Abstract
GB virus B (GBV-B) is a recently discovered hepatotropic flavivirus that is distantly related to hepatitis C virus (HCV). We show here that translation of its polyprotein is initiated by internal entry of ribosomes on GBV-B RNA. We analyzed the translational activity of dicistronic RNA transcripts containing wild-type or mutated 5′ nontranslated GBV-B RNA (5′NTR) segments, placed between the coding sequences of two reporter proteins, in vitro in rabbit reticulocyte lysate and in vivo in transfected BT7-H cells. We related these results to a previously proposed model of the secondary structure of the GBV-B 5′NTR (M. Honda, et al. RNA 2:955–968, 1996). We identified an internal ribosome entry site (IRES) bounded at its 5′ end by structural domain II, a location analogous to the 5′ limit of the IRES in both the HCV and pestivirus 5′NTRs. Mutational analysis confirmed the structure proposed for domain II of GBV-B RNA, and demonstrated that optimal IRES-mediated translation is dependent on each of the putative RNA hairpins in this domain, including two stem-loops not present in the HCV or pestivirus structures. IRES activity was also absolutely dependent on (i) phylogenetically conserved, adenosine-containing bulge loops in domain III and (ii) the primary nucleotide sequence of stem-loop IIIe. IRES-directed translation was inhibited by a series of point mutations predicted to stabilize stem-loop IV, which contains the initiator AUG codon in its loop segment. A reporter gene was translated most efficiently when fused directly to the initiator AUG codon, with no intervening downstream GBV-B sequence. This finding indicates that the 3′ limit of the GBV-B IRES is at the initiator AUG and that it does not require downstream polyprotein-coding sequence as suggested for the HCV IRES. These results show that the GBV-B IRES, while sharing a common general structure, differs both structurally and functionally from other flavivirus IRES elements.
GB virus B (GBV-B) is a recently identified member of the family Flaviviridae that has yet to be classified within a specific genus (23). Its genome was molecularly cloned from material taken from a tamarin that had been experimentally infected with a putative hepatitis agent (the GB agent) that had been passaged serially in this species. Although the inoculum used to infect the tamarins was originally derived from a human patient who was considered to have viral hepatitis (3), there have been no reports of natural GBV-B infections in humans, tamarins, or, for that matter, any other animal species. However, cotton-top tamarins can be infected experimentally, and these animals develop an acute hepatitis consistent with molecular evidence that the virus is hepatotropic (21, 23). The sequence of the GBV-B genome exhibits a putative genome organization similar to that of other members of the Flaviviridae, especially hepatitis C virus (HCV) (10, 23). Moreover, its NS3 proteinase has been shown to share substrate specificity with the HCV proteinase (20). GBV-B has a close phylogenetic relationship to HCV, although there is only ∼28% amino acid identity with HCV across its entire open reading frame (10). Because the host range of GBV-B extends to small New World primates that are not permissive for HCV infection, these features make GBV-B a particularly interesting virus to study.
Cap-independent translation of the HCV and pestivirus genomes is driven by an internal ribosome entry site (IRES) located within the 5′ nontranslated RNA (5′NTR) (12, 16, 27, 29). This IRES activity is critically dependent on the structure of the highly ordered 5′NTR in these viruses (15, 16, 28, 30). The 5′NTR of GBV-B is predicted to fold into an RNA structure that is similar to that described for the 5′NTRs of HCV and the pestiviruses bovine viral diarrhea virus and classical swine fever virus (also known as hog cholera virus) (Fig. 1) (1, 5, 6, 26). Thus, it is likely to contain an IRES that is functionally similar to those in these other flaviviruses and to translate its genome by a cap-independent process.
FIG. 1.
Predicted models of the secondary structures of the 5′NTRs of GBV-B (a) and HCV (b). (a) Structure proposed for the GBV-B 5′NTR by Honda et al. (5, 6). Major predicted structural domains are labeled I to IV, while individual stem-loops are labeled Ia and are analogous to similarly labeled structures in the HCV 5′NTR. Base-pair interactions involving the loop sequence of stem-loop IIIf that are predicted to result in a putative RNA pseudoknot are drawn as solid lines. Lightly shaded boxes represent AUG triplets located within the 5′NTR, while the solid black box represents the polyprotein translation initiation site. The open box indicates a helical segment at the base of domain II, and open circles represent individual nucleotides that were subjected to site-directed mutagenesis in the studies described here. Unpaired bases within domain II that are conserved in domain II of the HCV structure are shown in boldface type. The 5′ and 3′ limits of the GBV-B IRES, as determined in this study, are indicated by the arrows. (B) Structure proposed for the HCV 5′NTR (3, 5, 6). There are numerous similarities with the GBV-B structure but no predicted stem-loop structures analogous to the Ib, IIb, and IIc stem-loops of GBV-B.
Despite the similarities in the predicted structure of the 5′NTR of GBV-B and that of HCV and pestiviruses, there are some distinct differences (6) (compare Fig. 1a and b) (17). First, there is substantial divergence from the HCV structure near the 5′ end of the GBV 5′NTR. The sequence in this region has been predicted to form two stem-loops in GBV-B (Fig. 1a, stem-loops Ia and Ib) but only a single stem-loop in the HCV structure (Fig. 1b, stem-loop I). This feature of the predicted GBV-B structure thus more closely resembles the structures predicted for the 5′NTRs of the pestiviruses (1, 4). A second major difference is that the 5′NTR of GBV-B is 445 nucleotides (nt) in length and thus significantly longer than the 5′NTRs of either HCV or the pestiviruses (341 to 384 nt). The greater length of the GBV-B 5′NTR appears to be due to the presence of two additional stem-loops (Fig. 1a, stem-loops IIb and IIc) that are not present in the other flaviviral 5′NTRs (6). These two stem-loops are located within domain II of the predicted GBV-B structure and likely within the putative GBV-B IRES, given the location of the IRES in the other flaviviruses (9). It is of interest to determine whether these stem-loop structures are required for activity of the GBV-B IRES, since they are absent in the other flaviviral IRES elements. Finally, a third difference is that the GBV-B sequence, like the HCV sequence, is predicted to form a stem-loop containing the initiator AUG codon at the 5′ end of the open reading frame within its loop segment (Fig. 1a, domain IV) (6). This structure is not present in the pestivirus sequences and has been suggested to play a regulatory role in HCV translation (6). Its presence in the GBV-B structure is intriguing since it has been shown to be nonessential for HCV IRES activity.
In addition to these differences in their proposed structures, very little primary nucleotide sequence is conserved among the 5′NTRs of GBV-B, HCV, and the pestiviruses. However, despite substantial variation in the nucleotide sequence of base-paired segments, the short unpaired loop sequences of the complex stem-loop forming domain II of the GBV-B structure are identical to the homologous loop segments in both HCV and the pestiviruses (5). In addition, four unpaired adenosine residues, A253, A392, A271, and A272, which are located within internal loops of domain III, are entirely conserved between GBV-B, HCV (Fig. 1), and the pestiviruses. Furthermore, the primary sequence of one small hairpin, stem-loop IIIe, is identical in GBV-B, HCV, and the pestivirus 5′NTRs. The conservation of these elements in these different viruses suggests an essential function in either replication or translation.
In an effort to better understand the functional correlates of these conserved 5′NTR elements, as well as the functional importance of differences in the structures proposed for these 5′NTRs, we have carried out a detailed mutational analysis of the GBV-B sequence. We show that this 5′NTR contains an IRES with activity approximating that of the HCV IRES and compare this novel translational control element with the other flavivirus IRES elements.
MATERIALS AND METHODS
Cells.
BT7-H cells, which constitutively express T7 RNA polymerase (31), were grown in Dulbecco's modified eagle medium (Gibco BRL) supplemented with 10% fetal calf serum and 500 μg (active compound) of geneticin (Gibco BRL) per ml.
Plasmids.
pLuc-GBVB-CAT contains a cDNA insert representing nt 1 to 445 of the GBV-B sequence and was a generous gift from John Simons and Isa Mushahwar of Abbott Laboratories. To construct a dicistonic reporter plasmid with which we could assess the ability of the GBV-B 5′NTR to direct internal initiation of translation, the firefly luciferase fragment in pLuc-wt-CAT (R. C. A. Rijnbrand, P. J. Bredenbeek, P. C. J. Haasnoot, W. J. M. Spaan, and S. Lemon. unpublished data) was replaced by a PCR-amplified DNA fragment containing the coding sequence for renilla luciferase (rLuc). The HCV 5′NTR in the resulting construct was then replaced by a fragment amplified from pLuc-GBVB-CAT that contained the entire 5′NTR and AUG initiation codon of the GBV-B sequence to generate pRLuc-GBB+3-CAT. For convenience, this reporter construct is referred to as p-wt in the experiments described below (Fig. 2a). It contains a T7 transcriptional unit in which the rLuc and chloramphenicol acetyltransferase (CAT) coding sequences are separated by the GBV-B 5′NTR sequence in the intercistronic space. A series of deletion mutants and additional plasmids containing point mutations within the GBV-B sequence were subsequently constructed from p-wt by PCR-directed mutagenesis or site-directed mutagenesis (QuickChange; Statagene) carried out on isolated restriction digestion fragments. The sequence of the mutated fragments was confirmed to exclude additional unwanted mutations before they were reinserted into the background of p-wt. Monocistronic reporter plasmids lacking the upstream rLuc coding sequence and the 5′ 284 nt of the GBV-B 5′NTR were constructed by digestion of p-wt (or its derivatives) with SacII (located upstream of rLuc) and AgeI (located within the GBV-B 5′NTR), followed by blunting of the ends by T4 DNA polymerase and religation.
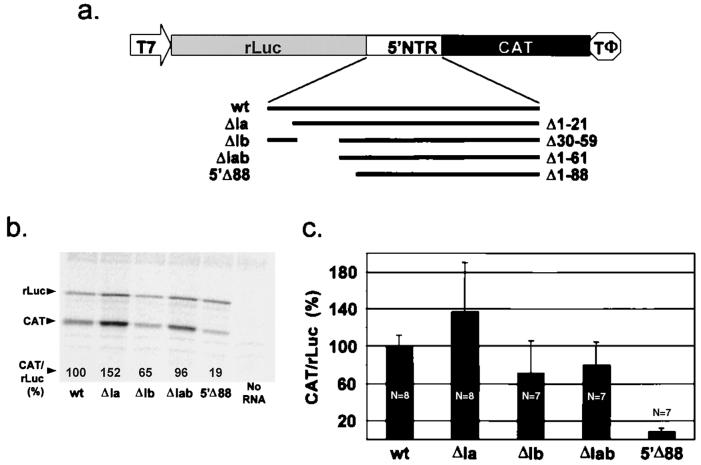
FIG. 2.
Demonstration of the GBV-B IRES and mapping of its 5′ limit. (a) Diagram showing the organization of the transcriptional unit in plasmid used to characterize the IRES. T7 and TΦ represent the T7 RNA polymerase promoter and terminator sequences. The locations of specific deletion mutations created within the 5′NTR of GBV-B in derivatives of p-wt are shown, with the nucleotides deleted listed at the right. (b) SDS-PAGE of products of in vitro translation reactions programmed with the indicated RNA transcripts. CAT/rLuc is a numeric ratio calculated from the PhosphorImager analysis of the gel, relative to an arbitrary value of 100% for p-wt transcripts. (c) Relative translational activity (CAT/rLuc) of the indicated mutants following their transfection as DNA into BT7-H cells; a value of 100% was arbitrarily assigned to cells transfected with p-wt. The number of replicate transfection assays (N) is shown for each mutant. Error bars indicate standard deviations. The results indicate that the 5′ limit of the IRES is located between nt 61 and 88, or close to the 5′ limit of structural domain II (Fig. 1a).
Recombinant DNA techniques were performed by standard procedures (19). Restriction endonucleases, DNA polymerases, and T4 DNA ligase were obtained from Promega and New England Biolabs. Oligonucleotides were purchased from Genosys and Gibco-BRL.
In vitro translations.
Plasmid DNA was transcribed in vitro from p-wt and its mutated derivatives by using T7 RNA polymerase, and the integrity of the resulting RNA transcripts was confirmed in agarose gels. In vitro translation reactions were carried out in rabbit reticulocyte lysates (Flexi-Lysate; Promega) programmed with these RNA transcripts (125 ng/10 μl) and containing 80 or 125 mM KCl. Translation products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5% gels and quantified by analysis with a PhosphorImager (Molecular Dynamics).
DNA transfection of BT7-H cells.
Nearly confluent BT7-H cells grown in 60-mm-diameter plastic dishes were transfected with plasmid DNA mixed with FuGENE 6 (Boehringer Mannheim). In a polypropylene tube, 100 μl of OptiMEM (Gibco-BRL) and 6 μl of FuGENE reagent were incubated for 10 min at room temperature prior to the addition of plasmid DNA (2 μg) that had been purified on a QIAprep spin column (Qiagen). The mixture was incubated for an additional 15 min at room temperature, and 100 μl was added directly to cells fed previously with 2 ml of growth medium. The cells were incubated for 20 to 24 h posttransfection at 37°C, then washed twice with phosphate-buffered saline, and harvested with a plastic scraper into 1 ml of TEN (40 mM Tris-HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl). The suspended cells were collected by low-speed centrifugation at room temperature, resuspended in 125 μl of 0.25 M Tris-HCl (pH 8.0), and lysed by three cycles of freeze-thawing. Cellular debris was removed by additional centrifugation immediately after the final freeze-thaw cycle.
Measurement of reporter protein activities.
rLuc activity was measured in 20-μl aliquots of cell lysate, using rLuc reagents from the Dual Luciferase reporter assay system (Promega). For measurement of CAT activity, 10 μl of lysate was incubated at 60°C for 10 min to inactivate endogenous acetylase activity, then added to a mixture of 5 μl of n-butyryl coenzyme A (200 μg/ml), 100 μl of 0.25 M Tris-HCl (pH 8.0), and 10 μl of labeled d-threo[dichloroacetyl-1,2-14C]chloramphenicol (50 to 60 mCi/mmol), and reincubated for 1 h at 37°C (30). The activity was determined by liquid scintillation counting following xylene phase extraction.
RESULTS
Demonstration of the GBV-B IRES and mapping of its 5′ border.
To confirm the existence of an IRES within the 5′NTR of GBV-B, the 5′NTR sequence was placed between the rLuc and CAT coding sequences to create vector p-wt (Fig. 2a). A series of mutants derived from this plasmid contained large deletions that removed putative stem-loop structures near the 5′ terminus of the GBV-B sequence (pΔIa, pΔIb, or pΔIab) or the first 88 nt of the 5′NTR (pΔ1-88). The CAT coding region in the RNAs transcribed from these plasmids can be translated only by internal initiation, a process dependent on the presence of an IRES within the intercistronic space. Thus, measurement of CAT expression from these transcripts allowed us to determine whether the GBV-B 5′NTR contains an IRES and, if so, whether the 5′ 88 nt of the 5′NTR are necessary for this IRES activity. Figure 2b shows the results of in vitro translation reactions carried out in rabbit reticulocyte lysates programmed with these RNAs; Fig. 2c shows the level of CAT expression relative to rLuc expression in transfected BT7-H cells. These cells constitutively express T7 RNA polymerase and produce uncapped cytoplasmic transcripts from plasmids containing T7 transcriptional units (31). The level of CAT expression was examined in relation to the quantity of rLuc expressed from the upstream cistron of the transcripts in this and most of the experiments that follow, as rLuc provided a convenient measure of the quantity of RNA available for translation.
The results shown in Fig. 2b and c show that the GBV-B 5′NTR contains an IRES. The activity of this IRES was comparable to that of the HCV IRES when the HCV 5′NTR was inserted in a similar context between the rLuc and CAT coding sequences, with the CAT sequence fused directly to the HCV initiator AUG codon (data not shown). Both in vitro and in vivo, the removal of the most 5′ stem-loop within the GBV-B 5′NTR (stem-loop Ia) resulted in a slight enhancement of translation of the downstream CAT cistron relative to the upstream rLuc cistron (compare wt [wild type] with ΔIa in Fig. 2b and 2c). These results are reminiscent of a similar increase in HCV and pestiviral IRES activity that has been observed upon removal of the analogous structure from these 5′NTRs (7, 8, 15, 16). In contrast, removal of either hairpin Ib or the combination of Ia and Ib resulted in a minimal (20 to 30%) decrease of translation in BT7-H cells (Fig. 2c, ΔIb and ΔIab) while having a similar effect on CAT synthesis in vitro (Fig. 2b). These results indicate that sequences located 5′ of nt 61 are not necessary for activity of the GBV-B IRES. However, deletion of nt 1 to 88 almost completely eliminated translation of CAT from the downstream reading frame in BT7-H cells (Fig. 2c, Δ1-88) and reduced translation to 19% of the wt level in reticulocyte lysate (Fig. 2b). Thus, the 5′ limit of the IRES is located between nt 61 and 88. As shown in Fig. 1a, this segment includes the putative helix at the base of the large complex stem-loop forming domain II of the GBV-B 5′NTR. These data are therefore consistent with the involvement of the domain II structure in the GBV-B IRES.
Each of the stem-loop structures in domain II is required for efficient GBV-B IRES activity.
The most striking difference between the GBV-B 5′NTR that of HCV is the presence of a large insertion that has been proposed to form two additional hairpin structures (stem-loops IIb and IIc) within domain II (6) (Fig. 1a). Although the upstream stem-loop IIa of GBV-B shares significant structural similarities and even limited primary sequence identity with the most 5′ stem-loops in the IRESs of HCV and the pestiviruses (5), these two hairpins in GBV-B have no counterparts in the 5′NTRs of these other viruses. To determine their role in IRES-mediated translation, we evaluated the translational efficiency of dicistronic transcripts with deletions of hairpin IIa, IIb, or IIc (Fig. 3a, ΔIIa, ΔIIb, and ΔIIc, respectively). In addition, we evaluated a mutant that lacks both hairpin IIb and IIc and that thus possesses a putative structure that superficially looks much like that found in HCV (Fig. 3a, ΔIIbc). Each of these mutant transcripts showed significantly reduced IRES activity, both in vitro in reticulocyte lysates (Fig. 3c) and in vivo in transfected BT7-H cells (Fig. 3d). In BT7-H cells, the activity of the IRES was reduced to approximately 15% of the wt level with the ΔIIa and ΔIIb transcripts and to about 30% of the wt level with ΔIIc. The mutant lacking both IIb and IIc (ΔIIbc) translated at approximately 20% of the wt level. The inhibition of translation following removal of stem-loops IIb and IIc was somewhat less pronounced in the in vitro translation reaction (Fig. 3c). However, in aggregate, these results indicate that all three hairpin structures are required for optimal IRES activity.
FIG. 3.
Mutational analysis of domain II of the GBV-B 5′NTR. (a) Diagram showing mutations introduced into the sequence of domain II to evaluate the IRES requirement for specific stem-loop structures in this domain. (b) Expanded view of the helix at the base of domain II (see boxed sequences in Fig. 1a). Nucleotides at which mutations were introduced by site-directed mutagenesis are indicated by open circles. (c) In vitro translation of the domain II mutants. See the legend to Fig. 2b for details. (d) Translational activity of domain II mutants in transfected BT7-H cells. See the legend to Fig. 2c for details. The results of these experiments provide confirmation of the predicted helical structure at the base of domain II and demonstrate that each of the three subsidiary stem-loops of domain II contributes to efficient IRES activity.
To determine the requirement for base pairing within the putative helix located at the base of domain II (Fig. 1a), we made a series of point mutations in the involved RNA segments (Fig. 3b). Mutations altering the sequence between nt 63 and 68 (mutant IIL) or nt 231 to 236 (mutant IIR) substantially disrupt this helix (Fig. 3b). The combination of these mutations in mutant IILR fully restores the potential for base pairing in this helix, with a predicted free energy that is virtually identical to that of the wt helix. The translational activity of the IRES in these mutants was significantly reduced both in vitro and in vivo (Fig. 3c and d). In BT7-H cells, the IRES activity of the IIL transcripts was approximately 50% of that of the wt, while IIR activity was only 10% of the wt activity. The combination of the two sets of mutations in IILR resulted in IRES activity that was approximately 65% that of wt. Thus, the insertion of the complementary IIL mutations into IIR resulted in an eightfold increase in the IRES activity of IIR. Similar results were obtained for reticulocyte lysates (Fig. 3c), although the level of complementation was somewhat greater. These results confirm the existence of the helix at the base of domain II and demonstrate its importance for IRES-mediated translation. They are remarkably similar to results obtained in mutational analyses of the HCV IRES. With the HCV IRES, mutations involving only the 5′ segment of the analogous helix at the base of domain II had a lesser effect on translation than mutations involving the 3′ segment of the helix (5).
IRES requirement for conserved domain III sequences.
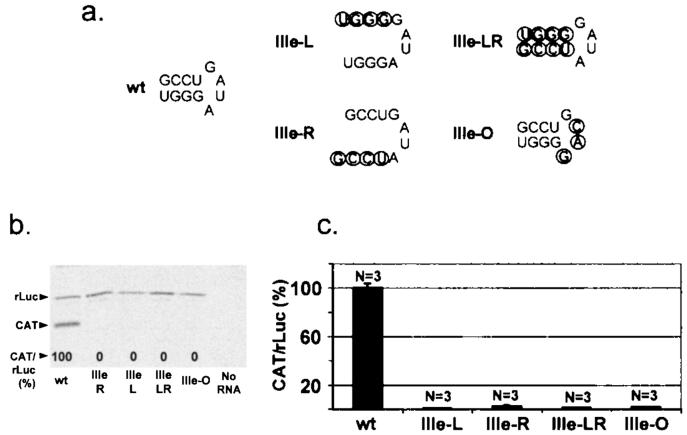
Although there is only limited primary sequence relatedness between the 5′NTRs of GBV-B and HCV (10), the structure of domain III appears remarkably conserved between these viral IRESes (compare Fig. 1). This makes it likely that the stem-loops that contribute to domain III are essential for GBV-B IRES-mediated initiation of translation, as the analogous structures have been shown to be for HCV (7, 15). However, previous studies of the HCV IRES did not address the requirements for stem-loop Ille. Interestingly, the primary nucleotide sequence of this hairpin structure is absolutely conserved between these two viruses, both in the stem and in the loop (compare the GBV-B and HCV structures in Fig. 1a and b). This makes it unique among the hairpin structures in the GBV-B IRES. To determine whether this reflects a requirement for IRES-mediated translation, we created mutations in the stem and loop sequences of IIIe within the dicistronic GBV-B construct. The nucleotide sequences of the 5′ and 3′ strands of the helix were individually altered to disrupt base pairing in the IIIe-L and IIIe-R mutants, while the reversal of the sequences of both strands in IIIe-LR restored the potential for base-pair formation (Fig. 4a). Both in vitro and in vivo, each of these mutants was devoid of significant IRES activity (Fig. 4b and c). Similar results were obtained with a mutant involving the stem-loop IIIe loop sequence, in which there were substitutions of three of the four loop nucleotides (Fig. 4a, IIIe-O). The IRES activity of this mutant was at background levels (Figs. 4b and c). These data demonstrate that domain III is critical for activity of the GBV-B IRES, as it is with HCV (7, 15). However, they also indicate that the primary sequence of both the stem and loop of IIIe are essential for IRES activity. The absence of translational activity in the IIIe-LR mutant, in which the potential for stem formation is preserved (Fig. 4a), may explain the absence of variation in the sequence of this putative helix among different flaviviruses, in contrast to other helical structures in domain II and III (1).
FIG. 4.
The phylogenetically conserved stem-loop IIIe is critically important for translational activity of the GBV-B IRES. (a) Changes made in the nucleotide sequences of stem and loop regions of stem-loop IIIe. Altered nucleotides are encircled, with effects on the predicted structure as shown. Also shown is the translational activity of the stem-loop IIIe mutants in rabbit reticulocyte lysate (b) or transfected BT7-H cells (c). See the legend to Fig. 2 for details.
Another remarkable similarity between the GBV-B and HCV and pestivirus IRESes is the preservation of four unpaired adenosine residues within domain III, at positions 253, 271, 272, and 392 of the GBV-B structure (Fig. 1a and b). Interestingly, the unpaired adenosines at positions 253 and 392 are flanked by conserved base pairings on both sides and thus form a small, well-conserved internal loop that is in close proximity to the pseudoknot. To test whether this sequence conservation is driven by requirements of the IRES, we altered the conserved adenosines and, in addition, the conserved, base-paired nucleotides that flank them in the wt dicistronic GBV-B construct. The adenosines at positions 253 and 392 were individually changed to uridines in the mutants IIIA-L and IIIA-R, respectively, in each case replacing the internal loop with either a U-A or an A-U base pair (Fig. 5a). The combination of both mutations (IIIA-LR) resulted in the apposition of two uridines at this locus that, like the wt adenosines, are unable to form a conventional Watson-Crick base pair and thus likely to recreate an internal loop (Fig. 5a). The IRES activity of all three mutants was at background levels, both in vitro and in vivo (Fig. 5c and d).
FIG. 5.
Four phylogenetically conserved, unpaired adenosine residues are critically important for GBV-B IRES activity. Changes were made in the context of p-wt to test the effect of the conserved adenosine residues at positions 253 and 392 (a) and positions 271 and 272 (b). Nucleotides that have been changed by site-directed mutagenesis are encircled. (c and d) Translational activities of the indicated mutants in rabbit reticulocyte lysate (c) and transfected BT7-H cells (d). All of the substitutions at these sites were lethal for translation, as were substitutions involving base pairs flanking the internal loop shown in panel a.
Similar results were obtained with substitutions involving the conserved base pairs (G252:C393 and G254:U391) that flank these adenosine residues. The change of G252→C in combination with G254→U (Fig. 5a, IIIs-L) or C393G in combination with U391→G (Fig. 5a, IIIs-R) should result in a severe distortion of the stem and further open the internal loop (Fig. 5a). Not surprisingly, both mutations severely impaired IRES-directed translation in vitro and in vivo (Fig. 5c and d). The combination of these two sets of mutations restores the potential for base pairing adjacent to the unpaired adenosines (Fig. 5a, IIIs-LR) but resulted in only a marginal restoration of IRES activity (Fig. 5c and d). These results are not surprising, since these base pairs are highly conserved among different flaviviruses. However, we observed a similar effect when we altered the base pair G256:C289, which is not conserved in the HCV and pestivirus structures (1, 6) (compare Fig. 1a and b). This mutant was not evaluated in BT7-H cells, but it had negligible IRES activity in rabbit reticulocyte lysates (Fig. 5c, NC). These results indicate that both the internal loop formed by the unpaired adenosines and the primary sequence of the flanking base-paired segments at the base of domain III are critical for activity of the GBV-B IRES.
Similarly, the unpaired adenosines at nt 271 and 272 of the GBV-B sequence, which are conserved in HCV (compare Fig. 1a and b) and the pestiviruses, are also essential for IRES activity. An additional mutant in which these two adenosine residues were substituted with U and C, respectively (Fig. 5b, IIIa-ΔAA), had little more than background translational activity, both in vitro and in vivo (Fig. 5c and d).
GBV-B IRES activity is not dependent on polyprotein-coding sequence.
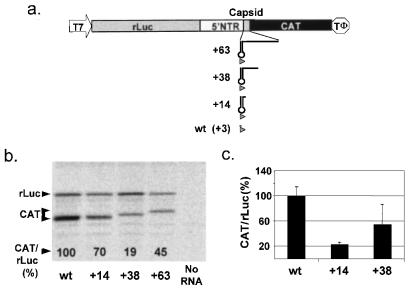
Reynolds et al. (13) suggested that approximately 32 nt of the HCV polyprotein-coding sequence is required downstream of the initiator AUG for efficient HCV IRES-mediated translation. If this is correct, the strong functional and structural similarities that are evident between the HCV and GBV-B 5′NTRs suggest that this might also be true for GBV-B. To assess this possibility, we altered the cistron downstream of the GBV-B IRES in p-wt to include 14, 38, or 63 nt of the GBV-B polyprotein-coding sequence, fused in frame at the 5′ end of the CAT sequence (Fig. 6a, +14, +38, and +63). Since the wt construct contains an AUG codon in the same position as the AUG of the GBV-B open reading frame, it should be considered to contain 3 nt of the coding sequence in these experiments. With each of these constructs, both rLuc and CAT translation were readily detectable in rabbit reticulocyte lysates (Fig. 6b) and in transfected BT7-H cells (Fig. 6c).
FIG. 6.
GBV-B IRES-mediated translation does not require polyprotein-coding sequences. (a) Bicistronic RNAs expressing CAT under the control of the GBV-B IRES with different lengths of coding sequence between the IRES and the reporter sequence. Arrowheads represent the AUG at the translation initiation site, with stem-loop IV depicted in the +14 transcript and transcripts with longer lengths of coding sequence, 38 and 63 nt. Transcripts from p-wt have 3 nt of the coding sequence, the AUG codon only. The translational activities of the transcripts shown in panel a were monitored in vitro in rabbit reticulocyte lysate (b) and in transfected BT7-H cells (c). See the legend to Fig. 2 for details.
However, both in vitro and in vivo, GBV-B IRES activity was greatest in the absence of any GBV-B coding sequence other than the initiator AUG codon. The activity of the IRES within the context of RNAs containing 14, 38, or 63 nt of the GBV-B coding sequence was less than that observed with the wt construct (Fig. 6b and c). We did not determine whether the amino-terminal fusion of short GBV-B polypeptide sequences to CAT could have reduced the specific activity of the reporter enzyme expressed from the +14 and +38 constructs in BT7-H cells (Fig. 6b). However, such an effect would not explain the reductions observed with these constructs in IRES-mediated translation of 35S-labeled CAT protein in reticulocyte lysates (Fig. 6b). These results indicate that the 3′ limit of the minimal GBV-B IRES is located at the initiator AUG codon. Contrary to what has been claimed for HCV (13), the GBV-B IRES has no specific requirement for polyprotein-coding sequence.
Stem-loop IV is inhibitory for IRES-directed translation.
The preceding experiment demonstrated that RNA transcripts containing 14 nt of the GBV-B coding sequence downstream of the IRES were translated with reduced efficiency, compared with those lacking any polyprotein-coding sequence other than the AUG. Interestingly, the +14 RNA transcripts contain a complete domain IV stem-loop, while this structure is lacking in the wt transcripts that do not contain any GBV-B coding sequence other than the AUG (Fig. 1). It is likely that the presence of this stem-loop accounts for the reduction in the translational activity of the +14 transcripts compared to the wt transcripts (Fig. 6). We recently demonstrated that slight increases in the predicted stability of stem-loop IV in the HCV sequence drastically reduce IRES efficiency (6). The GBV-B initiator codon appears to be located within the loop segment of a very similar hairpin (compare Fig. 1a and b). Although the putative GBV-B stem-loop IV has a predicted stability lower than that calculated for stem-loop IV of HCV (−3.1 kcal/mol versus −5.4 kcal/mol), it was of interest to determine whether a similar relationship exists with respect to the GBV-B IRES. Significantly, this structure is not present in the pestiviral sequences (6, 25).
To determine whether the stability of structure IV in the GBV-B sequence influences the translational efficiency of the IRES, we introduced into the background of the +14 construct additional nucleotide changes that were predicted to either increase or decrease the stability of the stem-loop [Fig. 7a, IV(+), −12.9 kcal/mol, and IV(−), −1.4 kcal/mol] or to generate an alternative structure with only a slightly increased predicted stability [Fig. 7a, IV(±), −3.4 kcal/mol]. Since the experiment shown in Fig. 6 demonstrated that the 3′ limit of the IRES is located at or upstream of the AUG codon, each of these stem-loop IV substitutions was introduced 3′ of the AUG codon. In addition, the substitutions were made at least 6 nt into the coding sequence in an effort to minimally perturb the context of the AUG codon. In the in vitro translation reaction, the activity of the IRES of the IV(+) mutant was reduced to background levels (Fig. 7b, left panel). In contrast, mutant IV(−), in which stem-loop IV was ablated, demonstrated IRES activity that was approximately equivalent (71%) to that of the wt sequence. The third mutant, IV(±), in which stem-loop IV was replaced with structure of slightly greater stability, had 28% of the wt translational activity in vitro (Fig. 7b, left panel). A reduction in translation was also observed with the IV(+) construct in transfected BT7-H cells (Fig. 7c), while the IV(±) construct had greater translation activity than the wt sequence (+14). These results are reminiscent of what has been described previously for the HCV IRES (6). Stem-loop IV is not required for efficient GBV-B translation, but translation is substantially inhibited by mutations that are predicted to increase its stability.
FIG. 7.
The stability of stem-loop IV influences translational activity of the GBV-B IRES. (a) Three different mutants were created to evaluate the effect of altering the stability of stem-loop IV. RNA structures and predicted free energies are from the MFOLD program of Michael Zucker, with folding energies of Turner, on the Washington University server (http://mfold/wustl.edu/∼folder/). Nucleotides that differ from the +14 construct sequence are encircled. The initiator AUG codon is highlighted in each transcript. (b) For the left panel, translational activities of the dicistronic transcripts depicted in panel a were assessed in vitro in rabbit reticulocyte lysates. To facilitate comparisons between the stem-loop IV mutants, the relative translational activity (amount of CAT expressed per unit of rLuc) of the +14 transcripts was arbitrarily assigned a value of 100%. The right panel shows CAT expressed in reticulocyte lysates programmed for translation with 5′-truncated, monocistronic RNAs containing the indicated stem-loop IV mutations. These truncated RNAs lack the 5′ 284 nt of the GBV-B 5′NTR and are translated by a ribosome scanning mechanism. (c) The translational activities of dicistronic transcripts depicted in panel a were assessed in transfected BT7-H cells. To facilitate comparisons between the stem-loop IV mutants, the relative translational activity (amount of CAT expressed per unit of rLuc) of the +14 transcripts was arbitrarily assigned a value of 100%.
To rule out the unlikely possibility that the reduction in translation observed with the IV(+) transcripts was due to the inability of scanning 40S ribosomes to penetrate the stabilized stem-loop IV structure and reach the AUG codon, we analyzed the translational activity of stem-loop IV mutants in the context of monocistronic RNA transcripts in which the 5′ 284 nt of the GBV-B sequence (that is, the sequence up to stem-loop IIIa) was deleted [Fig. 7b, right panel, Δ5′+14, Δ5′IV(±), and Δ5′IV(+)]. This 5′ deletion removes most of the IRES structure from these transcripts, preventing internal entry of the ribosome on these RNAs. However, even though these monocistronic RNAs retain two additional AUG codons upstream of the authentic initiator codon (at nt 355 and 365 [Fig. 1a]), the CAT reading frame is translated at a detectable level, probably by a mechanism involving the bypass of upstream AUG codons by scanning ribosomes. In rabbit reticulocyte lysates, we observed no differences in the translational activities of the Δ5′+14, Δ5′IV(±), and Δ5′IV(+) transcripts (Fig. 7b, right panel). The quantity of CAT produced in reticulocyte lysates programmed with the Δ5′IV(+) transcripts was 85% of that produced in lysate programmed with the Δ5′+14 RNA. These results contrast with those shown in Fig. 7b, left panel, and 7c, and they indicate that the stabilized stem-loop IV in the Δ5′14(+) transcript is readily penetrated by a scanning 40S subunit. The stem-loop IV(+) mutation effectively eliminated IRES-directed translation (Fig. 7b, left panel, and 7c) but had no impact on translation initiated by ribosomes scanning from the 5′ end of the truncated Δ5′IV(+) transcripts (Fig. 7b, right panel). Taken together, these data indicate that the 40S subunit does not scan into the stem-loop IV segment of the GBV-B sequence from an upstream point of contact during IRES-directed translation. The strong inhibition of IRES-mediated translation by stabilization of stem-loop IV indicates instead that the 40S subunit forms an important contact with GBV-B RNA directly at the site of the initiator AUG codon.
DISCUSSION
GBV-B is a recently discovered, incompletely characterized virus belonging to the family Flaviviridae and phylogenetically closely related to HCV (10, 24). Like HCV, it is hepatotropic and capable of causing acute hepatic injury in infected primates. Although recovered from an experimentally infected tamarin, only a single example of this virus has yet been identified, and its natural host species remains uncertain. The close relationship of GBV-B to HCV, a major cause of chronic liver disease in humans, makes it a particularly interesting virus to study, especially in the absence of good experimental systems for HCV. In contrast to the more distantly related flaviviruses GB virus A and GB virus C (otherwise known as hepatitis G virus), the 5′NTR of GBV-B has significant structural homology to HCV and its genome encodes a readily identifiable capsid protein (6, 9, 22). The experiments described here show that GBV-B, like HCV and the pestiviruses, translates its genome by means of an efficient IRES element located within its 5′NTR.
Our results indicate that the GBV-B IRES has a number of structural and functional features in common with the HCV IRES, but they also demonstrate some impressive differences between these IRES elements. The experiments depicted in Fig. 2 and 6 demonstrate that the IRES spans domains II and III of the GBV-B 5′NTR structure and thus occupies a position that is analogous to the position of the HCV IRES within the 5′NTR of that virus. Placed within the same reporter sequence context, these two flaviviral sequences have approximately equal translational activities (data not shown). The GBV-B and HCV structures are remarkably similar, despite the fact that there is very little conservation of primary nucleotide sequence between these viruses (5, 6, 9, 10).
The most impressive difference between the two predicted structures is the inclusion of a large, approximately 97-nt insertion within domain II of the GBV-B 5′NTR. Computer modeling suggests the inserted sequence forms two extended stem-loop structures (Fig. 1a, stem-loops IIb and IIc) that are absent from the IRESes of HCV and the pestiviruses (6, 9). Although the deletion of these two stem-loops is likely to result in a structure with greater superficial similarity to the structure of the HCV IRES, we demonstrated that the retention of the two stem-loops is essential for optimal GBV-B IRES activity (Fig. 3). The deletion of these stem-loops from the GBV-B sequence resulted in less impairment of translation in reticulocyte lysates than in BT7-H cells, but the effect was evident in both systems. These results are of interest with respect to a possible evolutionary relationship between the GBV-B and HCV IRES elements. It is intriguing to speculate that the GBV-B structure may be more closely related to the structure of a common ancestral IRES. Evolutionary modifications to the HCV structure appear to have allowed it to overcome the deletion of stem-loops IIb and IIc and to evolve toward a smaller, more compact sequence with approximately equivalent internal ribosome entry activity.
The analysis of compensatory mutations that were created within the extended helix at the base of domain II of the GBV-B structure (Fig. 3) provides support for the secondary structure model we had proposed previously (6). Substitutions of the nucleotide sequence between positions 231 and 236 resulted in a strong decrease in translation. The translational activity of this mutant was significantly restored by introduction of the complementary changes into the sequence between nt 63 and 68 (Fig. 3). This observation confirms the presence of the predicted helical segment at the base of domain II (Fig. 1a). There was less reduction in translation that was observed with the disruption of the helix due to substitutions within its 5′ strand, between nt 63 and 68, than in its 3′ strand, between nt 231 and 236 (Fig. 3, compare the translational activity of IIL with that of IIR). This indicates that retention of the primary nucleotide sequence is more important in the 3′ segment of this helix than in the 5′ segment. This may reflect stochastic alternative base pairing that allows the partial retention of IRES activity with the 5′ mutations, but it is interesting that we have observed a similar effect with analogous mutations of domain II of the HCV IRES (5). Thus, this appears to be a general feature of the domain II structures of the flavivirus IRESes.
In general, mutations in domain III of the GBV-B IRES had much more profound inhibitory effects on translation than mutations in domain II (for example, compare Fig. 3 with Fig. 4 and 5). We identified several base-paired helical segments in domain III within which the primary nucleotide sequence was critically important and could not be replaced with alternative base-pair arrangements. In contrast to the helix at the base of domain II (Fig. 3), replacing the stem element of stem-loop IIIe with complementary base-paired sequences led to nearly complete loss of translation (Fig. 4). This hairpin structure is unique in that its primary nucleotide sequence is conserved in both its stem and loop segments between the IRES elements of GBV-B, HCV, and the pestiviruses (9). Similarly, we found that the nearby nonpaired A253 and A392 residues could not be replaced either singly or as a pair by uridine residues and that base pairs flanking the internal bulge created by these unpaired adenosines could not be replaced by alternative base pairs (Fig. 5). These results are consistent with a recent report by Pestova et al. (11), who found that a deletion of 4 nt that included an adenosine homologous to the A253 in GBV-B resulted in a loss of ribosome binding by a pestivirus IRES. Together, these data indicate a strict requirement for conservation of the primary nucleotide sequence in this part of the IRES. This strict conservation for sequence is likely to reflect a stringent requirement for an RNA structure that is conducive to interactions with the 40S ribosome subunit or a protein translation factor (11). An alternative interpretation of the results shown in Fig. 5 might be that the model structure is incorrect in this region of the IRES and that the compensatory mutations we created in the GBV-B sequence do not involve actual base pairs. This is unlikely to be the case, however, because there is very strong experimental evidence for the existence of the pseudoknot in these structures and substantial natural sequence covariation in stem-loop IIId, structures that flank this region (Fig. 1) (9, 16, 28).
A similar explanation is likely for the strong inhibition of translation that we observed following the modification of the broadly conserved A271 and A272 residues in the GBV-B IRES (Fig. 5, mutant IIIa-ΔAA). Eucaryotic translation initiation factor 3 has been shown to interact with the apical segment of the domain III structures of both HCV and pestivirus IRES elements (2, 11, 25). Since these two adenosines are located just upstream of stem-loop IIIa (Fig. 1a), it is possible that these substitutions interfered with an interaction between the GBV IRES and eucaryotic initiation factor 3. This initiation factor is essential for translation of the HCV polyprotein (11).
All of the foregoing results show that the GBV-B IRES has a number of features in common with the IRES elements of HCV and the pestiviruses. Because there has been considerable controversy about the 3′ limits of the IRES in HCV and the pestiviruses, we examined this aspect of the GBV-B IRES in detail. Reynolds et al. (13) reported that approximately 32 nt of HCV polyprotein-coding sequence must be present downstream of the HCV 5′NTR to ensure optimal translation and have suggested an important role for the primary nucleotide sequence of this segment in the process of internal ribosome entry. However, the HCV IRES does function efficiently when the initiator AUG is fused directly to some reporter protein sequences (Fig. 2) (27, 29), and other results suggest that the constraints on downstream sequence are more related to a requirement for unstructured RNA rather than a requirement for specific sequence (6). The results of the experiments depicted in Fig. 6 and 7 strongly support this latter hypothesis.
Taken together, our results indicate that the 3′ border of the GBV-B IRES element is located at or 5′ of the AUG codon (strictly speaking, between stem-loop IIIe and the AUG codon, or between nt 403 and 446) (Fig. 4 and 6). The inclusion of 14, 38, or 63 nt of GBV-B polyprotein-coding sequences between the AUG codon and the CAT reporter coding sequence in the +14, +38, and +63 constructs significantly reduced IRES activity compared to the wt construct (Fig. 6). The reduction in translation upon inclusion of the 5′ 14 nt of the GBV-B capsid protein coding sequence was due to the inclusion of the complete stem-loop IV sequence in these transcripts. This stem-loop, which contains the AUG initiator codon within its loop sequence (Fig. 1a), thus appears to be inhibitory to initiation of translation. As with the analogous stem-loop in the HCV sequence (6), we found that mutations predicted to enhance the stability of this stem-loop substantially reduced the activity of the upstream IRES (Fig. 7).
However, RNA transcripts with a large 5′-terminal deletion and containing similar stabilizing mutations in the stem-loop IV sequence translated as well as transcripts with the wt stem-loop IV (Fig. 7). Thus, this structure is inhibitory only for ribosomes that are entering internally on the RNA and not for ribosomes that are scanning toward it from a more 5′ point of contact. These data provide strong evidence for the absence of any scanning in the normal internal initiation of translation on GBV-B RNAs and, as for the RNAs of HCV and the pestiviruses, suggest that the 40S subunit makes an important primary contact at the site of the initiator AUG codon (6, 14, 18). The presence of stem-loop IV in both the GBV-B and HCV sequences (Fig. 1) remains puzzling. Its location and its suppressive effect on cap-independent translation suggest that stem-loop IV may plan an important role in regulating translation (6). However, no data have yet confirmed this hypothesis. The presence of this stem-loop in GBV-B and HCV, but not the pestiviruses which have been increasingly used as surrogates for the study of HCV (6), highlights the potential importance of further characterizing the GBV-B virus and its genome.
ACKNOWLEDGMENTS
We thank Isa Mushahwar and John Simons for the gift of plasmid pLuc-GBVB-CAT, and we thank E. R. Surriga and S. Smith for sequencing of plasmid DNAs.
This work was supported in part by grant U19-AI40035 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Brown E A, Zhang H, Ping L-H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratti E, Tisminetzky S, Zotti M, Baralle F E. Functional analysis of the interaction between HCV 5′UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179–3187. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deinhardt F, Holmes A W, Capps R B, Popper H. Studies on the transmission of human viral hepatitis to marmoset monkeys. I. Transmission of disease, serial passages, and description of liver lesions. J Exp Med. 1967;125:673–688. doi: 10.1084/jem.125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng R, Brock K V. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analyses. Nucleic Acids Res. 1993;21:1949–1957. doi: 10.1093/nar/21.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honda M, Beard M R, Ping L H, Lemon S M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 7.Honda M, Ping L-H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosomal entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 8.Kamoshita N, Tsukiyama-Kohara K, Kohara M, Nomoto A. Genetic analysis of internal ribosomal entry site on hepatitis C virus RNA: implication for involvement of the highly ordered structure and cell type-specific transacting factors. Virology. 1997;233:9–18. doi: 10.1006/viro.1997.8600. [DOI] [PubMed] [Google Scholar]
- 9.Lemon S M, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 10.Muerhoff A S, Leary T P, Simons J N, Pilot-Matias T J, Dawson G J, Erker J C, Chalmers M L, Schlauder G G, Desai S M, Mushahwar I K. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J Virol. 1995;69:5621–5630. doi: 10.1128/jvi.69.9.5621-5630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds J E, Kaminiski A, Kettinen H J, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds J E, Kaminski A, Carroll A R, Clarke B E, Rowlands D J, Jackson R J. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 15.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 16.Rijnbrand R, van der Straaten T, Van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rijnbrand R, Lemon S M. IRES mediated translation in hepatitis C virus replication. Curr Top Microbiol Immunol. 1999;242:85–116. doi: 10.1007/978-3-642-59605-6_5. [DOI] [PubMed] [Google Scholar]
- 18.Rijnbrand R C A, Abbink T E M, Haasnoot P C J, Spaan W J M, Bredenbeek P J. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Scarselli E, Urbani A, Sbardellati A, Tomei L, De Francesco R, Traboni C. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. J Virol. 1997;71:4985–4989. doi: 10.1128/jvi.71.7.4985-4989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlauder G G, Dawson G J, Simons J N, Pilot-Matias T J, Gutierrez R A, Heynen C A, Knigge M F, Kurpiewski G S, Buijk S L, Leary T P, Muerhoff A S, Desai S M, Mushahwar I K. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- 22.Simons J N, Desai S M, Schultz D E, Lemon S M, Mushahwar I K. Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications on genome organization. J Virol. 1996;70:6126–6135. doi: 10.1128/jvi.70.9.6126-6135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 24.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, Van Sant C L, Mushahwar I K. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sizova D V, Kolupaeva V G, Pestova T V, Shatsky I N, Hellen C U. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P-L, Simmonds P International HCV Collaborative Study Group. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 27.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Le S, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whetter L E, Day S P, Elroy-Stein O, Brown E A, Lemon S M. Low efficiency of the 5′ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J Virol. 1994;68:5253–5263. doi: 10.1128/jvi.68.8.5253-5263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]