐1 is dramatically upregulated in CNE‐2L2 human nasopharyngeal carcinoma cells with reduced malignancy (AS cells) and is proposed to be a candidate tumor suppressor gene. We therefore examined the effect of BCSC‐1 expression on malignant behaviors of CNE‐2L2 cells. Growth in vitro and tumorigenesis in nude mice of wild‐type CNE‐2L2 cells (W cells) were inhibited by ectopic BCSC‐1, and those of AS cells were promoted by BCSC‐1 suppression. The tumor suppressor function of BCSC‐1 was further confirmed by a study showing that intratumor BCSC‐1 injection caused growth suppression of the tumor from W cells inoculated in nude mice. Immunohistochemistry exhibited marked reduction of BCSC‐1 expression in 11 of 39 human nasopharyngeal carcinoma specimens. Because BCSC‐1 expression was as rich as that in normal cells in the rest of the carcinoma specimens and was poor in CNE‐2L2 cells, HNE‐1 human nasopharyngeal carcinoma cells with rich BCSC‐1 expression were used as a control in the study. No effect of BCSC‐1 transfection on growth of the cells was observed. The data suggest that BCSC‐1 suppression might play roles in tumorigenesis of some nasopharyngeal carcinomas and that BCSC‐1 might be a potential gene therapy target in nasopharyngeal carcinomas with poor BCSC‐1 expression. Enhanced aggregation of cells together with increased E‐cadherin and α‐catenin expression and reduced Wnt signaling might be involved in the mechanisms of tumor suppressor function of BCSC‐1. (Cancer Sci 2009; 100: 1817–1822)
We previously observed that α‐mannosidase Man2c1 suppression causes profound inhibition of malignant activities of the human nasopharyngeal carcinoma‐derived cell line CNE‐2L2.( 1 ) In order to explore possible mechanisms of malignancy inhibition, mRNA differential display analysis was carried out on CNE‐2L2 cells with Man2c1 suppression (called AS cells) and CNE‐2L2 cells transfected with mock (called M cells). Of the 1069 genes examined, 28 were upregulated and 31 were downregulated in AS cells. LOH11CR2A, also called BCSC‐1 (GenBank no. NM_014622), was identified as the most dramatically upregulated gene. BCSC‐1 cDNA was originally cloned and proposed to be a candidate tumor suppressor gene (TSG) by Martin et al.( 2 ) Therefore, association of BCSC‐1 with malignant behaviors of CNE‐2L2 cells was examined in this study.
It was observed that growth and tumorigenesis of wild‐type CNE‐2L2 cells (W cells) were inhibited by ectopic BCSC‐1 whereas those of AS cells were promoted by BCSC‐1 suppression, indicating that BCSC‐1 is a TSG in CNE‐2L2 cells. The tumor suppressor function was further confirmed by a study showing that intratumor BCSC‐1 injection results in growth inhibition of tumors from W cells inoculated in nude mice. BCSC‐1 expression was examined by means of immunohistochemistry in human nasopharyngeal carcinomas. Because only some carcinoma specimens showed marked reduction of BCSC‐1 expression and BCSC‐1 expression was poor in CNE‐2L2 cells, the HNE‐1 nasopharyngeal carcinoma cell line with rich BCSC‐1 expression was used as a control in the present study. The data obtained suggest that BCSC‐1 suppression might play roles in tumorigenesis of some nasopharyngeal carcinomas and BCSC‐1 might be a potential target in gene therapy of those nasopharyngeal carcinomas with poor BCSC‐1 expression.
TSG have been reported to exhibit multiple inhibitive effects on tumorigenesis.( 3 ) Tumorigenesis, including that of nasopharyngeal carcinoma,( 4 ) is a very complicated process and is regulated by a number of factors.( 5 , 6 ) Therefore, it could be postulated that the suppressive effect of BCSC‐1 on malignant behaviors of CNE‐2L2 cells would be at multiple points. Because reduced cell adhesion caused by decreased E‐cadherin expression( 7 ) and activation of the Wnt signaling pathway( 8 ) were found in nasopharyngeal carcinoma, the effect of BCSC‐1 expression on cell aggregation, E‐cadherin expression, and Wnt signaling was examined. It was observed that ectopic BCSC‐1 resulted in enhanced cell aggregation associated with increased expression of E‐cadherin/α‐catenin, and BCSC‐1 suppression promoted Wnt signaling.
Materials and Methods
Cell lines, tissue sections, and animals. The CNE‐2L2 human nasopharyngeal carcinoma cell line was developed by Yu and Gao.( 9 ) The HNE‐1 human nasopharyngeal carcinoma cell line was developed by Yao et al.( 10 ) CNE‐2L2 cells with Man2c1 suppression (AS cells) and CNE‐2L2 cells with mock transfection (M cells) were developed by Yue et al.( 1 ) GP‐293 and Hek293 cells were obtained from the Cell Culture Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China). Culture medium RPMI‐1640, DMEM, and FCS were purchased from Gibco‐BRL (Frederick, MD, USA). Biopsy specimen sections (5 µm) of human nasopharyngeal tissue and nasopharyngeal carcinoma were provided by the Department of Pathology, Cancer Hospital, Chinese Academy of Medical Sciences (Beijing, China). Female 8‐week‐old BALB/c mice and female 8‐week‐old BALB/c (nu/nu) mice were purchased from the Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences (Beijing, China).
Microarray analysis. mRNA differential display analysis on a BioStarH‐I DNA chip (oligonucleotide array for 1069 human genes; BioStar Genechip, Shanghai, China) was carried out to examine the effect of Man2c1 suppression on gene expression of CNE‐2L2 cells (Supporting information: Materials and methods).
Development of the anti‐BCSC‐1 protein mAb. A BCSC‐1 fusion protein expressed by pET‐30a‐BCSC‐1 in Escherichia coli was purified and used for immunization of mice and screening of hybridomas (Supporting information: Materials and Methods). Immunization of 8‐week‐old BALB/c mice, cell fusion, and hybridoma selection were conducted according to procedures described previously.( 11 ) Isotype detection was carried out using Isostrip Monoclonal Antibody Isotyping Kit (Roche Shanghai, Shanghai, China). Ascites were produced in BALB/c mice and mAb purification from ascites was carried out using ammonium sulfate and then caprylic acid following procedures described previously.( 11 )
ELISA. ELISA was carried out on the BCSC‐1 fusion protein. April fusion protein expressed by pET‐30a‐April (kindly provided by Ms Ling Zhang, Department of Immunology, Peking Union Medical College, Beijing, China) was used as a negative control.
Western blot analysis. Western blotting was carried out following procedures described previously.( 1 ) The mAb anti‐BCSC‐1, mAb anti‐hE‐Cadherin (clone SHE78‐7; Zymed, South San Francisco, CA, USA), and mAb anti‐hα‐catenin or hβ‐catenin (Santa Cruz, Santa Cruz, CA, USA) were used as the primary antibodies. Goat anti‐mouse IgG conjugated with horseradish peroxidase (BD Biosciences, San Jose, CA, USA) was used as the secondary antibody. β‐Actin was used as an internal control. The protein bands were visualized using SuperSignal West Pico Trial Kit (Pierce, Rockford, IL, USA).
Stable transfection of cells with BCSC‐1. The PCR‐amplified product from pUCm‐T‐BCSC‐1 using BCSC upstream 1 and downstream 1 primers (Supporting information: Table 1) was inserted into EcoRI‐ and XbaI‐digested pcDNA4/myc‐His A (Invitrogen, Carlsbad, CA, USA). pcDNA4/myc‐His A or pcDNA4/myc‐His A‐BCSC‐1 was transfected into W cells upon the action of Lipofectamine (Gibco‐BRL). Zeocin‐resistant cell pools (BCSC‐1 and mock cells) were obtained after 7 days of culture in medium containing zeocin (50 µg/mL).
Suppression of BCSC‐1 by siRNA. Four siRNA oligonucleotides specific to BCSC‐1 (Supporting information: Table 1) were used in the present study. siRNA was annealed and inserted into MluI‐ and XhoI‐digested pRNA‐H1.1/Retro (Genscript, Piscataway, NJ, USA). A control plasmid generating shRNA with non‐silencing RNA was also constructed. Generation of recombinant retrovirus was carried out according to the procedures introduced by Liu et al.( 12 ) When GP‐293 cell culture reached 70–80% confluence in six‐well plates, recombinant plasmids and pVSVG mixed with Lipofectamine were applied to the cell culture according to the manufacturer's instructions. The recombinant retrovirus produced was used to infect AS cells. After 12 h, the culture medium was replaced with fresh medium supplemented with hygromycine (400 µg/mL) (Sigma, St Louis, MO, USA). After 7–10 days, hygromycine‐resistant cell pools called si‐1, si‐2, si‐3, si‐4, and Retro infected with the virus harboring the control plasmid were developed.
RT‐PCR and real‐time PCR. Total RNA was extracted from cells with Trizol reagent (Gibco‐BRL). cDNA was synthesized using the Reverse Transcription System (Promega, Madison, WI, USA). PCR was carried out using 2 µL of cDNA in a 50‐µL reaction system at 94°C for 5 min, then 94°C for 45 s, 58°C for 45 s, and 72°C for 90 s for 30 cycles, followed by 72°C for 5 min for the final extension. PCR products were examined by electrophoresis on a 1% agarose gel containing ethidium bromide. Real‐time PCR was run using the SYBR Premix Ex Taq Kit (Takara Beijing, Beijing, China) according to the instructions, using the BCSC upstream 2 and downstream 2 primers (Supporting information: Table 1). PCR microplate was the product of Axygen (Union City, CA, USA). The data for BCSC‐1 was standardized to β‐actin. BCSC‐1 mRNA/β‐actin mRNA for W cell (Fig. 1a, Fig. 2a left panel) and for AS cell (Fig. 2a right panel) was normalized as 1.
Figure 1.
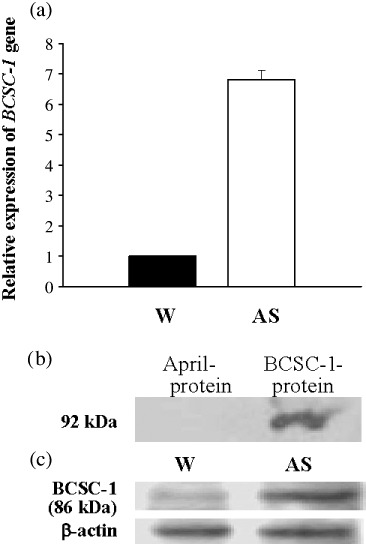
Upregulation of BCSC‐1 in AS cells. (a) Real‐time PCR using the SYBR Premix Ex Taq Kit. The data for BCSC‐1 was standardized to β‐actin. For W cells the level of β‐actin mRNA was set at 1. (b,c) Western blotting carried out on recombinant proteins (b) and proteins extracted from W and AS cells (c) with mAb anti‐BCSC‐1 as a probe. The deduced molecular weight of the BCSC‐1 protein is 86 kDa. W and AS represent wild‐type CNE‐2L2 cells and CNE‐2L2 cells with Man2c1 suppression respectively.
Figure 2.
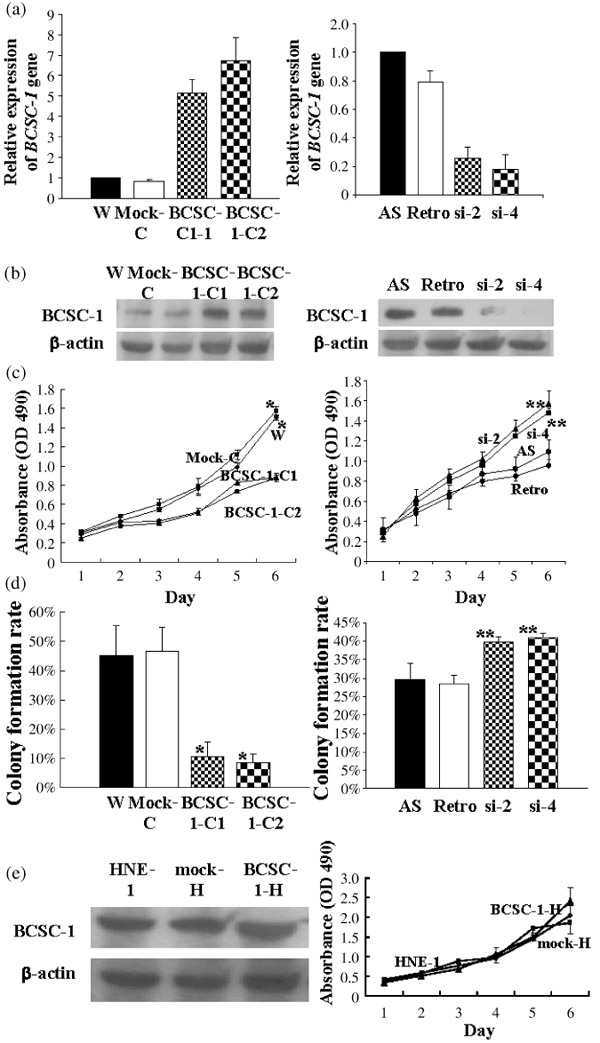
Suppressive effect of BCSC‐1 on the growth of CNE‐2L2 cells but not on that of HNE‐1 cells. BCSC‐1 expression was examined by (a) real‐time PCR and (b) western blotting with the designated mAb anti‐BCSC‐1 protein as a probe (b, the left panel of e). For cell growth test, 500 cells per well were seeded into 96‐well plates. (c, the right panel of e) Cell growth curves. Colony formation assay was carried out in soft agar. (d) Colony formation rate. The data are expressed as the means of three experiments. W, mock‐C, mock‐H, and Retro represent wild‐type CNE‐2L2 cells, CNE‐2L2 cells transfected with pcDNA4/myc‐His A, HNE‐1 cells transfected with pcDNA4/myc‐His A, and CNE‐2L2 cells infected with retrovirus harboring a control plasmid, respectively. BCSC‐1‐1 and BCSC‐1‐2 are two W cell pools with ectopic BCSC‐1. si‐2 and si‐4 are two AS cell pools with BCSC‐1 suppression. HNE‐BCSC‐1 is HNE‐1 cells transfected with BCSC‐1. *The difference between W or mock and BCSC‐1‐1 or BCSC‐1‐2 is statistically significant (paired t‐test, P < 0.01). **The difference between AS or Retro and si‐2 or si‐4 was statistically significant (paired t‐test, P < 0.05).
Cell proliferation assay. Cell proliferation was determined using the CellTiter 96 AQueous One Solution Cell Proliferation Assay Kit (Promega). Five hundered cells per well in 200 µL DMEM (Gibco‐BRL) containing 10% FCS were seeded into 96‐well plates (Corning, Coening, NY, USA). On days 1, 2, 3, 4, 5, and 6, 20 µL of assay solution was applied to each well and the plates were incubated at 5% CO2, 37°C for 2.5 h. The absorbance was measured at 490 nm. The assays were carried out three times.
Colony formation. Colony formation was carried out in soft agar according to procedures described previously.( 1 ) Suspensions containing 1 × 104 cells were seeded into the top of the agar. After 14 days incubation at 5% CO2, 37°C, the number of colonies with a diameter >500 µm was counted and the colony formation rate was calculated. Counts were expressed as the number of colonies per plate on average from three independent experiments.
Cell aggregation assay. The cell aggregation assay was carried out according to procedures described previously.( 1 ) Cell aggregation was examined under a reverse microscope.
Tumorigenicity in nude mice. Cells (1 × 106) in DMEM without serum (0.2 mL) were subcutaneously inoculated into the right armpit of nude mice with seven mice in each group. The longest (a) and shortest (b) diameters of tumors were measured weekly for 8 weeks. Tumor volume was calculated according to the formula: volume = 4/3πa × b 2. Tumors were excised under anesthesia and weighed at the end of week 8.
Construction of recombinant adenovirus vector and preparation of adenovirus stocks. EcoRI‐ and SalI‐excised BCSC‐1 from pET‐30a‐BCSC‐1 was inserted into the shuttle plasmid pDC316 (Microbix Biosystems, Toronto, Canada). Adenovirus (Ad5‐BCSC‐1) was developed by the Gene Technology Company (Beijing, China). Briefly, pDC316‐BCSC‐1 was cotransfected with the genomic plasmid pBHGlox_E1, 3Cre (Microbix Biosystems) into Hek293 cells by calcium phosphate precipitation. The adenovirus (Ad5‐BCSC‐1) was purified by means of cesium chloride gradient ultracentrifugation. Viral titer was determined as infectious units (IFU) using TCIG50 end‐point dilution. The final titer of the purified viral vectors was 5.7 × 109 IFU/mL. The virus stocks were aliquoted and stored at –80°C. Ad5‐egfp was purchased from the Gene Technology Company.
Adenovirus‐mediated gene therapy in nude mice. Each BALB/c mouse was subcutaneously inoculated with 2 × 106 W cells at the left armpit. The experiment was divided into three groups with six mice in each group. When tumors reached the size of 50–100 mm3, 0.1 mL PBS, 0.1 mL Ad5‐egfp (4 × 108 IFU), or 0.1 mL Ad5‐BCSC‐1 (4 × 108 IFU) was injected into the tumor in each mouse for groups 1, 2, or 3 respectively. The injection was repeated 1 week later. A further 2 weeks later, tumors were excised under anesthesia, size‐measured, and weighed. Tumor volume was calculated according to the formula mentioned above.
Immunohistochemistry. Immunohistochemistry was carried out using the Biotin SP‐HRP (DAB) Kit (Tianjin Bomeike Biotech, Tianjin, China) following the manufacturer's protocol. The mAb anti‐BCSC‐1 was used as the primary antibody.
Multiple protein sequence alignments. The homologous proteins were searched from the NCBI database using the HomoloGene program. Multiple protein sequence alignments were carried out using the CLUSTALW program.
Results
BCSC‐1 protein is highly conserved during evolution. Homologene presents nine homologues of BCSC‐1 in Homo sapiens (NM_014622), Pan troglodytes (XP_001137734.1), Canis lupus familiaris (XP_546453.2), Bos Taurus (XP_600048.3), Mus musculus (NP_766355.2), Rattus norvegicus (NP_942050.1), and Danio rerio (XP_691937.3, XP_001336637.2 and XP_001920968.1). These proteins exhibit high homology of sequence (Supporting information Fig. 1), indicating that BCSC‐1 is a highly conserved protein in evolution.
Upregulation of BCSC‐1 in CNE‐2L2 cells with Man2c1 suppression. To explore possible mechanisms of reduced malignancy of CNE‐2L2 cells by Man2c1 suppression, mRNA differential display analysis between cells with Man2c1 suppression (AS cells) and cells transfected with mock (M cells) was carried out. Of the 1069 genes examined, 28 were upregulated and 31 were downregulated in AS cells. LOH11CR2A, also called BCSC‐1 (GenBank no. NM_014622), was identified as the most dramatically upregulated gene (Supporting information: Table 2). Real‐time PCR validated the result obtained from microarray analysis (Fig. 1a).
In order to further examine BCSC‐1 expression in cells, the mAb anti‐BCSC‐1 was developed. BCSC‐1 and April fusion proteins were used for screening of hybridoma supernatants. The supernatants positive for BCSC‐1 but negative for April in ELISA were rescreened by western blotting. Figure 1(b) shows a supernatant reactive to BCSC‐1 fusion protein but not to April fusion protein (the deduced molecular weight of BCSC‐1 protein is 86 kDa). Western blotting in Figure 1(c) shows a mAb with much stronger reactivity to the ~86 kDa band in AS cells compared with W cells, further confirming enhanced BCSC‐1 expression in AS cells and indicating that the mAb also recognizes natural BCSC‐1. The mAb was used in immunohistochemistry. Supporting information: Figure 2 shows that the staining with the mAb on the tumor from AS cells was much stronger than on the tumor from W cells inoculated in nude mice.
Suppressive effect of BCSC‐1 on growth of CNE‐2L2 cells with poor BCSC‐1 expression but not on HNE‐1 cells with rich BCSC‐1 expression. The human nasopharyngeal carcinoma‐derived cell lines CNE‐2L2 (with poor BCSC‐1 expression) and HNE‐1 (with rich BCSC‐1 expression) were used in the present study. BCSC‐1‐C1 and BCSC‐1‐C2 were two cell pools from wild‐type CNE‐2L2 cells (W cells) transfected with BCSC‐1. BCSC‐1‐H was a cell pool from HNE‐1 cells transfected with BCSC‐1. Mock‐C and mock‐H were the cell pools from W cells and HNE‐1 cells transfected with mock respectively. BCSC‐1 expression was markedly enhanced in BCSC‐1‐C1 and BCSC‐1‐C2 cells (Fig. 2a,b, left panel), but not in BCSC‐1‐H cells (Fig. 2e, left panel). Growth in culture (Fig. 2c, left panel) and colony formation (Fig. 2d, left panel) of BCSC‐1‐C1 and BCSC‐1‐C2 cells were markedly reduced. However, no effect of BCSC‐1 transfection on HNE‐1 cell growth was observed (Fig. 2e, right panel). si2 and si4 were two cell pools infected with the virus harboring siBCSC‐1‐2 and siBCSC‐1‐4, respectively, and showed profound suppression of BCSC‐1 expression (Fig. 2a,b, the right panels). BCSC‐1 suppression promoted cell growth in culture (Fig. 2c, the right panel) and colony formation (Fig. 2d, the right panel) of AS cells, further confirming the suppressor function of BCSC‐1 on CNE‐2L2 cell growth.
Tumorigenesis suppression of CNE‐2L2 cells by BCSC‐1. Tumor growth curves showed that the tumors from BCSC‐1‐C1 or BCSC‐1‐C2 cells inoculated in nude mice grew much slower than those from controls (Fig. 3a). The average weights of the tumors at the end of the study were 4.05 ± 2.70, 3.07 ± 1.90, 0.65 ± 0.28, and 1.10 ± 0.70 g for W cells, M cells, BCSC‐1‐C1 cells, and BCSC‐1‐C2 cells, respectively. The difference between W or M and BCSC‐1‐C1 or BCSC‐1‐C2 was statistically significant (paired t‐test, P < 0.01). In contrast, BCSC‐1 suppression promoted tumorigenesis of AS cells (Fig. 3b). All but one of the mice inoculated with si‐2 cells and four mice inoculated with si‐4 cells died of tumor growth in the middle of the experiment. The tumor in the mouse inoculated with si‐2 cells remaining alive was 60.9 cm3 in size and 4.03 g in weight. The average weights of the tumors at the end of the study were 2.52 ± 0.23, 2.28 ± 0.42, and 4.79 ± 1.27 g for AS cells, Retro cells, and si‐4 cells respectively. The difference between AS or Retro and si‐4 was statistically significant (paired t‐test, P < 0.01).
Figure 3.
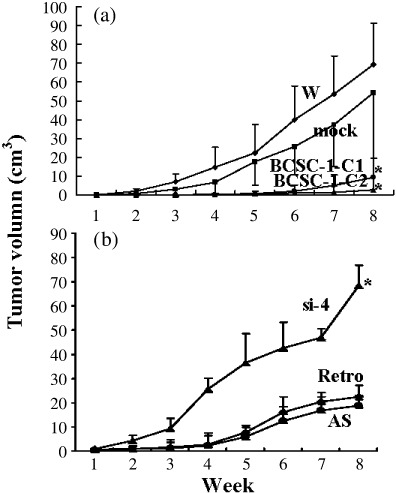
Suppression of tumorigenesis of CNE‐2L2 cells by BCSC‐1. A total of 5 × 105 cells were subcutaneously inoculated into the right armpit of nude mice with seven mice in each group. The longest (a) and shortest (b) diameters of the tumors were measured weekly for 8 weeks. The volume of the tumors was calculated according to the formula: volume = 4/3πa × b 2. (a) (b) Growth curves of tumors. *The difference between wild type (W) or Mock and BCSC‐1‐1 or BCSC‐1‐2 and between AS or Retro and si‐4 was statistically significant (paired t‐test, P < 0.01).
To further confirm the tumor suppressor function of BCSC‐1, experimental tumor gene therapy with BCSC‐1 was carried out. When tumors from inoculated W cells in nude mice reached the size of 50–100 mm3, Ad5‐BCSC‐1, Ad5‐egfp, or PBS was injected into tumors. Two weeks after the second virus injection, the average tumor size of the PBS group, Ad5‐egfp group, and Ad5‐BCSC‐1 group was 3.14 ± 0.85, 3.61 ± 0.89, and 0.80 ± 0.41 cm3 respectively. The difference between PBS or Ad5‐egfp and Ad5‐BCSC‐1 was significant (paired t‐test, P < 0.01). A tumor size comparison is shown in Figure 4.
Figure 4.
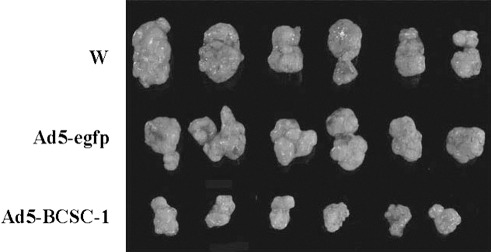
Growth suppression of CNE‐2L2 tumors by intratumor BCSC‐1 injection. A suspension containing 1 × 106 wild‐type CNE‐2L2 cells (W) was subcutaneously inoculated into each nude mouse. When tumors grew to 50–100 mm3, tumors in each mouse of groups 1, 2, and 3 were injected with 0.1 mL PBS, 0.1 mL Ad5‐egfp (4 × 108 infectious units [IFU]) or 0.1 mL Ad5‐BCSC‐1 (4 × 108 IFU), respectively. The injection was repeated 1 week later. Two weeks after the second injection, tumors were excised. The figure shows a size comparison of the tumors.
Effect of BCSC‐1 on cell aggregation, E‐cadherin/α‐catenin/β‐catenin expression, and Wnt signaling. Ectopic BCSC‐1 promoted aggregation of CNE‐2L2 cells (Fig. 5a) and E‐cadherin and α‐catenin expression (Fig. 5b). BCSC‐1 suppression resulted in upregulation of Wnt3, Wnt4, Wnt5a, Wnt6, Wnt10b, Fzd4, and Tcf3 (Fig. 5c) and enhanced expression of β‐catenin (Fig. 5d). Expression of the remainder of the Wnt signaling pathway was not affected by BCSC‐1 suppression. Expression of Wnt family members was assayed by RT‐PCR using the primers listed in Supporting information: Table 1.
Figure 5.
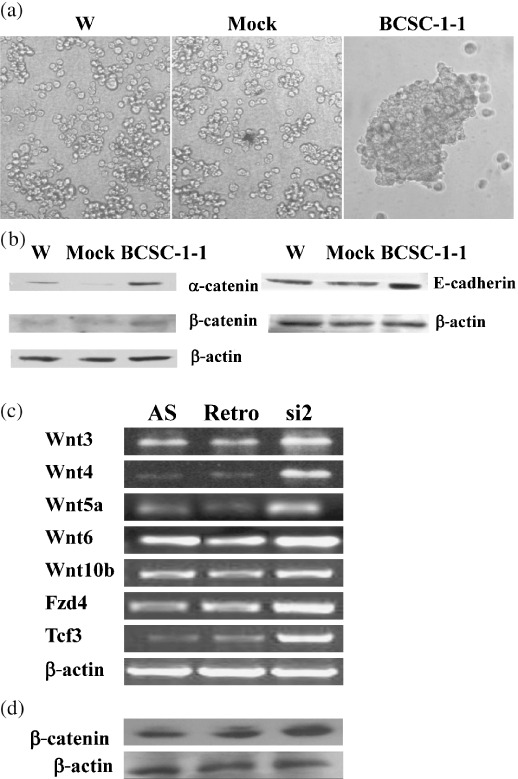
Effect of BCSC‐1 expression on cell aggregation, expression of E‐cadherin and α‐catenin and β‐catenin, and Wnt family members. A total of 1 × 105 cells were suspended in 1 mL HBSS‐Ca2+ and incubated for 60 min at 5%CO2, 37°C with intermittent gentle shaking. (a) Cell aggregation was examined using a reverse microscope. (b,d) Western blot analysis was carried out with mAb anti‐E‐cadherin, anti‐α‐catenin, or anti‐β‐catenin as the primary antibody. RT‐PCR for Wnt family members was carried out with the primers listed in Supporting information: Table 1C. (a) and (b) shows that ectopic BCSC‐1 results in enhanced agression of and increased E‐cadherin. (c) and (d) show that BCSC‐1 suppression causes enhanced Wnt‐signaling and β‐cat enin in AS cell respectively.
BCSC‐1 expression in human nasopharyngeal carcinomas. Immunohistochemistry was carried out to examine BCSC‐1 expression in three human normal nasopharyngeal tissue specimens and 39 human nasopharyngeal carcinoma specimens. Normal nasopharyngeal epithelial cells in the three specimens showed rich BCSC‐1 expression. Some lymphoid cells in the interstitial tissues also showed rich BCSC‐1 expression. Eleven of 39 carcinoma specimens (28.2%) exhibited marked reduction of BCSC‐1 expression whereas BCSC‐1 expression in the rest was as rich as that in normal cells. Supporting information: Figure 3 shows examples of BCSC‐1 expression in normal nasopharyngeal tissues and in nasopharyngeal carcinomas assayed by immunohistochemistry.
Discussion
BCSC‐1 (NM_014622) is a newly characterized candidate TSG located at chromosome 11q23 encoding a 786‐amino acid protein with a predicted molecular mass of 86 kDa.( 2 ) Sequence alignment of nine BCSC‐1 protein homologues shows high homology of the sequences, indicating that BCSC‐1 is a highly conserved protein in evolution. Based on the proposal by Martin et al. that BCSC‐1 is a candidate TSG( 2 ) and the microarray data showing enhanced BCSC‐1 expression in the CNE‐2L2 cells with reduced malignancy caused by α‐mannosidase Man2c1 suppression, the effect of BCSC‐1 expression on malignant behaviors of human nasopharyngeal carcinoma cells was examined in the present study. BCSC‐1 was found to be a TSG in CNE‐2L2 cells.
Tumorigenesis is a very complicated process and is regulated by a number of factors.( 4 , 5 , 6 ) TSG have been found to exhibit multiple inhibitive effects on tumorigenesis.( 3 ) For example, the retinoblastoma (RB) gene was reported to both govern the passage of cells through the G1 phase‐restriction point( 13 ) and promote terminal differentiation.( 14 ) In addition, Rb has also been observed to play roles in ensuring proper chromosome functions such as mitotic progression, faithful chromosome segregation,( 15 ) checkpoint control,( 16 ) and chromatin remodeling,( 17 ) suggesting that Rb is involved in the maintenance of genome integrity. Tumorigenesis of nasopharyngeal carcinoma is also a very complicated process and is associated with a number of factors.( 4 ) Because activation of the Wnt signaling pathway( 8 ) and reduced cell adhesion caused by decreased E‐cadherin( 7 ) were reported in nasopharyngeal carcinoma, the effect of BCSC‐1 expression on Wnt signaling, E‐cadherin expression, and aggregation of CNE‐2L2 cells were examined. It was observed that ectopic BCSC‐1 caused enhanced aggregation of cells together with increased expression of E‐cadherin/α‐catenin. In epithelial cells, cell adhesion is mainly mediated by E‐cadherin,( 18 ) which functions through complex formation with catenins.( 19 ) Loss or reduction of E‐cadherin expression was reported to be correlated with reduced aggregation and dedifferentiation of many carcinomas.( 20 , 21 ) Therefore, enhanced aggregation of cells associated with increased expression of E‐cadherin/α‐catenin would be one of the mechanisms associated with the suppressive effect of BCSC‐1 on the malignant behaviors of CNE‐2L2 cells. The Wnt pathway is one of the cell proliferation signaling pathways, enhancement of which plays important roles in tumorigenesis.( 22 , 23 ) The Wnt pathway is either β‐catenin‐dependent or β‐catenin‐independent.( 23 ) In the present study expression of all members of the Wnt family together with their cell surface receptors was assayed by RT‐PCR and that of β‐catenin was examined by western blotting in whole cells. Upregulation of Wnt3, Wnt4, Wnt5a, Wnt6, Wnt10b, Fzd4, and Tcf3 together with β‐catenin in si‐2 cells suggests that BCSC‐1 suppression would enhance Wnt signaling, which might play roles in the malignancy recovery of the cells.
With regard to possible mechanisms associated with enhanced BCSC‐1 expression induced by Man2c1 suppression, as few papers dealing with the possible functions of Man2c1 can be found in the literature,( 24 ) it is impossible to figure out an exact picture. Based on the fact that the major function of α‐mannosidase is to trim mannoses in N‐glycans( 25 ) and most proteins, including transcription factors, are N‐glycosylated,( 26 , 27 ) we imagine that Man2c1 suppression might result in N‐glycosylation modification of some transcription factors, consequently influencing transcription of genes including BCSC‐1.
Supporting information
Supporting information: Materials and Methods
Fig. S1. Sequence alignments for eight homologues of BCSC‐1 proteins.
Fig. S2. Man2c1 suppression caused upregulation of BCSC‐1 in CNE‐2L2 cells.
Fig. S3. BCSC‐1 expression in human nasopharyngeal carcinomas.
Table S1. List of the oligonucleotide sequences used
Table S2. Genes differentially expressed in AS relative to wild‐type CNE‐2L2 cells
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Acknowledgments
This work was supported by The National 863 Program of China (Grant no. 2002BA711A03 and 2002BA711A01) and The National Key Basic Research Program of China (Grant no. 2001CB510004).
References
- 1. Yue W, Jin YL, Shi GX et al . Suppression of 6A8 α‐mannosidase gene expression reduced the potentiality of growth and metastasis of human nasopharyngeal carcinoma. Int J Cancer 2004; 108: 189–95. [DOI] [PubMed] [Google Scholar]
- 2. Martin ES, Cesari R, Pentimalli F et al . The BCSC‐1 locus at chromosome 11q23‐q24 is a candidate tumor suppressor gene. Proc Natl Acad Sci USA 2003; 100: 11 517–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zheng L, Lee WH. The retinoblastoma gene: a prototypic and multifunctional tumor suppressor. Exp Cell Res 2001; 264: 2–18. [DOI] [PubMed] [Google Scholar]
- 4. Chou J, Lin Y‐C, Kim J et al . Nasopharyngeal carcinoma – review of the molecular mechanisms of tumorigenesis. Clin Rev 2008; 30: 946–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yokota J. Tumor progression and metastasis. Carcinogenesis 2000; 21: 497–503. [DOI] [PubMed] [Google Scholar]
- 6. Cairns RA, Khokha R, Hill RP. Molecular mechanisms of tumor invasion and metastasis: an integrated view. Curr Mol Med 2003; 3: 659–71. [DOI] [PubMed] [Google Scholar]
- 7. Li Z, Ren Y, Lin S, Liang Y, Liang H. Association of E‐cadharin and β‐catenin with metastasis in nasopharyngeal carcinoma. Chin Med J (Engl) 2004; 117: 1232–9. [PubMed] [Google Scholar]
- 8. Zeng ZY, Zhou YH, Zhang WL et al . Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Human Pathol 2007; 38: 120–33. [DOI] [PubMed] [Google Scholar]
- 9. Yu DN, Gao J. Study on the invasive properties of cloned human nasopharyngeal carcinoma CNE‐2L2 variants using organ culture in vitro . Acta Acad Med Sin (Chi) 1994; 16: 317–21. [PubMed] [Google Scholar]
- 10. Yao KT, Zhang HY, Zhu HC et al . Establishment and characterization of two epithelial tumor cell lines (HNE‐1 and HONE‐1) latently infected with Epstein–Barr virus and derived from nasopharyngeal carcinomas. Int J Cancer 1990; 45: 83–9. [DOI] [PubMed] [Google Scholar]
- 11. Harlow E, Lane D. Antibody: A Laboratory Manual. New York: Cold Spring Harbor Laboratory, 1988. [Google Scholar]
- 12. Liu CM, Liu DP, Dong WJ, Liang CC. Retrovirus vector‐mediated stable gene silencing in human cell. Biochem Biophys Res Comm 2004; 313: 716–20. [DOI] [PubMed] [Google Scholar]
- 13. Sherr CJ. Cancer cell cycles. Science 1996; 274: 1672–7. [DOI] [PubMed] [Google Scholar]
- 14. Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev 1996; 10: 2794–804. [DOI] [PubMed] [Google Scholar]
- 15. Zheng L, Chen Y, Riley DJ, Chen PL, Lee WH. Retinoblastoma protein enhances the fidelity of chromosome segregation mediated by hsHeclp. Mol Cell Biol 2000; 20: 3529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flatt PM, Tang LJ, Scatena LD, Szak ST, Pletenpol JA. p53 regulation of G2 checkpoint is retinoblastoma protein dependent. Mol Cell Biol 2000; 20: 4210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrera RE, Chen F, Weinberg RA. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb‐deficient fibroblasts. Proc Natl Acad Sci USA 1996; 93: 11 510–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science 1991; 251: 1452–5. [DOI] [PubMed] [Google Scholar]
- 19. Beavon IRG. The E‐cadherin–catenin complex in tumor metastasis: structure, function and regulation. Eur J Cancer 2000; 36: 1607–20. [DOI] [PubMed] [Google Scholar]
- 20. Birchmeier W, Bejerems J. Cadherin expression in carcinomas: role in the formation of cell junction and the prevention of invasiveness. Biochem Biophys Acta 1994; 1198: 11–26. [DOI] [PubMed] [Google Scholar]
- 21. Tamura S, Shiozaki H, Miyata M et al . Decreased E‐cadherin expression is associated with haematogenous recurrence and poor prognosis in patients with squamous cell carcinoma of the esophagus. Br J Surg 1996; 83: 1608–14. [DOI] [PubMed] [Google Scholar]
- 22. Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol 2003; 129: 199–221. [DOI] [PubMed] [Google Scholar]
- 23. Kikuchi A, Yamamoto H. Tumor formation due to abnormalities in the β‐catenin‐independent pathway of Wnt signaling. Cancer Sci 2008; 99: 202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suzuki T, Hara I, Nakano M et al . Man2C1, an α‐mannosidase, is involved in trimming free oligosaccharides in the cytosol. Biochem J 2006; 400: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Daniel PF, Winchester B, Warren CD. Mammalian α‐mannosidase – multiple forms but a common purpose? Glycobiology 1994; 4: 113–25. [DOI] [PubMed] [Google Scholar]
- 26. Kukuruzinska MA, Lennon K. Protein N‐glycosylation: molecular genetics and functional significance. Crit Rev Oral Biol Med 1998; 9: 415–48. [DOI] [PubMed] [Google Scholar]
- 27. Kane R, Murtagh J, Finlay D et al . Transcription factor NFIC undergoes N‐glycosylation during early mammary gland involution. J Biol Chem 2002; 277: 25 893–903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information: Materials and Methods
Fig. S1. Sequence alignments for eight homologues of BCSC‐1 proteins.
Fig. S2. Man2c1 suppression caused upregulation of BCSC‐1 in CNE‐2L2 cells.
Fig. S3. BCSC‐1 expression in human nasopharyngeal carcinomas.
Table S1. List of the oligonucleotide sequences used
Table S2. Genes differentially expressed in AS relative to wild‐type CNE‐2L2 cells
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item


