Abstract
CD24 is a glycosylphosphatidylinositol‐anchored membrane protein reported to be overexpressed in human tumorigenesis and progression. Our purpose was to determine the role of CD24 in the proliferation of colorectal cancer cells and the potential mechanisms in this process. Our data showed that CD24 promoted cell growth and induced activation of extracellular signal‐regulated kinases, Raf‐1, and p38 mitogen‐activated protein kinase. Furthermore, suppression of extracellular signal‐regulated kinases and p38 mitogen‐activated protein kinase activity by their specific inhibitors, U0126 and SB203580, abrogated CD24‐induced proliferation in vitro. By tumorigenicity assay in female BALB/c nude mice, we further demonstrated that CD24 promoted tumor growth in vivo. Immunohistochemical analysis revealed that CD24 expression occurred in 92.5% of human colorectal cancer tissue, and increased with tumor progression. More importantly, the stainings of phospho‐extracellular signal‐regulated kinases and phospho‐p38 mitogen‐activated protein kinase were strongly correlated with CD24 expression. Taken together, our data suggest that CD24‐dependent extracellular signal‐regulated kinases and p38 mitogen‐activated protein kinase activations are required for colorectal cancer cell proliferation in vitro and in vivo. The linkage of CD24 and the mitogen‐activated protein kinase pathway may unravel a novel mechanism in the regulation of colorectal cancer proliferation. (Cancer Sci 2009; 00: 000–000)
CD24, a GPI‐anchored membrane protein, consists of a small protein core comprising 27 amino acids with several potential O‐ or N‐linked glycosylation sites, which leads to the molecular weight ranging between 38 and 70 kDa.( 1 ) CD24, referred to as a B cell differentiation marker at first, plays crucial roles in lymphocyte maturation,( 2 , 3 , 4 ) neuronal development,( 5 ) and tissue renewal homeostasis under physiological conditions.( 6 ) Recently, CD24 was reported to be upregulated in human cancers such as breast cancer,( 7 ) prostate cancer,( 8 ) and cholangiocarcinoma,( 9 ) and its overexpression is usually associated with poor prognosis,( 7 , 8 , 9 , 10 , 11 , 12 ) suggesting that this gene may play a key role in tumorigenesis. Recent studies showed that ectopic overexpression of CD24 in a rat carcinoma cell line increased cell proliferation and adhesion,( 13 ) and downregulation of CD24 expression in human carcinoma cell lines resulted in growth inhibition, decreased clonogenicity, changes in the actin cytoskeleton, and induction of apoptosis,( 14 ) suggesting that CD24 has key effects on biological aspects of cancer cells. In CRC, CD24 is commonly upregulated and cytoplasmic CD24 expression remains a significant independent prognostic marker.( 15 , 16 ) Furthermore, downregulation of CD24 expression induces growth inhibition in CRC and pancreatic cancer, and this effect is augmented in combination with classic chemotherapies.( 17 ) Taken together, CD24 plays an important role in the development and progression of malignant tumor, including CRC. However, the underlying mechanisms remain largely unclear.
Mitogen‐activated protein kinases are serine/threonine kinases that play an important role in signal transduction from the cell surface to the nucleus. The mammalian MAPK can be divided into ERK, JNK, and p38 MAPK. In response to a wide range of extracellular stimuli, the MAPK cascades determine cell fate, including cell growth, differentiation, and apoptosis.( 18 ) A previous study showed that CD24 expression is negatively associated with p‐MAPK in cholangiocarcinoma,( 9 ) suggesting that MAPK pathways may be involved in CD24‐induced tumorigenesis.
In our present study, we determined the role of CD24 in the proliferation of CRC cells and attempted to elucidate the potential role of the MAPK pathway in this process. Our data showed that CD24‐dependent ERK1/2 and p38 MAPK activation is required for CRC cell proliferation in vitro and in vivo. The linkage of CD24 and the MAPK pathways may unravel a novel mechanism in the regulation of CRC proliferation.
Materials and Methods
Reagents and antibodies.
RPMI‐1640 medium and FBS were purchased from Gibco (Purchase, NY, USA). G418 and SB203580 were obtained from Calbiochem (Darmstadt, Germany). U0126, ERK1/2 (137F5) antibody, p38 MAPK antibody, SAPK/JNK antibody, phospho‐SAPK/JNK (Thr183/Tyr185) antibody, and phospho‐p38 MAPK antibody were purchased from Cell Signaling Technology (Beverly, MA, USA). CD24 (C‐20) polyclonal antibody, Raf‐1 (C‐12) antibody, p‐Raf‐1 (Ser338), p‐ERK (E‐4), and β‐actin (C‐4) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). CD24 Ab‐1 (Clone SN3) mouse monoclonal antibody was from Neomarker (Lab Vision Corporation, Fremont, CA, USA).
Tissue samples.
Formalin‐fixed, paraffin‐embedded tissue samples from 106 primary human CRC were randomly obtained at surgery and had been processed by routine clinical histopathological methods. The patients comprised 65 men and 41 women, with a mean age of 59.6 years (range 26–87 years). The pathological tumor stages were determined according to the TNM classification of the American Joint Committee on Cancer Criteria, and the World Health Organization (WHO) classification of tumors was used to determine the histological grade. The study was carried out in accordance with the institutional ethical guidelines and had been approved by the medical ethics committee of Southern Medical University. Informed consent was obtained from all patients.
Cells culture and treatments.
SW1116, SW480, SW620, HCT8, LoVo, and Colo205 cell lines were obtained from American Type Culture Collection (Rockville, MD, USA). Cells were maintained in RPMI‐1640 medium supplemented with 10% FBS (complete medium) at 37°C with an atmosphere of 5% CO2 until confluency and subcultured using 0.05% trypsin. For treatment with pharmacological inhibitors, cells were kept in complete medium containing 10 μm U0126 or 20 μm SB203580. Medium containing inhibitor was renewed every day.
Construction of the CD24 expression plasmid and stable transfection.
A full‐length human CD24 cDNA corresponding to bases 111–353 of Genbank accession no. NM_013230 was amplified by PCR from a normal intestinal cDNA library (a gift of Professor Jide Wang). The primer pairs for CD24 were 5′‐TAGGTACCACTATGGGCAGAGCAATGG‐3′ (forward) and 5′‐CCGGAATTCCGTTAAGAGTAGAGATGC‐3′ (reverse). The PCR product was cloned into the pcDNA3.1(+) vector (Invitrogen, Purchase, NY, USA) to create the pcDNA3.1(+)‐CD24 plasmid. The orientation of the insert and the sequence of cDNA were verified by sequencing. Cultured SW480 cells were transfected with the recombinant plasmids and the empty vector using the lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Transfectants were selected in medium containing 600 μg/mL G418. After selection and isolation of stably transfected clones, the clones were analyzed for CD24 expression using RT‐PCR and FCM.
RT‐PCR.
Total RNA was extracted from cells using Trizol (Invitrogen). RNA samples (2 μg) were subjected to reverse transcription using a RevertAid First Strand cDNA Synthesis Kit (Fermentas, MBI, Vilnius, Lithuania). The primer pairs for CD24 were 5′‐GACATGGGCAGAGCAATGGTGGC‐3′ (forward) and 5′‐GAGTGAGACCACGAAGAGACTGGC‐3′ (reverse).( 19 ) The PCR was initiated by 5 min incubation at 94°C, ended after a 10 min extension at 72°C, 32 cycles for denaturation at 94°C for 30 s, annealing at 55°C, and extension at 72°C for 1 min using a PCR kit (SBS Genetech Co., Beijing, China). RT‐PCR was repeated twice and GADPH mRNA was amplified simultaneously as an internal control.
Flow cytometry.
Cells were harvested and resuspended in PBS/5% FBS at a density of 2×106/mL. CD24 monoclonal antibodies (Ab‐1; Neomarker) were applied at 5 μg/mL (1:100) in a 200‐μL cell suspension. After incubation for 60 min on ice, the cells were washed with PBS/5% FBS twice and incubated for a further 40 min with FITC‐conjugated secondary antibody (1:200; Santa Cruz Biotechnology) on ice. The cells were washed with PBS/5% FBS twice again and analyzed by FCM. As negative controls, cells were stained with the isotype‐matched control antibodies.
Preparation of siRNA and transfection.
A siRNA duplex targeting CD24 and a negative control sequence were designed and synthesized by GenePharma Co. (Shanghai, China). The target sequence of the CD24 siRNA duplex was 5′‐GAUUUAUUCCAGUGAAACATT‐3′. BLAST research ensured that the sequence had no significant homology with other human genes. Freeze‐dried siRNA was reconstituted with RNase‐free water to prepare a 20 μm stock solution. The siRNA and negative control sequences were transfected into SW620 cells using lipofectamine 2000 reagent according to the manufacturer’s instructions.
Western blotting.
Protein extraction and immunoblotting have been described before.( 20 ) Actin was detected as an internal control. All experiments were repeated twice and blots were analyzed using quantity one 1‐D analysis software (Bio‐Rad, Hercules, CA, USA).
Proliferation assays.
Cell growth was evaluated using a WST‐8 assay (Beyotime Institute Biotech, Shanghai, China). The cells were seeded into 96‐well culture plates at a density of 2000 cells, and the medium was replaced with 100 μL fresh medium and 10 μL WST‐8 reagent in each well at 24, 48, 72, and 96 h. After the plate was returned to the incubator for 1–2 h, the absorbance at 450 nm was measured using an enzyme‐linked immunosorbent assay plate reader.
Tumorigenicity assay in nude mice.
Animal experiments were approved by the Animal Care and Use Committee (approval number SCXK 2003‐0002 2008A053) and all the protocols were in agreement with the requirements of the committee. Eight female BALB/c nude mice (5 weeks old) were obtained from Guangdong Province Medical Experimental Animal Center, and were maintained under specific pathogen‐free conditions in the Experiment Animal Center of Nanfang Hospital (Guangzhou, China). Mice were injected subcutaneously with 4×106 cells into the left (SW480vec cells) or right (SW480CD24+1 cells) flanks, and were then monitored once every 5 days for 5 weeks. The size of the tumor was determined by caliper measurement of the subcutaneous tumor mass. Tumor volume was calculated according to the formula: tumor volume = length × (width)2/2.
Immunohistochemical analysis and evaluation.
The immunohistochemical methods have been described before.( 21 ) Sections were incubated with primary antibodies in appropriate dilutions overnight at 4°C (CD24 antibody, 1:400 dilution; p‐ERK antibody, 1:100 dilution; p‐p38 antibody, 1:100 dilution). Scoring of tissue slides was carried out independently by two investigators; the percentage of positive cells and the intensity of staining were scored from 0 to 3: 0, less than 10% of cells stained; 1, 10–50% of cells stained; 2, 50–75% of cells stained; 3, more than 75% of cells stained as described previously.( 22 )
Statistical analysis.
The ANOVA was used to determine whether the results of proliferation assays had statistical significance. The non‐parametric Mann–Whitney or Kruskal–Wallis tests were applied to test the significance of differences among groups of clinicopathological parameters, whereas the correlations among p‐ERK1/2, p‐p38 MAPK, and CD24 were evaluated by the Spearman rank‐order correlation coefficient. The level of significance was defined as P < 0.05. SPSS 13.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
Results
Expression of CD24 was cell type‐dependent in CRC cell lines.
FCM analysis was used to detect CD24 expression at the protein level in the CRC cell lines. Of the six cell lines, SW1116 maintained moderate expression of CD24, SW620 and Colo205 contained higher expression, but the expression was scarcely observed in SW480, HCT8, and LoVo cell lines (Fig. 1A). To investigate CD24 mRNA at the level of transcription, a standard RT‐PCR experiment was carried out. As seen in Figure 1(B), the result was consistent with FCM analysis. These results showed that expression of CD24 was cell type‐dependent in CRC cell lines.
Figure 1.
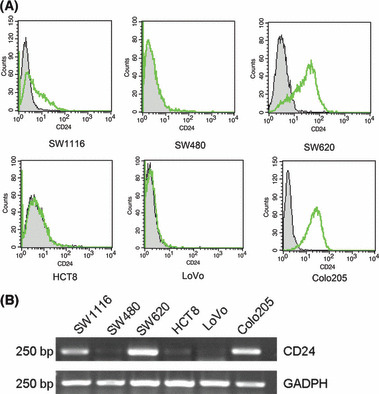
Expression of CD24 was cell type‐dependent in colorectal cancer cell lines. (A) Flow cytometric analysis of different colorectal cancer cell lines at the protein level. The cells were stained with anti‐CD24 monoclonal antibodies and FITC‐conjugated secondary antibodies. Abscissa, fluorescence intensity (log scale); ordinate, cell number (linear scale). Filled trace, background staining; open trace, staining with CD24. (B) Differentiated levels of CD24 mRNA were detected in colorectal cancer cell lines by RT‐PCR.
Overexpression of CD24 induced proliferation in vitro and in vivo.
The CD24 recombinant plasmid pcDNA3.1(+)‐CD24 was constructed using the expression vector pcDNA3.1(+), and was then stably transfected into the SW480 cells. Two clones, named as SW480CD24+1 and SW480CD24+4, exhibited dramatically increased expression of CD24 at the mRNA transcript level compared with the parental and vector control cells (Fig. 2A). Expression of CD24 at the protein level in these clones was confirmed by FCM analysis (Fig. 2B). The growth ability of SW480CD24+1 cells in vitro was assessed by WST‐8 assay, and the results showed that these cells proliferated more quickly than SW480vec cells, which were transfected with the vector alone, and the parental SW480 cells (Fig. 2C).
Figure 2.
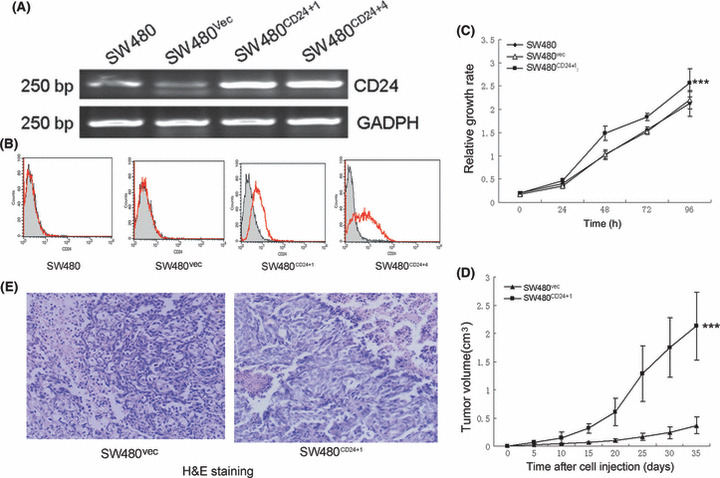
Effect of CD24 on the growth of SW480 cells. (A) CD24 mRNA was detected in cell clones of SW480 using RT‐PCR. (B) Flow cytometry confirmed the expression of CD24 at the protein level. Abscissa, fluorescence intensity (log scale); ordinate, cell number (linear scale). Filled trace, background staining; open trace, staining with CD24. (C) Increased growth ability was detected in SW480CD24+1 cells compared with SW480vec cells and parental SW480 cells using the WST‐8 assay (***P < 0.001). Ordinate, absorbance at 450 nm. (D) CD24 promoted tumor growth in nude mice (***P < 0.001). BALB/c nude mice were injected subcutaneously with 4×106 cells and tumor volume was measured once every 5 days. (E) Histological analysis of implanted tumors with H&E staining (×200).
To examine whether overexpression of CD24 could promote tumorigenicity of SW480 in nude mice, the growth of a xenograft tumor model was applied. As shown in Figure 2(D), the SW480CD24+1 cells developed tumors with larger volumes than SW480vec cells (2.13±0.60 vs 0.37±0.15 at the fifth week, P < 0.001). All tumors were confirmed by H&E staining (Fig. 2E).
Overexpression of CD24 induced activation of ERK1/2, Raf‐1, and p38 MAPK, but had no effect on JNK1/2.
To elucidate the potential mechanisms of CD24‐induced proliferation, we examined the activation of the MAPK pathway. As shown in Figure 3(A,B), ERK1/2 and p38 MAPK were activated in the SW480CD24+1 and SW480CD24+4 cells compared with SW480vec cells and the parental SW480 cells. However, no activation of JNK1/2 was observed (Fig. 3C). To determine the role of CD24 in the Raf–ERK pathway, we further detected the activity of Raf‐1, the upstream ERK1/2 activator, and our data showed that Raf‐1 protein levels and kinase activity were elevated after the SW480 cells were overexpressed with CD24 (Fig. 3D).
Figure 3.

The activities of ERK1/2, Raf‐1, p38 MAPK, and JNK in SW480CD24+1 and SW480CD24+4 cells were assessed by western blotting. Overexpression of CD24 induced the activation of (A) ERK1/2, (B) p38 MAPK, and (D) the upstream molecule of ERK1/2, Raf‐1. (C) However, no activation was observed in JNK1/2. β‐Actin expression was used to normalize for equal loading. (E) Immunohistochemical analysis of implanted tumors with antibodies to CD24, p‐ERK, and p‐p38 (×400).
Subsequently, we checked the effect of CD24 on ERK1/2 and p38 MAPK in implanted tumors of mice. Immunostaining showed that CD24, p‐ERK1/2, and p‐p38 MAPK were scarcely detectable in tumors derived from SW480vec cells but were highly expressed in tumors derived from SW480CD24+1 cells (Fig. 3E).
Suppressing the activation of ERK1/2 and p38 MAPK abrogated CD24‐induced proliferation in the SW480CD24+1 cells.
To further evaluate the contribution of ERK1/2 and p38 MAPK to the CD24‐induced proliferation, specific pharmacological inhibitors suppressing ERK1/2 or p38 MAPK activation were used. The starved cells were treated either with an inhibitor of the upstream ERK1/2 activator MEK1/2 (U0126), or the p38 MAPK inhibitor SB203580. DMSO was used as a control. The cells were treated separately with 10 μm U0126 or 20 μm SB203580 and observed at 24 and 72 h after treatment followed by WST‐8 assay for cell viability. In the meantime, parallel cultures were analyzed for total phosphorylation changes by western blotting, and the result confirmed that the activation of ERK1/2 (Fig. 4A) and p38 MAPK (Fig. 4C) was significantly inhibited. The WST‐8 assays showed that there were significant decreases of cell viability in the cells treated with U0126 (Fig. 4B) and SB203580 (Fig. 4D) after 72 h.
Figure 4.

Activation of ERK1/2 and p38 MAPK was required for CD24‐induced proliferation. Western blotting analysis confirmed that (A) ERK1/2 and (C) p38 MAPK activation was suppressed when cells were treated with 10 μm U0126 or 20 μm SB203580. The cells were incubated with RPMI‐1640 medium containing 1% FBS for 12 h. Starved cells were treated with U0126 or SB203580 2 h before complete medium was added. The total protein and phosphorylation levels of ERK1/2 and p38 MAPK expression were determined by western blotting analysis 30 min later. Cell growth in the presence of (B) U0126 or (D) SB203580 was assessed after 24 and 72 h treatment by WST‐8 assay. All experiments were repeated twice. Ordinate, absorbance at 450 nm.
Downregulation of CD24 inhibited cell growth and reduced activation of ERK1/2, Raf‐1, and p38 MAPK.
To confirm the effect of CD24 on growth and MAPK pathway activation, the strategy of siRNA‐based knockdown of CD24 in SW620 cells was used. To develop siRNA for CD24, we initially tested three individual duplexes and identified one siRNA that efficiently reduced CD24 expression for further experiments. As shown in Figure 5(A,B), the cells treated with the siRNA showed a decrease in the mRNA and protein levels compared with the parental SW620 and negative control cells. Following siRNA‐mediated depletion of CD24 in SW620 cells, we observed a significant decrease (P < 0.001) in cell number during a 96‐h period, suggesting that CD24 knockdown induced cell growth inhibition (Fig. 5C). Western blotting showed that downregulation of CD24 reduced activation of ERK1/2, Raf‐1, and p38 MAPK in SW620 cells (Fig. 5D), which further validates the association between CD24 and the MAPK pathway.
Figure 5.
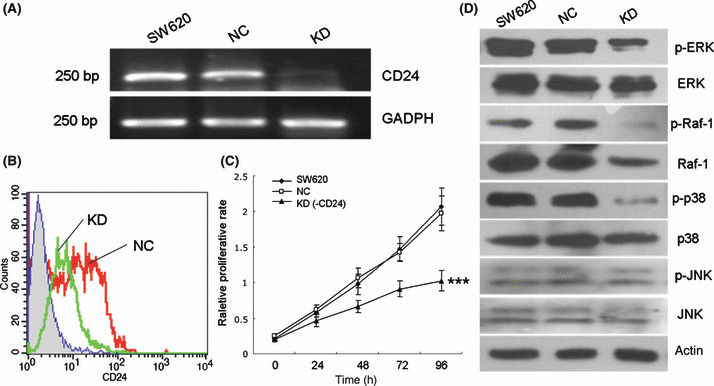
Knockdown of CD24 inhibited cell growth and reduced the activation of ERK1/2, Raf‐1, and p38 MAPK. (A) CD24 mRNA was detected in parental SW620 and treated cells using RT‐PCR. (B) Flow cytometry confirmed downregulation of CD24 at the protein level. Abscissa, fluorescence intensity (log scale); ordinate, cell number (linear scale). Filled trace, background staining; open trace, staining with CD24. (C) WST‐8 assay showed that knockdown of CD24 inhibited cell growth in SW620 cells (***P < 0.001). (D) Western blotting showed that downregulation of CD24 reduced the activation of ERK1/2, Raf‐1, and p38 MAPK, but had no effect on JNK1/2. NC, SW620 cells transfected with negative control sequence; KD, SW620 cells treated with siRNA duplex targeting CD24.
Immunostaining of p‐ERK1/2 and p‐p38 MAPK strongly correlated with CD24 expression in human CRC tissue.
To investigate the correlations among p‐ERK1/2, p‐p38 MAPK, and CD24 in vivo, immunohistochemical staining of the three molecules was carried out in serial sections of CRC tissues. CD24 staining was present mainly in the membrane and/or cytoplasm; 92.5% cases showed positive staining and 57.6% cases were moderately or strongly positive, whereas its expression in adjacent normal mucosa was either absent or barely detectable on cell membranes. There was a significantly positive correlation between CD24 expression and nodal status (r = 0.206, P = 0.034), distant metastasis (P = 0.025), and tumor stage (r = 0.242, P = 0.012), while no correlation was observed between CD24 expression and age, sex, localization, depth of invasion, or grade (Fig. 6; Table 1), showing that CD24 expression increased with tumor progression.
Figure 6.
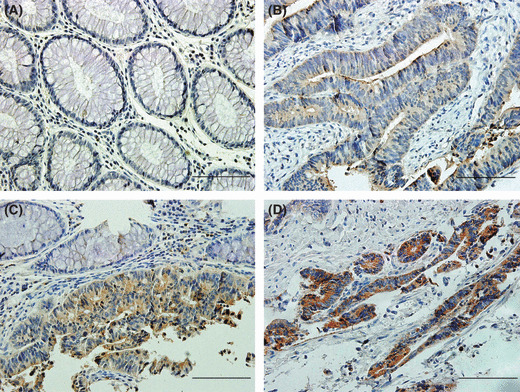
CD24 expression increased with colorectal cancer progression. (A) Normal mucosa showing negative expression of CD24. (B) Weak expression in a stage I tumor. (C) An invasive stage II carcinoma showing moderate staining. Note an obvious interface between normal mucosa and cancer. (D) Strong expression in a stage III colorectal cancer. The scale of scale bars: 10 μm.
Table 1.
Association between CD24 expression and clinicopathological parameters
| Variable | n | CD24 | P‐value | |||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||
| n (%) | n (%) | n (%) | n (%) | |||
| All cases | 106 | 8 (7.5) | 37 (34.9) | 43 (40.6) | 18 (17.0) | |
| Gender | ||||||
| Male | 65 | 4 (6.2) | 24 (36.9) | 25 (38.5) | 12 (18.5) | 0.817† |
| Female | 41 | 4 (9.8) | 13 (31.7) | 18 (43.9) | 6 (14.6) | |
| Age (years) | ||||||
| ≤60 | 48 | 2 (4.2) | 17 (35.4) | 23 (47.9) | 6 (12.5) | 0.861† |
| >60 | 58 | 6 (10.3) | 20 (34.5) | 20 (34.5) | 12 (20.7) | |
| Tumor location | ||||||
| Right colon | 19 | 2 (10.5) | 8 (42.1) | 4 (21.1) | 5 (26.3) | 0.545‡ |
| Left colon | 24 | 1 (4.2) | 11 (45.8) | 10 (41.7) | 2 (8.3) | |
| Rectum | 63 | 5 (7.9) | 18 (28.6) | 29 (46.0) | 11 (17.5) | |
| Depth of invasion | ||||||
| T1 | 6 | 0 (0) | 0 (0) | 6 (100) | 0 (0) | 0.849§ |
| T2 | 15 | 3 (20) | 4 (26.7) | 5 (33.3) | 3 (20.0) | |
| T3 | 59 | 4 (6.8) | 24 (40.7) | 21 (35.6) | 10 (16.9) | |
| T4 | 26 | 1 (3.8) | 9 (34.6) | 11 (42.3) | 5 (19.2) | |
| Nodal status | ||||||
| N0 | 55 | 6 (10.9) | 23 (41.8) | 21 (38.2) | 5 (9.1) | 0.034§ |
| N1 | 31 | 2 (6.5) | 6 (19.4) | 12 (38.7) | 11 (35.5) | |
| N2 | 20 | 0 (0) | 8 (40.0) | 10 (50.0) | 2 (10.0) | |
| Distant metastasis | ||||||
| M0 | 90 | 8 (8.9) | 33 (36.7) | 37 (41.1) | 12 (13.3) | 0.025† |
| M1 | 16 | 0 (0) | 4 (25.0) | 6 (37.5) | 6 (37.5) | |
| Tumor stage | ||||||
| I | 16 | 1 (6.3) | 3 (18.8) | 11 (68.8) | 1 (6.3) | 0.012§ |
| II | 37 | 5 (13.5) | 20 (54.1) | 9 (24.3) | 3 (8.1) | |
| III | 37 | 2 (5.4) | 10 (27.0) | 17 (45.9) | 8 (21.6) | |
| IV | 16 | 0 (0) | 4 (25.0) | 6 (37.5) | 6 (37.5) | |
| Grade | ||||||
| 1 | 23 | 3 (13.0) | 8 (34.8) | 9 (39.1) | 3 (13.0) | 0.887§ |
| 2 | 70 | 5 (7.1) | 21 (30.0) | 29 (41.4) | 15 (21.4) | |
| 3 | 13 | 0 (0) | 8 (61.5) | 5 (38.5) | 0 (0) | |
†Determined using the two‐tailed Mann–Whitney test; ‡determined using the two‐tailed Kruskal–Wallis test; §determined using the Spearman rank‐order correlation coefficient.
Observation using the serial sections showed that p‐ERK1/2 was located mainly in the cytoplasm and p‐p38 MAPK was disseminated in both the cytoplasm and nucleus of the same cancer cells. Expression of p‐ERK1/2 and p‐p38 MAPK was observed in 84.0% and 79.0% cases, respectively, while their staining in adjacent normal mucosa was weak or absent and had no association with any clinicopathological parameters (data not shown).
More importantly, as shown in Figure 7, strong correlations were found between CD24 expression and immounostaining of p‐ERK1/2 or p‐p38 MAPK in CRC (r = 0.618, P < 0.001; and r = 0.536, P < 0.001 respectively), suggesting that activation of ERK1/2 and p38 MAPK strongly correlated with CD24 expression in CRC tissue.
Figure 7.
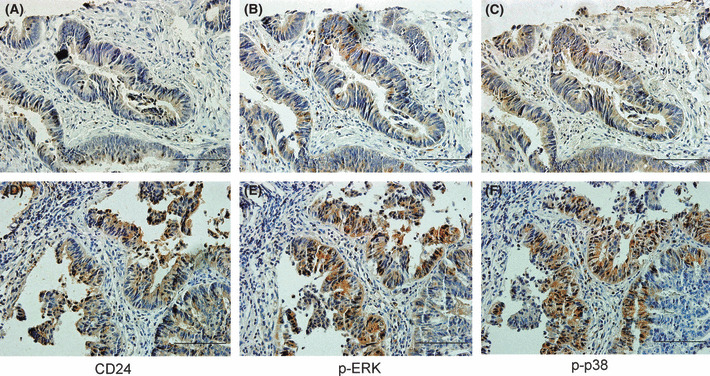
Activation of ERK1/2 and p38 MAPK in human colorectal cancer strongly correlated with cytoplasmic CD24 expression. (A–C) Serial sections of a stage II tumor showing weak expression for (A) CD24, (B) p‐ERK1/2, and (C) p‐p38 MAPK. (D–F) Serial sections of a stage III tumor demonstrating moderate immunostaining for (D) CD24, (E) p‐ERK1/2 and (F) p‐p38 MAPK. The scale of scale bars: 10 μm.
Discussion
In the present study, we evaluated the role of CD24 in the proliferation of CRC cells and the potential mechanisms. Our data showed that CD24 expression occurred in 92.5% of human CRC and increased with tumor progression. CD24 could induce proliferation of CRC cells in vitro and in vivo. In addition, we showed for the first time that CD24‐induced proliferation was dependent on the activation of ERK1/2 and p38 MAPK in CRC. Moreover, activation of ERK1/2 and p38 MAPK also appeared to be strongly correlated with CD24 expression in CRC tissue. Our finding provided a novel interpretation for the mechanism of CD24‐induced proliferation in CRC.
To date, published studies have mainly concentrated on the expression and function of CD24 in epithelial cancers, but literature about the molecular mechanism of CD24 contributing to malignant transformation is very limited. Recent studies by microarray analysis at the mRNA level have revealed that genes in the Ras pathway, MAPK, and Bcl‐2 family are frequently downregulated when CRC cells are subjected to anti‐CD24 mAb or shRNA.( 17 ) Our present results showed that ERK1/2 and p38 MAPK were activated by CD24 and were required for CD24‐induced proliferation. However, JNK1/2, which play important roles in controlling programmed cell death or apoptosis,( 23 , 24 ) did not participate in the process. To our knowledge, this is the first detailed analysis at the protein level of the MAPK family and their contribution to CD24‐induced proliferation in CRC, even in solid tumors. In addition, CD24 is a member of the glycosylated GPI‐linked cell surface protein family organizing the functional microdomains known as lipid rafts, which are proposed to function as platforms for the attachment of proteins when membranes are moved around inside the cell and during signal transduction.( 25 , 26 ) Crosslinking of CD24 induces rapid Tyr phosphorylation of several cellular proteins of the Src family in a cell‐type depending manner,( 27 , 28 ) along with subsequent activation of MAPK in Burkitt’s lymphoma cells.( 2 ) Src family members are key molecules of non‐receptor tyrosine kinases involved in signal transduction from membrane to cytoplasm.( 29 ) Therefore, these studies suggest that CD24 could evoke an intracellular signaling pathway through interacting with src kinases or other molecules, although it has no cytoplasmic portion to transduce signals.
The Raf–ERK pathway is widely expressed and is involved in the regulation of mitosis and the proliferative rate of tumor cells,( 23 ) and targeting this cascade for the treatment of cancer has been the subject of intense research and pharmaceutical scrutiny.( 30 ) Blockade of the pathway suppresses growth of colon tumors in vivo,( 31 ) and the selective inhibitor of MEK1/ERK44/42, PD98059, augments sulindac sulfide‐induced apoptosis in human colon carcinoma cells.( 32 ) In our study, immunohistochemical analysis showed that the ERK1/2 pathway was activated in CRC and the MEK1/2 inhibitor U0126 abrogated CD24‐induced proliferation, suggesting that the ERK1/2 pathway is a potential target for therapy of CRC. Our data showed that CD24‐induced proliferation was not only ERK1/2‐dependent but also p38 MAPK‐dependent. p38 MAPK is generally activated by inflammatory cytokines and environmental stresses and may contribute to diseases like asthma and autoimmunity;( 24 ) it also negatively regulates cell proliferation and renewal in lung and liver cancer development processes.( 33 , 34 ) Consistent with recent studies, our data showed that p38 MAPK inhibitor is mainly responsible for inducing apoptosis in human colon cancer cells.( 35 , 36 ) Taken together, these data suggest that the role of p38 MAPK in the cellular response to stimulus is cell type‐ and context‐dependent and is pro‐oncogenic in CRC.
CD24 induced CRC cell proliferation through activating the Raf–ERK pathway and p38 MAPK, suggesting that the linkage of CD24 and the MAPK pathway may unravel a novel mechanism in the regulation of CRC proliferation. Moreover, our data showed that CD24 is overexpressed in CRC tissue, and increases with tumor progression. More importantly, a previous study has shown that cytoplasmic CD24 expression in colorectal cancer is associated with poor prognosis.( 15 ) Therefore, CD24 may be a new potential target for the prevention and treatment of CRC. Further investigation will be required to elucidate this question.
Abbreviations
- CRC
colorectal cancer
- ERK
extracellular signal‐regulated kinase
- FCM
flow cytometry
- GPI
glycosylphosphatidylinositol
- JNK
c‐Jun N‐terminal kinase
- MAPK
mitogen‐activated protein kinase
Acknowledgment
The authors thank Dr Jide Wang (Department of Digestive Medicine, Nanfang Hospital, Guangzhou, China) for providing the normal intestinal cDNA library.
References
- 1. Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin‐like adhesion molecule. J Mol Histol 2004; 35: 255–62. [DOI] [PubMed] [Google Scholar]
- 2. Taguchi T, Kiyokawa N, Mimori K et al. Pre‐B cell antigen receptor‐mediated signal inhibits CD24‐induced apoptosis in human pre‐B cells. J Immunol 2003; 170: 252–60. [DOI] [PubMed] [Google Scholar]
- 3. Li O, Zheng P, Liu Y. CD24 expression on T cells is required for optimal T cell proliferation in lymphopenic host. J Exp Med 2004; 200: 1083–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li O, Chang X, Zhang H et al. Massive and destructive T cell response to homeostatic cue in CD24‐deficient lymphopenic hosts. J Exp Med 2006; 203: 1713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nieoullon V, Belvindrah R, Rougon G, Chazal G. mCD24 regulates proliferation of neuronal committed precursors in the subventricular zone. Mol Cell Neurosci 2005; 28: 462–74. [DOI] [PubMed] [Google Scholar]
- 6. Nieoullon V, Belvindrah R, Rougon G, Chazal G. Mouse CD24 is required for homeostatic cell renewal. Cell Tissue Res 2007; 329: 457–67. [DOI] [PubMed] [Google Scholar]
- 7. Kristiansen G, Winzer KJ, Mayordomo E et al. CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res 2003; 9: 4906–13. [PubMed] [Google Scholar]
- 8. Kristiansen G, Pilarsky C, Pervan J et al. CD24 expression is a significant predictor of PSA relapse and poor prognosis in low grade or organ confined prostate cancer. Prostate 2004; 58: 183–92. [DOI] [PubMed] [Google Scholar]
- 9. Agrawal S, Kuvshinoff BW, Khoury T et al. CD24 expression is an independent prognostic marker in cholangiocarcinoma. J Gastrointest Surg 2007; 11: 445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chou YY, Jeng YM, Lee TT et al. Cytoplasmic CD24 expression is a novel prognostic factor in diffuse‐type gastric adenocarcinoma. Ann Surg Oncol 2007; 14: 2748–58. [DOI] [PubMed] [Google Scholar]
- 11. Lee HJ, Kim DI, Kwak C, Ku JH, Moon KC. Expression of CD24 in clear cell renal cell carcinoma and its prognostic significance. Urology 2008; 72: 603–7. [DOI] [PubMed] [Google Scholar]
- 12. Sano A, Kato H, Sakurai S et al. CD24 expression is a novel prognostic factor in esophageal squamous cell carcinoma. Ann Surg Oncol 2009; 16: 506–14. [DOI] [PubMed] [Google Scholar]
- 13. Baumann P, Cremers N, Kroese F, et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res 2005; 65: 10783–93. [DOI] [PubMed] [Google Scholar]
- 14. Smith SC, Oxford G, Wu Z et al. The metastasis‐associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res 2006; 66: 1917–22. [DOI] [PubMed] [Google Scholar]
- 15. Weichert W, Denkert C, Burkhardt M et al. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res 2005; 11: 6574–81. [DOI] [PubMed] [Google Scholar]
- 16. Sagiv E, Memeo L, Karin A et al. CD24 is a new oncogene, early at the multistep process of colorectal cancer carcinogenesis. Gastroenterology 2006; 131: 630–9. [DOI] [PubMed] [Google Scholar]
- 17. Sagiv E, Starr A, Rozovski U et al. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res 2008; 68: 2803–12. [DOI] [PubMed] [Google Scholar]
- 18. Chang L, Karin M. Mammalian MAP kinase signaling cascades. Nature 2001; 410: 37–40. [DOI] [PubMed] [Google Scholar]
- 19. Fukushima T, Tezuka T, Shimomura T, Nakano S, Kataoka H. Silencing of insulin‐like growth factor‐binding protein‐2 in human glioblastoma cells reduces both invasiveness and expression of progression‐associated gene CD24. J Biol Chem 2007; 282: 18. [DOI] [PubMed] [Google Scholar]
- 20. Wang X, Li M, Wang J et al. The BH3‐only protein, PUMA, is involved in oxaliplatin‐induced apoptosis in colon cancer cells. Biochem Pharmacol 2006; 71: 1540–50. [DOI] [PubMed] [Google Scholar]
- 21. Wang J, Wang X, Gong W, Mi B, Liu S, Jiang B. Increased expression of β‐catenin, phosphorylated glycogens synthase kinase 3β, cyclin D1, and c‐myc in laterally spreading colorectal tumors. J Histochem Cytochem 2009; 57: 363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deutsch AJ, Aigelsreiter A, Steinbauer E et al. Distinct signatures of B‐cell homeostatic and activation‐dependent chemokine receptors in the development and progression of extragastric MALT lymphomas. J Pathol 2008; 215: 431–44. [DOI] [PubMed] [Google Scholar]
- 23. Tournier C, Hess P, Yang DD et al. Requirement of JNK for stress‐induced activation of the cytochrome c‐mediated death pathway. Science 2000; 288: 870–4. [DOI] [PubMed] [Google Scholar]
- 24. Johnson G, Lapadat R. Mitogen‐activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002; 298: 1911–2. [DOI] [PubMed] [Google Scholar]
- 25. Simons K, Ikonen E. Functional rafts in cell membranes. Nature 1997; 387: 569–72. [DOI] [PubMed] [Google Scholar]
- 26. Suzuki T, Kiyokawa N, Taguchi T, Sekino T, Katagiri YU, Fujimoto J. CD24 induces apoptosis in human B cells via the glycolipid‐enriched membrane domains/rafts‐mediated signaling system. J Immunol 2001; 166: 5567–77. [DOI] [PubMed] [Google Scholar]
- 27. Sammar M, Gulbins E, Hilbert K, Lang F, Altevogt P. Mouse CD24 as a signaling molecule for integrin‐mediated cell binding: functional and physical association with src‐kinases. Biochem Biophys Res Commun 1997; 234: 330–4. [DOI] [PubMed] [Google Scholar]
- 28. Zarn JA, Zimmermann SM, Pass MK, Waibel R, Stahel RA. Association of CD24 with the kinase c‐fgr in a small cell lung cancer cell line and with the kinase lyn in an erythroleukemia cell line. Biochem Biophys Res Commun 1996; 225: 384–91. [DOI] [PubMed] [Google Scholar]
- 29. Sandilands E, Frame MC. Endosomal trafficking of Src tyrosine kinase. Trends Cell Biol 2008; 18: 322–9. [DOI] [PubMed] [Google Scholar]
- 30. Roberts PJ, Der CJ. Targeting the Raf‐MEK‐ERK mitogen‐activated protein kinase cascade for the treatment of cancer. Oncogene 2007; 26: 3291–310. [DOI] [PubMed] [Google Scholar]
- 31. Sebolt‐Leopold JS, Dudley DT, Herrera R et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo . Nat Med 1999; 5: 810–6. [DOI] [PubMed] [Google Scholar]
- 32. Sun Y, Sinicrope FA. Selective inhibitors of MEK1/ERK44/42 and p38 mitogen‐activated protein kinases potentiate apoptosis induction by sulindac sulfide in human colon carcinoma cells. Mol Cancer Ther 2005; 4: 51–9. [PubMed] [Google Scholar]
- 33. Hui L, Bakiri L, Mairhorfer A et al. P38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK–c‐Jun pathway. Nat Genet 2007; 39: 741–9. [DOI] [PubMed] [Google Scholar]
- 34. Ventura JJ, Tenbaum S, Perdiguero E et al. P38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet 2007; 39: 750–8. [DOI] [PubMed] [Google Scholar]
- 35. Tsuchiya T, Tsuno NH, Asakage M et al. Apoptosis induction by p38 MAPK inhibitor in human colon cancer cells. Hepatogastroenterology 2008; 55: 930–5. [PubMed] [Google Scholar]
- 36. Lim SJ, Lee YJ, Lee E. P38MAPK inhibitor SB203580 sensitizes human SNU‐C4 colon cancer cells to exisulind‐induced apoptosis. Oncol Rep 2006; 16: 1131–5. [PubMed] [Google Scholar]


