Abstract
Alcohol dehydrogenase and aldehyde dehydrogenase are key enzymes in alcohol metabolism and therefore may be of importance to colorectal cancer development. The present case–control study was conducted to determine the influence of ADH2, ADH3 and ALDH2 polymorphisms in Fukuoka, Japan, with 685 incident cases of histologically confirmed colorectal adenocarcinomas and 778 community controls selected randomly from the study area. Alcohol use was ascertained by in‐person interview. Statistical adjustment was made for sex, age class, area, and alcohol use. Individuals with the allele 47Arg of the ADH2 polymorphism (slow metabolizers) had a statistically significant increase in risk, with an adjusted OR of 1.32 (95% CI = 1.07–1.63), compared with those having the ADH2*47His/His genotype. This association was not affected by the level of alcohol consumption. The ADH3 polymorphism showed no measurable association with the risk of colorectal cancer on either overall analysis or stratified analysis with alcohol use. The heterozygous ALDH2*487Glu/Lys genotype was not associated with an increase in the risk of colorectal cancer (adjusted OR 0.89, 95% CI = 0.71–1.13) compared with the ALDH2*487Glu/Glu genotype. Rather unexpectedly, the homozygous ALDH2*487Lys/Lys genotype was related to a statistically significantly decreased risk of colorectal cancer (adjusted OR 0.55, 95% CI = 0.33–0.93). It is unlikely that acetaldehyde metabolism determined by ALDH2 polymorphism contributes to the risk of colorectal cancer, whereas the role of ADH2 polymorphism deserves further investigation. (Cancer Sci 2007; 98: 1248–1253)
- Abbreviations: ADH
alcohol dehydrogenase
- ALDH
aldehyde dehydrogenase
- CI
confidence interval
- HDL
high‐density lipoprotein
- OR
odds ratio
- PCR–RFLP
polymerase chain reaction–restriction fragment length polymorphism.
Alcohol consumption has fairly consistently been related to an increased risk of colorectal cancer.( 1 ) In a pooled analysis of eight cohort studies in North America and Europe, a consumption of ≥45 g of alcohol per day was associated with a 1.4‐fold increase in the risk of colorectal cancer.( 2 ) A positive association between alcohol and colon or colorectal cancer has also been observed in Asian countries,( 3 , 4 , 5 , 6 , 7 ) with few exceptions.( 8 ) However, uncertainty remains as to the biological mechanisms for the association between alcohol use and colorectal cancer.
Ethanol is first oxidized to acetaldehyde by ADH, and acetaldehyde is further metabolized to acetate by ALDH. Human ADH exhibits several isoenzymes, and functional polymorphisms are known for the ADH2 and ADH3 genes.( 9 ) A polymorphism in exon 3 of the ADH2 gene, resulting in an arginine to histidine substitution in codon 47, affects the enzyme activity substantially. Individuals that are homozygous for the ADH2*47His allele (previously called ADH2*2) metabolize ethanol 40 times faster than those homozygous for the ADH2*47Arg allele (previously called ADH2*1).( 10 ) The enzyme activity of ADH2*47His/Arg genotype is in the intermediate range between the two homozygous genotypes.( 11 ) The polymorphic site for the ADH3 gene is Ile349Val in exon 8. Maximal velocity is 2.5‐fold greater in individuals homozygous for the ADH3*349Ile allele (previously called ADH3*1) than in those homozygous for the ADH3*349Val allele (previously called ADH3*2).( 10 ) The ADH2*47His allele is fairly common in Asian populations and rare in Caucasians, while the ADH3*349Val allele is more frequent in Caucasians than in Asians.( 12 ) ALDH2 is the gene encoding mitochondrial ALDH, which contributes the majority of acetaldehyde oxidation in human liver and contains a functional polymorphism of Glu487Lys, with the variant ALDH2*487Lys (previously called ALDH2*2) allele resulting in an inactive form. The ALDH2*487Lys allele is mainly found in Asian populations.( 12 , 13 )
Several studies have investigated the relation of genetic polymorphisms of these alcohol‐metabolizing enzymes to colorectal cancer and adenomas. As regards the ADH2 polymorphism and colorectal cancer, a moderate increase in the risk of colorectal cancer was observed for each of the Arg/His and Arg/Arg genotypes compared with the His/His genotype in Japan,( 14 ) but not in Spain.( 15 ) The ADH3 polymorphism was unrelated to colorectal cancer in two studies of Caucasians,( 16 , 17 ) but one of these suggested an effect modification of alcohol consumption.( 16 ) Two studies have examined the relation between ADH3 polymorphism and colorectal adenomas in Caucasians, producing inconsistent results.( 18 , 19 ) Although there was no difference in the distribution of ADH3 genotypes between adenoma cases and controls in these studies, one showed a moderate increase in the risk of adenoma in men and women with the ADH3*349Ile/Ile genotype compared with those with the ADH3*349Ile/Val or ADH3*349Val/Val genotypes when alcohol consumption was high,( 18 ) whereas the other reported an increased risk of adenoma associated with the ADH3*349Val allele for men with high alcohol consumption.( 19 ) Studies regarding the ALDH2 polymorphism and colorectal cancer or adenomas have all been done in Japan.( 14 , 20 , 21 , 22 , 23 ) An approximately 3‐fold increase in the risk of colorectal cancer has been observed for ALDH2*487Glu/Lys versus ALDH2*487Glu/Glu among alcoholics.( 20 ) Another study suggested a greater increase in the risk of colon cancer, not of rectal cancer, associated with high alcohol consumption among individuals with the ALDH2*487Glu/Lys genotype.( 21 ) A small case–control study showed a positive interaction between high alcohol consumption and the ALDH2*487Glu/Lys genotype, particularly on the risk of rectal cancer.( 22 ) In contrast, the ALDH2 polymorphism did not show any measurable association with either colorectal cancer or adenomas in recent studies.( 14 , 23 )
The present paper examines the relation of the ADH2, ADH3 and ALDH2 polymorphisms to colorectal cancer in a case–control study in Japan, focusing on effect modification of alcohol consumption and gene–gene interaction.
Materials and Methods
The Fukuoka Colorectal Cancer Study is a case–control study of incident cases and community controls, with Fukuoka City and three adjacent areas as the catchment area. Details have been reported previously.( 24 ) Described below are methods relevant to the present analysis. The study protocol was approved by the ethical committees of Kyushu University and of all but two of the participating hospitals. There was no ethical committee at the two hospitals, and the survey was done at these hospitals with permission from the director of each hospital.
Subjects. Cases comprised a consecutive series of patients with histologically confirmed incident colorectal adenocarcinomas who were admitted to two university hospitals or six affiliated hospitals for surgical treatment during the period from October 2000 to December 2003. Other eligibility criteria included the following characteristics: age of 20–74 years at the time of diagnosis, residence in the study area, no prior history of partial or total removal of the colorectum, familial adenomatous polyposis or inflammatory bowel disease. Of the total of 1053 eligible cases, 840 cases (80%) participated in the interview, and 685 (65%) gave informed consent to genotyping.
Eligibility criteria for controls were the same as described for cases except for two items, that is, having no diagnosis of colorectal cancer and age of 20–74 years at the time of selection. A total of 1500 persons were selected as control candidates using two‐stage random sampling from among residents living in 15 small areas. A total of 833 persons participated in the survey, and 778 gave informed consent to genotyping. Reasons for exclusion and non‐participation were death (n = 7), migration from the study area (n = 22), undelivered mail (n = 44), mental incompetence (n = 19), history of partial or total removal of the colorectum (n = 21), diagnosis of colorectal cancer after the survey (n = 5), no response (n = 158), and refusal (n = 391). After exclusion of the first six categories of outcomes (n = 118), net participation rates were calculated as 60% (833/1382) for the interview and 56% (778/1382) for genotyping.
Interview. Research nurses interviewed cases and controls in person regarding physical activity, smoking, alcohol use, and other factors using a uniform questionnaire. Habitual alcohol consumption at the time 5 years prior to the onset of disease in cases or the interview in controls was ascertained. Individuals reported the average number of days per week that alcohol was consumed and the average amount of alcohol per day of drinking. The amount of alcohol was expressed by the conventional unit; one go (180 mL) of sake, one large bottle (633 mL) of beer, and half a go (90 mL) of shochu were each expressed as one unit; and one drink (30 mL) of whisky or brandy and one glass (100 mL) of wine were each converted to half a unit. The reproducibility of the questionnaire was tested on 29 control subjects (14 men and 15 women) with an interval of approximately 1 year, and the reported alcohol intake was highly reproducible (Spearman's r = 0.82).
Genotyping. A venous blood sample of 5 mL was taken after the interview. DNA was extracted from the buffy coat using a commercial kit (QIAGEN GmbH, Hilden, Germany) and genotyping was performed using the PCR–RFLP method. The PCR was performed in a reaction mixture of 10 µL containing 0.5 IU of Taq and 1 µL of template DNA with a concentration of approximately 50–150 ng/µL. The ADH2 Arg47His and ADH3 Ile349Val genotypes were determined according to the methods described by Osier et al.( 25 ) Primers for the ADH2 Arg47His genotypes were 5′‐ATT CTA AAT TGT TTA ATT CAA GAA g‐3′ (sense) and 5′‐ACT AAC ACA GAA TTA CTG GAC‐3′ (antisense). PCR products were digested with 20 IU of MslI for 16 h at 37°C in a mixture of 20 µL, resulting in fragments of 443 bp and 242 bp for the 47His allele and 685 bp for the 47Arg allele. The ADH3 Ile349Val genotypes were determined using primers of 5′‐TTG TTT ATC TGT GAT TTT TTT TGT‐3′ (sense) and 5′‐CGT TAC TGT AGA ATA CAA AGC‐3′ (antisense). The PCR product of 378 bp fragments was digested with 5 IU of SspI in a reaction mixture of 20 µL for 3 h at 37°C, resulting in fragments of 274 bp and 104 bp for the 349Ile allele and 378 bp for the 349Val allele. The ALDH2 Glu487Lys genotypes were determined, as described by Goedde et al.,( 26 ) using primers that were 5′‐CAA ATT ACA GGG TCA ACT GCT‐3′ (sense) and 5′‐CCA CAC TCA CAG TTT TCT CTT‐3′ (antisense). The PCR product was digested with Ksp632I (10 IU) or EarI (10 IU) for 12 h at 37°C in a mixture of 20 µL, resulting in fragments of 112 bp for the ALDH2*487Glu allele and 135 bp for the ALDH2*487Lys allele. The digested PCR products were separated using electrophoresis on 3% agarose gels (NuiSieve GTG, Rockland, ME, USA), and visualized with ethidium bromide.
Statistical analysis. The association of the genetic polymorphisms with risk of colorectal cancer was examined using multiple logistic regression analysis including indicator variables for sex, 5‐year age class (the lowest class was <40 years), resident area (Fukuoka City or adjacent areas), and alcohol intake (0, 0.1–1.9, or ≥2.0 units per day) as covariates. Adjusted OR and 95% CI were obtained from the logistic regression coefficient and the standard error for the corresponding indicator variable. Statistical significance for the interaction was tested using the likelihood ratio test comparing the logistic models with and without interaction terms for the genotype and alcohol category. Statistical significance was concluded if the two‐sided P‐value was less than 0.05 or if the 95% CI did not include unity. All statistical analyses were performed using SAS version 8.2 (SAS Institute Inc., Cary, NC, USA).
Results
The number of men among the 685 cases and 778 controls was 426 (62%) and 490 (63%), respectively. The mean age of the cases was 60 years (range 27–74), and that of the controls was 59 years (range 22–75). More than half of the cases (61%) and controls (64%) were residents of Fukuoka City. All of the distributions of genotypes for the ADH2 Arg47His, ADH3 Ile349Val, and ALDH2 Glu487Lys polymorphisms were in agreement with the Hardy–Weinberg equilibrium in both cases and controls. The alcohol‐drinking pattern differed strikingly by ALDH2 polymorphism and slightly so with respect to the ADH2 polymorphism (Fig. 1). Alcohol use was progressively less frequent with increasing numbers of the ALDH2*487Lys allele, and was slightly more frequent with increasing numbers of the ADH2*47Arg allele. There was no variation in the proportion of alcohol drinking according to the ADH3 Ile349Val polymorphism (data not shown).
Figure 1.
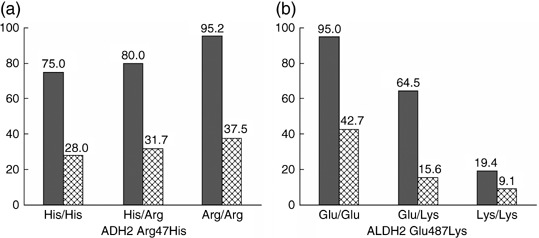
Proportions (%) of alcohol drinkers for men (gray bar) and women (hatched bar) according to the ADH2 Arg47His and ALDH2 Glu487Lys polymorphisms in the control group. Values shown at the top of each bar are percentages of alcohol drinkers. Trend P‐values were 0.03 in men and 0.35 in women for the ADH2 polymorphism, and <0.0001 in both sexes for the ALDH2 polymorphism. The trend P‐value was based to the Mantel‐Haenszel method with scores of 0, 1, and 2 assigned for the number of the variant allele.
Regarding the ADH2 polymorphism, the 47Arg allele was slightly more frequent in cases than in controls, and the adjusted OR for the Arg/His and Arg/Arg genotypes as compared with the His/His genotype were each greater than unity, the increase for the heterozygote being statistically significant (Table 1). The adjusted OR for those with the ADH2*47Arg allele compared with those without was 1.32 (95% CI = 1.07–1.63). There was no measurable difference in the distribution of ADH3 Ile349Val genotypes between cases and controls. The ALDH2*487Lys allele was less frequent in cases than in controls, and the adjusted OR of colorectal cancer for the Lys/Lys versus Glu/Glu genotype was statistically significantly lower than unity. Analysis by sex showed similar results for men and women. For instance, the adjusted OR for ADH2*47Arg/His and Arg/Arg genotypes combined were 1.34 (95% CI = 1.03–1.75) in men and 1.35 (95% CI = 0.95–1.91) in women. Regarding the ALDH2 polymorphism, the adjusted OR for the Glu/Lys and Lys/Lys genotypes compared with the Glu/Glu genotype in men were 0.77 (95% CI = 0.56–1.05) and 0.44 (95% CI = 0.22–0.91), respectively, while the corresponding values in women were 1.04 (95% CI = 0.71–1.53) and 0.67 (95% CI = 0.31–1.46), respectively.
Table 1.
Relation of ADH2, ADH3, and ALDH2 polymorphisms to colorectal cancer risk
| Genotype | Cases (n, %) | Controls (n, %) | Adjusted OR (95% CI) † |
|---|---|---|---|
| ADH2 Arg47His ‡ | |||
| His/His (fast) | 345 (50.8) | 452 (58.1) | 1.00 (referent) |
| Arg/His | 294 (43.3) | 289 (37.1) | 1.32 (1.06–1.64) |
| Arg/Arg (slow) | 40 (5.9) | 37 (4.8) | 1.36 (0.84–2.20) |
| ADH3 Ile349Val § | |||
| Ile/Ile (fast) | 609 (88.9) | 706 (90.9) | 1.00 (referent) |
| Ile/Val | 74 (10.8) | 68 (8.7) | 1.29 (0.91–1.83) |
| Val/Val (slow) | 2 (0.3) | 3 (0.4) | 0.77 (0.13–4.70) |
| ALDH2 Glu487Lys | |||
| Glu/Glu | 400 (58.4) | 416 (53.5) | 1.00 (referent) |
| Glu/Lys | 257 (37.5) | 309 (39.7) | 0.89 (0.71–1.13) |
| Lys/Lys (null activity) | 28 (4.1) | 53 (6.8) | 0.55 (0.33–0.93) |
Adjusted for sex, 5‐year age class, area, and alcohol use.
‡ Six cases were excluded because of undetermined genotype.
§ One control was excluded because of undetermined genotype. CI, confidence interval; OR, odds ratio.
Table 2 summarizes the results from the analysis regarding the interaction between alcohol intake and each genetic polymorphism. In this analysis, individuals heterozygous for the ADH2 or ADH3 polymorphism were each combined with those homozygous for the variant allele. Individuals homozygous for the variant allele of ALDH2 (487Lys/Lys) were excluded because alcohol use was almost null in this group. There was no appreciable effect modification of each polymorphism on the relation between alcohol and colorectal cancer. High alcohol consumption was related to a moderate increase in the OR of colorectal cancer regardless of the genotype. Adjusted OR for high alcohol use (≥2 units/day) versus no use after control for the ADH2, ADH3, and ALDH2 genotypes each were 1.34 (95% CI = 0.99–1.82), 1.37 (95% CI = 1.01–1.85) and 1.20 (95% CI = 0.85–1.69), respectively.
Table 2.
Combined effects of ADH2, ADH3, and ALDH2 polymorphisms with alcohol use on the risk of colorectal cancer
| Genotype | Alcohol intake (unit/day) † | |||
|---|---|---|---|---|
| 0 | <2 | ≥2 | ||
| ADH2 Arg47His | ||||
| His/His | No. ‡ | 142/192 | 109/167 | 94/93 |
| OR (95% CI) § | 1.00 (referent) | 0.97 (0.69–1.37) | 1.52 (1.02–2.25) | |
| Arg/His + Arg/Arg | No. | 128/119 | 109/123 | 97/84 |
| OR (95% CI) | 1.46 (1.04–2.03) Interaction P = 0.61 | 1.30 (0.92–1.85) | 1.69 (1.14–2.52) | |
| ADH3 Ile349Val | ||||
| Ile/Ile | No. | 235/281 | 199/266 | 175/159 |
| OR (95% CI) | 1.00 (referent) | 0.98 (0.74–1.28) | 1.44 (1.05–1.98) | |
| Ile/Val + Val/Val | No. | 37/30 | 22/23 | 17/18 |
| OR (95% CI) | 1.49 (0.89–2.50) Interaction P = 0.49 | 1.29 (0.70–2.40) | 1.27 (0.62–2.57) | |
| ALDH2 Glu487Lys ¶ | ||||
| Glu/Glu | No. | 98/103 | 150/171 | 152/142 |
| OR (95% CI) | 1.00 (referent) | 1.04 (0.71–1.53) | 1.24 (0.81–1.90) | |
| Glu/Lys | No. | 147/163 | 71/112 | 39/34 |
| OR (95% CI) | 1.02 (0.71–1.48) Interaction P = 0.30 | 0.74 (0.47–1.17) | 1.33 (0.74–2.39) | |
One unit of alcohol intake corresponded to 1 go (180 mL) of sake, 0.5 go (90 mL) of shochu, 1 large bottle (633 mL) of beer, 2 drinks (60 mL) of whiskey, or 2 glasses (200 mL) of wine.
‡ Numbers of cases/controls.
§ Adjusted for sex, 5‐year age class, and area.
¶ Individuals with 487Lys/Lys genotype were excluded, because alcohol drinkers were few. CI, confidence interval; OR, odds ratio.
The joint effects of the ADH2 or ADH3 polymorphism in combination with the ALDH2 polymorphism were examined among alcohol drinkers (Table 3). Individuals with the ADH2*47His/His genotype and the ALDH2*487Lys allele showed a statistically non‐significant, small decrease in the OR for colorectal cancer. No such decrease was noted on analysis of the non‐alcohol drinkers (data not shown). There was no clear interaction between the ADH3 and ALDH2 polymorphisms on the risk of colorectal cancer in either non‐alcohol drinkers or alcohol drinkers.
Table 3.
Combined effects of ADH2 or ADH3 polymorphism with the ALDH2 polymorphism on the risk of colorectal cancer in alcohol drinkers
| ADH2/ADH3 | ALDH2 Glu487Lys | ||
|---|---|---|---|
| Glu/Glu | Glu/Lys | ||
| ADH2 Arg47His | |||
| His/His | No. † | 153/170 | 50/84 |
| OR (95% CI) ‡ | 1.00 (referent) | 0.70 (0.46–1.07) | |
| Arg/His + Arg/Arg | No. | 148/143 | 57/62 |
| OR (95% CI) | 1.12 (0.81–1.55) Interaction P = 0.30 | 1.08 (0.70–1.67) | |
| ADH3 Ile349Val | |||
| Ile/Ile | No. | 274/283 | 100/135 |
| OR (95% CI) | 1.00 (referent) | 0.83 (0.60–1.14) | |
| Ile/Val + Val/Val | No. | 28/30 | 10/11 |
| OR (95% CI) | 1.00 (0.58–1.74) Interaction P = 0.76 | 0.98 (0.40–2.38) | |
Numbers of cases/controls.
‡ Adjusted for sex, 5‐year age class, area, and alcohol use low or high intake. CI, confidence interval; OR, odds ratio.
Discussion
The present study addressed the relation between genetic polymorphisms in alcohol metabolism and colorectal cancer. Individuals with the ADH2*47Arg allele (slow metabolizers) showed a modest, but statistically significant, increase in the risk of colorectal cancer. In contrast, individuals homozygous for the ALDH2 variant allele had a decreased risk of colorectal cancer. None of the polymorphisms affected the relation between alcohol and colorectal cancer. Because of the limited variation in the ADH3 polymorphism, the present study did not provide useful information regarding the role of ADH3 polymorphism in colorectal carcinogenesis.
Both ADH2 and ADH3 polymorphisms have been shown to affect the risk of various alcohol‐related conditions. The slow alcohol metabolism of ADH3 polymorphism has been related to increased risk of alcoholism and liver cirrhosis, elevated levels of serum HDL cholesterol, and decreased risk of myocardial infarction in Western populations.( 27 , 28 ) Studies in Japan have reported that the slow metabolizers with the ADH2 polymorphism had an increased risk of alcoholic liver disease,( 29 ) and of cerebral infarction.( 30 ) However, it remains uncertain whether these polymorphisms affect the risk of alcohol‐related cancers. In a meta‐analysis of seven case–control studies, no association between ADH3 polymorphism and upper aerodigestive cancers was found, nor was any interaction between ADH3 polymorphism and alcohol consumption on the risk of these cancers.( 12 ) A study of Japanese alcoholics has showed increased risks of oral, laryngeal, and esophageal cancer for the ADH2*47Arg/Arg genotype,( 31 ) while another study in Japan showed no effect modification of the ADH2 polymorphism on the association between alcohol and esophageal cancer.( 32 )
A recent Japanese study reported a positive association between the ADH2 polymorphism and colorectal cancer, showing a progressive increase in the risk with increasing numbers of the ADH2*47Arg allele.( 14 ) No such progressive increase in the risk was observed in the present study, but the authors’ findings are compatible with the previous observation in that the risk was elevated in individuals with the ADH2*47Arg allele. The authors have no clear explanation for the increased risk of colorectal cancer associated with the ADH2 polymorphism, but the consistency in the two independent studies warrants further investigation regarding the role of the ADH2 polymorphism in colorectal carcinogenesis.
The present study showed neither an increased risk of colorectal cancer associated with the ALDH2*487Lys allele nor interaction between the ALDH2*487Lys allele and alcohol consumption. These findings are at odds with results from previous studies of colorectal cancer,( 20 , 21 , 22 ) but are consistent with the recent observations on colorectal cancer,( 14 ) and adenomas.( 23 ) The statistically significant decrease in the risk of colorectal cancer associated with the ALDH2*487Lys/Lys genotype was rather unexpected and difficult to interpret. A decrease in the OR associated with the Lys/Lys genotype was also observed in the analysis confined to non‐drinkers of alcohol (n = 583), although the decrease was not statistically significant; adjusted OR for the Glu/Glu, Glu/Lys, and Lys/Lys genotypes were 1.00 (referent), 1.00 (95% CI = 0.68–1.46), and 0.64 (95% CI = 0.36–1.14), respectively. The decreased risk in individuals with the ALDH2487Lys/Lys genotype may have been due to residual confounding of lifestyle factors other than alcohol drinking. In the Fukuoka Colorectal Cancer Study, obesity and physical inactivity were related to increased risk,( 33 ) and there was a protective association with intake of n‐3 polyunsaturated fatty acids.( 34 ) With further adjustment for these factors as well as for dietary calcium and fiber using the variables and categories as defined previously,( 34 ) the adjusted OR for the Glu/Lys and Lys/Lys genotypes versus the Glu/Glu genotype were 0.90 (95% CI = 0.70–1.14) and 0.52 (95% CI = 0.31–0.88), respectively, in the analysis excluding four cases and two controls with a total calorie intake estimated to be >20 929 kJ/day. While a similar, inverse association was noted for colorectal adenomas,( 23 ) no such association was seen in another study of colorectal cancer in Japan.( 14 )
It is hypothesized that acetaldehyde is accumulated in individuals who are fast alcohol metabolizers and slow acetaldehyde metabolizers.( 29 ) Thus the authors hypothesized that the combination of ADH2*His/His and ALDH2*487Glu/Lys genotype might be related to an increased risk of colorectal cancer, but the risk of colorectal cancer was decreased, rather than increased in alcohol drinkers with such composite genotypes.
This finding on the gene–gene interaction, together with the above‐mentioned findings on the ALDH2 polymorphism, suggests that acetaldehyde metabolism in the liver is not measurably linked to colorectal carcinogenesis. Bacterial production of acetaldehyde in the colon is an alternative mechanism by which alcohol may enhance colorectal carcinogenesis. Human colonic contents and isolated colonic microbes are capable of producing acetaldehyde when incubated with ethanol in vitro.( 35 , 36 ) It has been demonstrated in piglets that high levels of acetaldehyde were produced in the colon during normal metabolism of alcohol.( 37 ) ALDH activity is much lower in the colonic mucosa than in the liver,( 38 , 39 ) and colonic epithelial cells are exposed to high concentrations of acetaldehyde in the colonic lumen. It is hypothesized that low folate status increases the risk of colorectal cancer by altering DNA methylation and DNA synthesis.( 40 , 41 ) Alcohol and acetaldehyde exert adverse effects on folate metabolism.( 42 ) High alcohol consumption results in inadequate folate status by decreasing intestinal absorption and increasing renal excretion. It is known that acetaldehyde rather than alcohol itself cleaves folate chemically. Ethanol ingestion has resulted in a substantial increase in the intracolonic concentration of acetaldehyde and decreased folate levels in the colonic mucosa in an experimental study of rats.( 43 )
The present study is probably the largest that has ever been reported regarding the ADH2 or ALDH2 polymorphism and colorectal cancer. Among the reported studies are those including 257 colorectal cancer cases and 771 controls,( 14 ) 270 cases and 121 controls,( 21 ) and 142 cases and 241 non‐cancer controls,( 22 ) in Japan. The size of a study is particularly important in investigating the role of rare genotypes in the gene–environment or gene–gene interaction. The participation rate in terms of genotyping was not so high in either cases (65%) or controls (56%). Because the ADH2 and ALDH2 polymorphisms affected alcohol drinking, a selection bias would be possible in the association with these polymorphisms if cases and controls participated in the study differentially with respect to alcohol drinking. Among those interviewed, however, the proportions of alcohol drinking in the cases and controls each did not differ by consent to genotyping.( 44 ) Alcohol consumption 5 years prior to the referent date was used, and the authors have no data as to how valid the recalled alcohol consumption in the past was, although it was found to be highly reproducible.
In summary, a case–control study in Japan showed an increased risk of colorectal cancer associated with the ADH2*47Arg allele, but not with the 487Lys allele of ALDH2 polymorphism. None of the polymorphisms affected the relation between alcohol consumption and colorectal cancer risk. It is unlikely that acetaldehyde metabolism determined by ALDH2 polymorphism contributes to the risk of colorectal cancer, whereas the role of ADH2 polymorphism deserves further investigation.
Acknowledgments
This work was supported by a Grant‐in‐Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan (18014022), and from a Grant‐in‐Aid in the year 2005, Fukuoka Cancer Society, Japan. The authors acknowledge support from Emeritus Professor Keizo Sugimachi; Professors Seiyo Ikeda, Takayuki Shirakusa, and Sumitaka Arima; and Drs Motonori Saku, Yoichi Ikeda, Soichiro Maekawa, Kazuo Tanoue, Kinjiro Sumiyoshi, and Shoichiro Saito in conducting the survey of cases. The following physicians kindly supervised the survey of controls at their clinics: Drs Hideaki Baba, Tomonori Endo, Hiroshi Hara, Yoichiro Hirokata, Motohisa Ikeda, Masayoshi Ishibashi, Fumiaki Itoh, Yasuhiro Iwanaga, Hideki Kaku, Shoshi Kaku, Minoru Kanazawa, Akira Kobayashi, Ryunosuke Kumashiro, Shinichi Matsumoto, Soukei Mioka, Umeji Miyakoda, Osamu Nakagaki, Nobuyoshi Nogawa, Nobuyuki Ogami, Toyoaki Okabayashi, Hironao Okabe, Nishiki Saku, Masafumi Tanaka, Masahiro Ueda, Bunichi Ushio, and Koheisho Yasunaga. The authors are grateful to research nurses: Nobuko Taguchi, Yuriko Moroe, Yuko Noda, Ryoko Tanaka, Hisako Nakagawa, and Yoko Mikasa; and research clerk Hiroko Mizuta; and the assistance of Masumi Koga and Akiko Koga, for their self‐sacrificing work.
References
- 1. World Cancer Research Fund and American Institute for Cancer Research . Food, nutrition and the prevention of cancer: a global perspective. Washington, DC: American Institute for Cancer Research, 1997: 216–51. [DOI] [PubMed] [Google Scholar]
- 2. Cho E, Smith‐Warner SA, Ritz J et al . Alcohol intake and colorectal cancer: a pooled analysis of 8 cohort studies. Ann Intern Med 2004; 140: 603–13. [DOI] [PubMed] [Google Scholar]
- 3. Shimizu N, Nagata C, Shimizu H et al . Height, weight, and alcohol consumption in relation to the risk of colorectal cancer in Japan: a prospective study. Br J Cancer 2003; 88: 1038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Otani T, Iwasaki M, Yamamoto S et al . Alcohol consumption, smoking, and subsequent risk of colorectal cancer in middle‐aged and elderly Japanese men and women: Japan Public Health Center‐based prospective study. Cancer Epidemiol Biomarkers Prev 2003; 12: 1492–500. [PubMed] [Google Scholar]
- 5. Wakai K, Kojima M, Tamakoshi K et al . Alcohol consumption and colorectal cancer risk: findings from the JACC Study. J Epidemiol 2005; 15: S173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ji BT, Dai Q, Gao YT et al . Cigarette and alcohol consumption and the risk of colorectal cancer in Shanghai, China. Eur J Cancer Prev 2002; 11: 237–44. [DOI] [PubMed] [Google Scholar]
- 7. Ho JW, Lam TH, Tse CW et al . Smoking, drinking and colorectal cancer in Hong Kong Chinese: a case‐control study. Int J Cancer 2004; 109: 587–97. [DOI] [PubMed] [Google Scholar]
- 8. Chen K, Jiang Q, Ma X et al . Alcohol drinking and colorectal cancer: a population‐based prospective cohort study in China. Eur J Epidemiol 2005; 20: 149–54. [DOI] [PubMed] [Google Scholar]
- 9. Yoshida A, Hsu LC, Yasunami M. Genetics of human alcohol‐metabolizing enzymes. Prog Nucl Acid Res Mol Biol 1991; 40: 255–87. [DOI] [PubMed] [Google Scholar]
- 10. Bosron W, Li TK. Genetic polymorphism of human liver alcohol and aldehyde dehydrogenase, and their relationship to alcohol metabolism and alcoholism. Hepatology 1986; 6: 502–10. [DOI] [PubMed] [Google Scholar]
- 11. Yin SJ, Bosron WF, Magnes LJ, Li TK. Human liver alcohol and dehydrogenase. Purification and kinetic characterization of the β2 β2, β2 β1, αβ2, and β2γ1 ‘Oriental’ isoenzymes. Biochemistry 1984; 23: 5847–53. [DOI] [PubMed] [Google Scholar]
- 12. Brennan P, Lewis S, Hashibe M et al . Pooled analysis of alcohol dehydrogenase genotype and head and neck cancer: a HuGE Review. Am J Epidemiol 2004; 159: 1–16. [DOI] [PubMed] [Google Scholar]
- 13. Takeshita T, Morimoto K, Mao XQ, Hashimoto T, Furuyama J. Characterization of three genotypes of low Km aldehyde dehydrogenase in a Japanese population. Hum Genet 1994; 94: 217–23. [DOI] [PubMed] [Google Scholar]
- 14. Matsuo K, Wakai K, Hirose K et al . A gene–gene interaction between ALDH2 Glu487Lys and ADH2 His47Arg polymorphisms regarding the risk of colorectal cancer in Japan. Carcinogenesis 2006; 27: 1018–23. [DOI] [PubMed] [Google Scholar]
- 15. Landi S, Gemignani F, Moreno V et al . A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet Genomics 2005; 15: 535–46. [DOI] [PubMed] [Google Scholar]
- 16. Chen J, Ma J, Stampfer MJ, Hines LM, Selhub J, Hunter DJ. Alcohol dehydrogenase 3 genotype is not predictive for risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev 2001; 10: 1303–4. [PubMed] [Google Scholar]
- 17. Van Der Logt EM, Bergevoet SM, Roelofs HM et al . Role of epoxide hydrolase, NAD(P)H: quinine oxidoreductase, cytochrome P450 2E1 or alcohol dehydrogenase genotypes in susceptibility to colorectal cancer. Mutat Res 2006; 593: 39–49. [DOI] [PubMed] [Google Scholar]
- 18. Tiemersma EW, Wark PA, Ocke MC et al . Alcohol consumption, alcohol dehydrogenese 3 polymorphism, and colorectal adenomas. Cancer Epidemiol Biomarkers Prev 2003; 12: 419–25. [PubMed] [Google Scholar]
- 19. Giovannucci E, Chen J, Smith‐Warner SA et al . Methylenetetrahydrofolate reductase, alcohol dehydrogenase, diet, and risk of colorectal adenomas. Cancer Epidemiol Biomarkers Prev 2003; 12: 970–9. [PubMed] [Google Scholar]
- 20. Yokoyama A, Muramatsu T, Ohmori T et al . Alcohol‐related cancers and aldehyde dehydrogenase‐2 in Japanese alcoholics. Carcinogenesis 1998; 19: 1383–7. [DOI] [PubMed] [Google Scholar]
- 21. Murata M, Tagawa M, Watanabe S, Kimura H, Takeshita T, Morimoto K. Genotype difference of aldehyde dehydrogenase 2 gene in alcohol drinkers influences the incidence of Japanese colorectal cancer patients. Jpn J Cancer Res 1999; 90: 711–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsuo K, Hamajima N, Hirai T et al . Aldehyde dehydrogenase 2 (ALDH2) genotype affects rectal cancer susceptibility due to alcohol consumption. J Epidemiol 2002; 12: 70–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirose M, Kono S, Tabata S et al . Genetic polymorphisms of methylenetetrahydrofolate reductase and aldehyde dehydrogenase2, alcohol use and risk of colorectal adenomas: Self‐Defense Forces Health Study. Cancer Sci 2005; 96: 513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kono S, Toyomura K, Yin G, Nagano J, Mizoue T. A case‐control study of colorectal cancer in relation to lifestyle factors and genetic polymorphisms. Design and conduct of The Fukuoka Colorectal Cancer Study. Asian Pac J Cancer Prev 2004; 5: 393–400. [PubMed] [Google Scholar]
- 25. Osier MV, Pakstis AJ, Soodyall H et al . A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet 2002; 71: 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goedde HW, Singh S, Agarwal DP, Fritze G, Stapel K, Paik YK. Genotyping of mitochondrial aldehyde oligonucleotides: comparison with phenotyping in hair roots. Hum Genet 1989; 81: 305–7. [DOI] [PubMed] [Google Scholar]
- 27. Cichoz‐Lach H, Partycka J, Nesima I, Celinski K, Slomka M, Wojcierowski J. Genetic polymorophism of alcohol dehydrogenase 3 in alcohol liver cirrhosis and in alcohol chronic pancreatitis. Alcohol Alcohol 2006; 41: 14–17. [DOI] [PubMed] [Google Scholar]
- 28. Hines LM, Stampfer MJ, Ma J et al . Genetic variation in alcohol dehydrogenase and the beneficial effect of moderate alcohol consumption on myocardial infarction. N Engl J Med 2001; 344: 549–55. [DOI] [PubMed] [Google Scholar]
- 29. Tanaka F, Shiratori Y, Yokosuka O, Imazeki F, Tsukada Y, Omata M. High incidence of ADH2*1/ALDH2*1 genes among Japanese alcohol dependents and patients with alcoholic liver disease. Hepatology 1996; 23: 234–9. [DOI] [PubMed] [Google Scholar]
- 30. Suzuki Y, Fujisawa M, Ando F, Niino N, Ohsawa I. Alcohol dehydrogenase 2 variant is associated with cerebral infarction and lacunae. Neurology 2004; 63: 1711–13. [DOI] [PubMed] [Google Scholar]
- 31. Yokoyama A, Muramatsu T, Omori T et al . Alcohol and aldehyde dehydrogenases gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis 2001; 22: 433–9. [DOI] [PubMed] [Google Scholar]
- 32. Yang CX, Matsuo K, Ito H et al . Esophageal cancer risk by ALDH2 and ADH2 polymorphisms and alcohol consumption: exploration of gene‐environment and gene–gene interactions. Asian Pac J Cancer Prev 2005; 6: 256–62. [PubMed] [Google Scholar]
- 33. Isomura K, Kono S, Moore MA et al . Physical activity and colorectal cancer: The Fukuoka Colorectal Cancer Study. Cancer Sci 2006; 97: 1099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimura Y, Kono S, Toyomura K et al . Meat, fish and fat intake in relation to subsite‐specific risk of colorectal cancer: The Fukuoka Colorectal Cancer Study. Cancer Sci 2007; 98: 590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jokelainen K, Roine RP, Vaananen H, Farkkila M, Salaspuro M. In vitro acetaldehyde formation by human colonic bacteria. Gut 1994; 35: 1271–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jokelainen K, Siitonen A, Jousimies‐Somer H, Nosova T, Heine R, Salaspuro M. In vitro alcohol dehydrogenase‐mediated acetaldehyde production by aerobic bacteria representing the normal colonic flora in man. Alcohol Clin Exp Res 1996; 20: 967–72. [DOI] [PubMed] [Google Scholar]
- 37. Jokelainen K, Matysiak‐Budnik T, Mäkisalo H, Höckerstedt K, Salaspuro M. High intracolonic acetaldehyde values produced by a bacteriocolonic pathway for ethanol oxidation in piglets. Gut 1996; 39: 100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koivisto T, Salaspuro M. Aldehyde dehydrogenases of the rat colon: Comparison with other tissues of the alimentary tract and the liver. Alcohol Clin Exp Res 1996; 20: 551–5. [DOI] [PubMed] [Google Scholar]
- 39. Visapää JP, Tillonen J, Salaspuro M. Microbes and mucosa in the regulation of intracolonic acetaldehyde concentration during ethanol challenge. Alcohol Alcohol 2002; 37: 322–6. [DOI] [PubMed] [Google Scholar]
- 40. Giovannucci E. Alcohol, one‐carbon metabolism, and colorectal cancer: recent insights from molecular studies. J Nutr 2004; 134: 2475S–81S. [DOI] [PubMed] [Google Scholar]
- 41. Kono S, Chen K. Genetic polymorphisms of methylenetetrahydrofolate reductase and colorectal cancer and adenoma. Cancer Sci 2005; 96: 535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mason JB, Choi SW. Effects of alcohol on folate metabolism: implications for carcinogenesis. Alcohol 2005; 35: 235–41. [DOI] [PubMed] [Google Scholar]
- 43. Homann N, Tillonen J, Salaspuro M. Microbially produced acetaldehyde from ethanol may increase the risk of colon cancer via folate deficiency. Int J Cancer 2000; 86: 169–73. [DOI] [PubMed] [Google Scholar]
- 44. Hagiwara T, Kono S, Yin G et al . Genetic polymorphism in cytochrome P450 7A1 and risk of colorectal cancer: The Fukuoka Colorectal Cancer Study. Cancer Res 2005; 65: 2979–82. [DOI] [PubMed] [Google Scholar]


