Abstract
Although many clinical and pathological prognostic factors such as tumor stage and lymph‐node involvement have been described, to date no reliable or clinically applicable marker or tumor aggressiveness has been identified for head and neck cancer. In an attempt to identify such a molecular prognostic marker, we analyzed the mRNA expression status of ING3 by quantitative reverse transcription–polymerase chain reaction. We also examined p53 mutation status and investigated its relationship with ING3, as well its clinicopathological characteristics. About half of the 71 tumor samples demonstrated downregulation of ING3 compared to their matched normal counterparts. Although most clinicopathological variables were not significantly related to ING3 downregulation or p53 mutation status, a significant relationship was detected in terms of overall survival between the cases with low and normal to high ING3 expression. At 5 years follow up, approximately 60% of the patients with normal to high ING3 expression survived, whereas this was 35% in the patients with low ING3 expression. Multivariate analysis also showed downregulation of ING3 as an independent prognostic factor for poor overall survival. These results reveal that ING3 would function as a potential tumor suppressor molecule and that low levels of ING3 may indicate an aggressive nature of head and neck cancer. (Cancer Sci 2008; 99: 531–538)
Head and neck squamous cell carcinoma (HNSCC) is the fifth most frequently occurring malignancy worldwide, representing a major international health problem. Despite improved detection and aggressive and multidisciplinary treatment approaches, including preoperative and postoperative chemotherapy or radiotherapy with surgery, head and neck cancers are still a great threat to human life and limited improvement in 5‐year survival has been gained over the last few decades.( 1 ) Moreover, current therapeutic agents are highly toxic, which decreases the quality of life of patients and has limited effect on survival. An ideal or best situation would be to find a single factor or several factors that could guide the clinician in choosing therapeutic options, as well as a treatment method that is effective and does not deteriorate the patients’ quality of life. Although many clinical and pathological prognostic factors, such as tumor stage, lymph‐node involvement, post‐surgical margin, and histological grade, have been described, to date no reliable marker of tumor aggressiveness has been identified for HNSCC.
Recent advances in the molecular biology of human cancers, including head and neck carcinomas, and technology have provided possible novel diagnostic and prognostic markers. These studies have already shown various molecular abnormalities in HNSCC, including activation of oncogenes,( 2 , 3 ) inactivation of tumor‐suppressor genes (TSG),( 4 , 5 , 6 , 7 ) expression of angiogenic factors,( 8 ) and loss of heterozygosity,( 5 , 6 , 7 , 9 ) at numerous chromosomal locations. Based on these molecular observations, recent studies have tried to find biomarkers that precisely identify patients at highest risk. These studies primarily had two purposes. The first was to find a reliable and easily applicable marker that can assist in the early diagnosis of cancer during carcinogenic stages. In such a study, 11 out of 35 oral premalignant lesions followed up from 1 to 16 years developed cancer and 7 of 11 cases had a p53 mutation.( 10 ) The second was that these molecular analyses would give information about the prognosis and behavior of the tumor, such as metastatic capacity, recurrence or aggressive phenotype, response to chemotherapy or radiotherapy, and survival of the patient. In fact, an increasing number of studies have been published in the literature recently that suggest that alterations of chromosomal loci and expression of cancer‐related genes could be used as prognostic or responsive markers for treatment. Most of these studies involved alterations in well‐known TSG and oncogenes, including p53, retinoblastoma (RB), p16, p21, p27, epidermal growth factor receptor (EGFR), cyclin D1, and Myc, and abnormalities of these loci or genes were detected in patients with poor prognosis.( 10 , 11 , 12 , 13 , 14 ) Thus, it is now well known that genetic abnormalities lead to cancer development and these alterations can predict prognosis either independently or combined with other pathologies.
We recently clarified the genomic structure of ING1, the prototype of the novel tumor suppressor ING‐family of genes, and showed for the first time its tumor‐suppressive role in a human cancer.( 5 ) Our continuous efforts led to identification of a novel member of this tumor‐suppressor ING family, ING3.( 6 ) In that study, frequent allelic loss of ING3 and its decreased expression in a limited number of HNSCC cases were shown. In the current study, we examined the mRNA expression of ING3 in a large population of HNSCC and compared the clinicopathological characteristics to evaluate the prognostic value of alterations in ING3 expression. Furthermore, we also examined the mutation status of the well‐known p53 TSG and tried to find out its prognostic association alone as well as together with ING3 in HNSCC.
Materials and Methods
Patients and samples. Paired normal and tumor samples were obtained from 71 patients with primary HNSCC between 1994 and 2005 at the department of otolaryngology, Okayama University Hospital after acquisition of informed consent from each patient. All tissues were frozen in liquid nitrogen immediately after surgery and stored at –80°C until the extraction of RNA. Patients included 61 men and 10 women with a mean age of 64 years (range, 40–81 years). The histological diagnosis of all tumor cases was squamous cell carcinoma. Normal tissue from each pair was also examined with hematoxylin–eosin staining to confirm their normal histology. The differentiation and diagnosis of the tumor was based on the surgical pathology reports of the hospital. All clinical information was obtained from the patient files, which included the initial diagnosis, treatment, and follow‐up data. The bioethics committee of the institution approved the study. The clinicopathological characteristics of the patients are shown in Table 1.
Table 1.
Clinicopathological characteristics of patients
| Criteria | n | % |
|---|---|---|
| Tumor site | ||
| Oral cavity | 31 | 43.7 |
| Oropharynx | 9 | 12.7 |
| Larynx | 13 | 18.3 |
| Hypopharynx | 11 | 15.5 |
| Maxilla | 7 | 9.8 |
| Age (mean ± SD) (years) | 64.3 ± 8.7 | |
| Sex | ||
| Male | 61 | 85.9 |
| Female | 10 | 14.1 |
| Tumor status | ||
| T1 | 6 | 8.4 |
| T2 | 23 | 32.4 |
| T3 | 21 | 29.6 |
| T4 | 21 | 29.6 |
| Node status | ||
| N0 | 32 | 45.1 |
| N1 | 10 | 14.1 |
| N2 | 29 | 40.8 |
| TNM stage | ||
| I | 3 | 4.2 |
| II | 12 | 16.9 |
| III | 18 | 25.4 |
| IV | 38 | 53.5 |
| Recurrence | ||
| None | 27 | 38 |
| Locoregional and/or distant | 44 | 62 |
| Differentiation † | 20 | 29.4 |
| Well | 20 | 29.4 |
| Moderate | 33 | 48.5 |
| Poor | 15 | 22.1 |
Three cases with unknown differentiation status were not included.
RNA isolation, cDNA preparation, and reverse transcription–polymerase chain reaction analysis. Total RNA was prepared using a modified acid guanidinium phenol chloroform method (Isogen; Nippon Gene, Tokyo, Japan). Total RNA was reverse transcribed with the Toyobo preamplification system (ReverTra Ace Kit; Toyobo, Osaka, Japan) starting with 2 µg of total RNA from each sample, according to the procedures provided by the supplier. ING3 mRNA expression in paired tumor and normal tissues was examined by duplex reverse transcription (RT)‐polymerase chain reaction (PCR). One microliter of each RT reaction was amplified in 50 µL of mixture containing 1.2 mM MgCl2, 1× PCR buffer, 200 µM of each deoxynucleotide triphosphate, 20 pmol of each primer, and 1 U recombinant thermus thermophilus (rTth) DNA polymerase XL (Applied Biosystems, Foster City, CA, USA). Thirty PCR cycles for the ING3 primers S2 (5′‐CAG CCT CTT CTA ACA ATG CCT A) and RAS1 (5′‐CTT CAT CAA ACA AAA GGA CCA CC), and 25 cycles for the β‐actin primers S1 (5′‐GGC CAA CCG CGA GAA GAT GAC) and AS1 (5′‐GCT CGT AGC TCT TCT CCA GGG) were used for amplification. The primers were designed using Genetyx‐Mac 10.1 software (Software Development, Tokyo, Japan). An initial denaturation step at 94°C for 3 min was followed by 30 cycles of a denaturation step at 94°C for 30 s, an annealing step at 60°C for 1 min, and an extension step at 72°C for 1 min. A final extension step at 72°C for 7 min was added. β‐Actin primers were added to each PCR tube at the end of the fifth cycle by holding the thermocycler at 94°C temporarily. Reproducibility was confirmed by processing all samples twice.
Quantification of the RT‐PCR products. Polymerase chain reaction products were separated through a 2% agarose gel and stained with ethidium bromide. As the sizes of the PCR products were 386 bp for β‐actin and 703 bp for ING3, they were readily distinguishable. The intensity of ethidium bromide staining of each band was measured using a CCD image sensor (Gel Print 2000/VGA; Toyobo), and analyzed by a computer program for band quantification (Quantity One; Toyobo). The value of tumor‐specific ING3 expression was determined by calculating the ratio of the expression levels in the tumor and in the matched normal sample, each of which was normalized for the corresponding β‐actin expression level (T, ING3/β‐actin expression ratio in tumor sample; N, ING3/β‐actin expression ratio in matched normal sample; T/N ratio, the relative ING3 expression in the tumor sample compared to its matched normal sample after normalization). Decreased and increased expression levels were determined as classes L and H when this ratio was less than 0.6 and greater than 1.4, respectively, as reported previously.( 6 ) Class N (normal expression) means that the value of the ratio was between 0.6 and 1.4.
Mutation analysis of p53. Each of the coding regions of exons 4–9 of the p53 gene were amplified by PCR with intron‐spanning primers designed using the Genetyx‐Mac 10.1 software: for exon 4, EX4‐S (5′‐ATC TAC AGT CCC CCT TGC CG) and EX4‐AS (5′‐GCA ACT GAC CGT GCA AGT CA); for exon 5, EX5‐S (5′‐GAC TTT CAA CTC TGT CTC CTT C) and EX5‐AS (5′‐AAC CAG CCC TGT CGT CTC TC); for exon 6, EX6‐S (5′‐ACC ATG AGC GCT GCT CAG AT) and EX6‐AS (5′‐AGT TGC AAA CCA GAC CTC AG); for exon 7, EX7‐S (5′‐CTT GGG CCT GTG TTA TCT CCT) and EX7‐AS (5′‐AGG GTG GCA AGT GGC TCC TG); for exon 8, EX8‐S (5′‐CCT TAC TGC CTC TTG CTT CTC) and EX8‐AS (5′‐TGA ATC TGA GGC ATA ACT GCA C); and for exon 9, EX9‐S (5′‐AGT TAT GCC TCA GAT TCA CTT TT) and EX9‐AS (5′‐GAT AAG AGG TCC CAA GAC TTA G). PCR amplification from genomic DNA and subsequent direct sequencing using the same primers was carried out as described previously.( 5 , 6 )
Statistical analysis. Pearson's χ2‐test and Student's t‐test were used to evaluate the correlation between ING3 mRNA expression and the clinicopathological characteristics of the patients. Survival curves were calculated according to Kaplan–Meier. For comparison of survival between low and normal to high expression of ING3, the log‐rank test was used. Overall survival in months was calculated from the day after surgery to the last follow‐up examination or death. The survival periods of the patients who were still alive were noted along with the date of the most recent follow‐up appointment. The duration of disease‐free survival (DFS) was determined from the day after surgery to the initial recurrence of the surgically resected cancer, evaluated by clinical examination. For multivariate analyses, we used the Cox proportional hazards model. All statistical manipulations were done using SPSS version 10 for the Windows software system (SPSS, Chicago, IL, USA) and a significant difference was identified when the probability was less than 0.05.
Results
mRNA expression analysis of ING3 in normal and tumor tissues. We analyzed the expression level of ING3 mRNA in tumor samples compared with the paired normal tissues. In 71 matched RNA samples, the expression levels of ING3 mRNA were compared by quantitative RT‐PCR using β‐actin mRNA as a control (Fig. 1; Table 2). PCR primers were designed to encompass the exon–intron junctions on the cDNA in order to eliminate the potential contamination of genomic DNA. Expression analysis demonstrated decreased or no expression of ING3 mRNA in 52.1% of primary tumors (37 out of 71) compared with matched normal samples, whereas 27% (19/71) of the samples showed a similar level of expression in normal and tumor tissues. Expression was not detectable in three samples (4%). In 21% (15/71) of the samples, increased expression of ING3 was detected in tumor tissues. For statistical analysis, normal and high expression groups were combined and ING3 expression was classified into two groups: low and normal to high expression.
Figure 1.
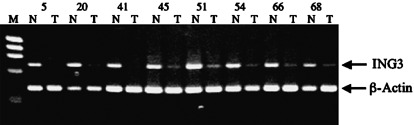
ING3 expression analysis in some of the matched tumor and normal samples. Representative raw data from the reverse transcription–polymerase chain reaction are shown. Lower band, β‐actin message (386 bp); upper band, ING3 message (703 bp); N, non‐tumor tissue; T, tumor tissue; M, size marker. Upper numbers (5, 20, 41, 45, 51, 54, 66, 68) show case numbers.
Table 2.
Inhibitor of growth (ING3) mRNA expression and p53 mutation status
| Case | Localization | ING3 | Exp † | p53 Mut | Case | Localization | ING3 | Exp † | p53 Mut |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Oropharynx | 1.68 | H | – | 37 | Hypopharynx | 1.12 | N | + |
| 2 | Oropharynx | 0.59 | L | + | 38 | Hypopharynx | 2.5 | H | – |
| 3 | Oral cavity | 0.49 | L | ND | 39 | Hypopharynx | 2.8 | H | – |
| 4 | Oropharynx | 1.25 | N | + | 40 | Hypopharynx | 0.3 | L | – |
| 5 | Larynx | 0 | L | – | 41 | Larynx | 0 | L | – |
| 6 | Maxilla | 3 | H | + | 42 | Maxilla | 0.34 | L | + |
| 7 | Oral cavity | 0.34 | L | – | 43 | Oral cavity | 0.38 | L | – |
| 8 | Larynx | 0.29 | L | – | 44 | Oral cavity | 1.93 | H | – |
| 9 | Oral cavity | 1.65 | H | – | 45 | Hypopharnx | 0.1 | L | + |
| 10 | Larynx | 1.7 | H | – | 46 | Oral cavity | 2.2 | H | – |
| 11 | Oral cavity | 0.44 | L | – | 47 | Oral cavity | 0.49 | L | – |
| 12 | Oral cavity | 0.49 | L | ND | 48 | Oral cavity | 0.5 | L | + |
| 13 | Maxilla | 1 | N | – | 49 | Oropharynx | 0.26 | L | – |
| 14 | Oral cavity | 6.8 | H | – | 50 | Hypopharynx | 0.71 | N | + |
| 15 | Maxilla | 2 | H | + | 51 | Oropharynx | 0.23 | L | – |
| 16 | Oral cavity | 0.7 | N | – | 52 | Hypopharynx | 0.4 | L | – |
| 17 | Oral cavity | 1 | N | – | 53 | Oral cavity | 1.2 | N | – |
| 18 | Larynx | 0.26 | L | – | 54 | Oral cavity | 0.16 | L | + |
| 19 | Maxilla | 0.34 | L | – | 55 | Larynx | 1.17 | N | – |
| 20 | Oral cavity | 0 | L | + | 56 | Oral cavity | 1.2 | N | – |
| 21 | Oral cavity | 1.7 | H | – | 57 | Oropharynx | 0.51 | L | – |
| 22 | Maxilla | 0.44 | L | – | 58 | Oral cavity | 0.47 | L | + |
| 23 | Oral cavity | 1 | N | + | 59 | Oral cavity | 0.58 | L | – |
| 24 | Oral cavity | 0.84 | N | – | 60 | Larynx | 1.48 | H | + |
| 25 | Hypopharynx | 0.72 | N | – | 61 | Larynx | 0.29 | L | + |
| 26 | Larynx | 1.61 | H | + | 62 | Larynx | 0.81 | N | + |
| 27 | Oral cavity | 0.51 | L | – | 63 | Oral cavity | 0.57 | L | – |
| 28 | Oropharynx | 0.96 | N | + | 64 | Hypopharynx | 0.34 | L | – |
| 29 | Maxilla | 1.24 | N | + | 65 | Larynx | 0.94 | N | + |
| 30 | Oropharynx | 0.35 | L | + | 66 | Oropharynx | 0.14 | L | – |
| 31 | Oral cavity | 0.54 | L | – | 67 | Hypopharynx | 0.36 | L | + |
| 32 | Oral cavity | 0.56 | L | + | 68 | Oral cavity | 0.12 | L | – |
| 33 | Hypopharynx | 1.3 | N | + | 69 | Oral cavity | 0.31 | L | + |
| 34 | Larynx | 0.57 | L | + | 70 | Oral cavity | 1.05 | N | – |
| 35 | Larynx | 1.7 | H | + | 71 | Oral cavity | 4.8 | H | + |
| 36 | Oral cavity | 1.09 | N | – |
Ratio of expression levels as described in Materials and Methods. H, high mRNA expression; L, low mRNA expression; N, normal mRNA expression; ND, not done.
Mutation analysis of the p53 gene. We analyzed genomic DNA from 69 patients with HNSCC for mutations in p53 by DNA direct sequencing and identified p53 mutations in 28 of them (40.6%; 28 of 69) (Table 3). Among these, four were nonsense mutations, one was both a nonsense and missense mutation, five had insertions or deletions leading to frameshifts, and one was mutated at the splicing junction; the remaining 17 were missense point mutations leading to amino acid substitutions (Table 3).
Table 3.
p53 mutation status
| Case | Codon change | Amino acid change | Exon | Case | Codon change | Amino acid change | Exon |
|---|---|---|---|---|---|---|---|
| 1 | – | 37 | 72. 2‐bp deletion | Frameshift | 4 | ||
| 2 | 306.CGA/TGA | Arg/Early stop | 8 | 38 | – | ||
| 3 | ND | 39 | – | ||||
| 4 | 168. ‐AG‐insert | Early stop | 5 | 40 | – | ||
| 5 | – | 41 | – | ||||
| 6 | 193.CAT/CTT | His/Leu | 6 | 42 | 173.GTG/ATG | Val/Met | 5 |
| 7 | – | 43 | – | ||||
| 8 | – | 44 | – | ||||
| 9 | – | 45 | 207. 1‐bp deletion | Frameshift | 7 | ||
| 10 | – | 46 | – | ||||
| 11 | – | 47 | – | ||||
| 12 | ND | 48 | 155.ACC/GCC, 213.CGA/TGA | Thr/Ala, Arg/Early stop | 5, 6 | ||
| 13 | – | 49 | – | ||||
| 14 | – | 50 | 191.CCT/CGT | Pro/Arg | 6 | ||
| 15 | Deletion | Frameshift | 7 | 51 | – | ||
| 16 | – | 52 | – | ||||
| 17 | – | 53 | – | ||||
| 18 | – | 54 | 175. CGC/CAC | Arg/His | 5 | ||
| 19 | – | 55 | – | ||||
| 20 | 193.CAT/CTT | His/Leu | 6 | 56 | – | ||
| 21 | – | 57 | – | ||||
| 22 | – | 58 | 237.ATG/ATA | Met/Ile | 7 | ||
| 23 | 502. 2 bp insert | Frameshift | 5 | 59 | – | ||
| 24 | – | 60 | 220.TAT/TGT | Tyr/Cys | 7 | ||
| 25 | – | 61 | 192.CAG/TAG | Gln/Early stop | 6 | ||
| 26 | 278.CCT/CTT | Pro/Leu | 8 | 62 | Splice junction | Int8‐Ex9 junction AG‐AT | 9 |
| 27 | – | 63 | – | ||||
| 28 | 192.Del & Ins | Frameshift | 6 | 64 | – | ||
| 29 | 175.CGC/CAC | Arg/His | 5 | 65 | 151.CCC/CGC | Pro/Arg | 5 |
| 30 | 278.CCT/TCT | Pro/Ser | 8 | 66 | – | ||
| 31 | – | 67 | 244.GGC/GTC | Gly/Val | 7 | ||
| 32 | 220.TAT/TGT | Tyr/Cys | 6 | 68 | – | ||
| 33 | 245.GGC/AGC | Gly/Ser | 7 | 69 | 220.TAT/AAT | Tyr/Asn | 6 |
| 34 | 135. ‐A‐insert | Early stop | 5 | 70 | – | ||
| 35 | 179.CAT/TAT | His/Tyr | 5 | 71 | 179.CAT/CGT | His/Arg | 5 |
| 36 | – |
Del, deletion; Ins, insertion; ND, not done.
We examined the relationship between p53 mutation and clinicopathological variables. Although a statistically significant relationship was not detected between p53 mutation status and any of the clinicopathological variables, a tendency of a higher rate of p53 mutation was shown in cases with smoking and previous cancer history (Table 4; P = 0.14, P = 0.12, respectively). We also did not find a statistically significant difference between p53 mutation ratio and primary tumor location. However, there was a tendency of lower rate of p53 mutation in oral cavity tumors compared with tumors from other locations. In this sense, the mutation ratio of p53 in maxillary cases, which may show different clinical and biological behavior, was similar to the mutation percentage of larynx, hypopharynx, and oropharynx tumors.
Table 4.
Relationship between ING3 mRNA expression and clinicopathological characteristics
| Characteristic | ING3 mRNA expression | p53 status | ||||
|---|---|---|---|---|---|---|
| Low (%) n = 37 (52.1) | Normal to high (%) n = 34 (47.9) | P‐value | Mutant (%) n = 28 (41.0) | Wild type (%) n = 41 (59.0) | P‐value | |
| Predictor | ||||||
| Sex ‡ | ||||||
| Male | 32 (86.5) | 29 (85.3) | 0.88 | 25 (89.3) | 35 (85.4) | 0.44 |
| Female | 5 (13.5) | 5 (14.7) | 3 (10.7) | 6 (14.6) | ||
| Age § | ||||||
| Mean ± SD (years) | 64.70 ± 8.9 | 64.0 ± 8.6 | 0.73 | 64.7 ± 7.5 | 64.3 ± 9.5 | 0.86 |
| Smoking ‡ | ||||||
| Yes | 29 (78.4) | 23 (67.6) | 0.31 | 23 (82.1) | 27 (65.9) | 0.14 |
| No | 8 (21.6) | 11 (32.4) | 5 (17.9) | 14 (34.1) | ||
| Alcohol consumption ‡ , ¶ | ||||||
| Yes | 22 (61.1) | 18 (54.5) | 0.58 | 17 (63.0) | 21 (52.5) | 0.40 |
| No | 14 (38.9) | 15 (45.5) | 10 (37.0) | 19 (47.5) | ||
| TNM stage † , ‡ | ||||||
| Early stage (I–II) | 7 (18.9) | 11 (32.4) | 0.19 | 6 (21.4) | 11 (26.8) | 0.61 |
| Late stage (III–IV) | 30 (81.1) | 23 (67.6) | 22 (78.6) | 30 (73.2) | ||
| Tumor stage † , ‡ | ||||||
| Early T (T1–T2) | 12 (32.4) | 17 (50.0) | 0.13 | 11 (39.3) | 16 (39.0) | 0.98 |
| Late T (T3–T4) | 25 (67.6) | 17 (50.0) | 17 (60.7) | 25 (61.0) | ||
| Nodal stage ‡ | ||||||
| N (0) | 16 (43.2) | 16 (47.1) | 0.75 | 11 (39.3) | 20 (48.8) | 0.44 |
| N (+) | 21 (56.8) | 18 (52.9) | 17 (60.7) | 21 (51.2) | ||
| p53 status ‡ | ||||||
| Wild type | 22 (61.8) | 19 (55.9) | 0.55 | – | – | |
| Mutated | 13 (38.2) | 15 (44.1) | – | – | ||
| Differentiation ‡ , ¶ | ||||||
| Well | 14 (37.8) | 6 (19.4) | 0.10 | 6 (23.1) | 14 (34.1) | 0.33 |
| Moderate to poor | 23 (62.2) | 25 (80.6) | 20 (76.9) | 27 (65.9) | ||
| Radiation therapy ‡ | ||||||
| Performed | 18 (48.6) | 19 (55.9) | 0.54 | 14 (50.0) | 19 (46.3) | 0.76 |
| Not performed | 19 (51.4) | 15 (47.1) | 14 (50.0) | 22 (53.7) | ||
| Chemotherapy ‡ | ||||||
| Performed | 14 (37.8) | 11 (32.4) | 0.63 | 10 (35.7) | 14 (34.1) | 0.89 |
| Not performed | 23 (62.2) | 23 (67.6) | 18 (64.3) | 27 (65.9) | ||
| Previous cancer history ‡ , ¶ | ||||||
| Exist | 5 (14.3) | 7 (22.6) | 0.38 | 7 (25.9) | 4 (10.8) | 0.12 |
| Not exist | 30 (85.7) | 24 (77.4) | 20 (74.1) | 33 (89.2) | ||
| Location ‡ | ||||||
| Oral cavity | 17 (46.0) | 14 (41.2) | 0.90 | 8 (28.5) | 21 (51.2) | 0.40 |
| Oropharynx | 6 (16.2) | 3 (08.8) | 4 (14.3) | 5 (12.2) | ||
| Larynx | 6 (16.2) | 7 (20.6) | 7 (25.0) | 6 (14.6) | ||
| Maxilla | 3 (8.1) | 4 (11.8) | 4 (14.3) | 3 (7.4) | ||
| Hypopharynx | 5 (13.5) | 6 (17.6) | 5 (17.9) | 6 (14.6) | ||
According to the International Union Against Cancer 1997 TNM classification system.
χ2‐test.
Student's t‐test.
¶ The cases with unknown situation for alcohol consumption (2), p53 mutation status (2), differentiation (3), and previous cancer history (5) were not included for evaluation.
Relationship between ING3 mRNA expression and clinicopathological factors. We examined the relationship between the level of ING3 mRNA expression and clinicopathological factors (Table 4). For statistical evaluation, tumor (T) stages and tumor–node–metastasis (TNM) stages were classified as early (T1 and T2; stage I and II) or late (T3 and T4; stage III and IV), nodal (N) stages were classified as N0 (without metastasis) or N+ (with metastasis). No correlation in terms of sex or age was displayed between the patients with low and normal to high ING3 mRNA expression.
With regard to smoking status, although no significant relationship was detected between smoking history and ING3 expression, a tendency of decreased ING3 expression was shown in the smoking group. Over 78% of the cases with low ING3 expression had smoked, whereas approximately 68% of the cases with normal to high ING3 expression were smokers. In other words, 56% (29/52) of the cases with smoking had ING3 mRNA downregulation, whereas 42% (8/19) of the cases without smoking showed decreased ING3 mRNA expression. Regarding alcohol consumption, no significant relationship was found with ING3 mRNA level.
A near‐significant relationship was shown between ING3 mRNA expression status and tumor differentiation or tumor size (T) (Table 4; P = 0.10, P = 0.13, respectively). When we classified them into two groups of well‐differentiated and moderately or poorly differentiated tumors, 70% (14/20) of well‐differentiated tumors had lower ING3 expression, whereas only 48% (23/48) of moderately or poorly differentiated tumors were related with decreased ING3 expression (Table 4; P = 0.10). With regard to T stage, 68% (25/37) of the tumors with low expression were at advanced stage, whereas only half of the tumor samples without ING3 downregulation were at late stage. In other words, 41% (12/29) of early stage tumors were related with decreased ING3 expression, whereas this value was 60% (25/42) in advanced tumors (Table 4; P = 0.13). Although less prominent and not statistically significant, we also detected a similar relationship between ING3 expression status and TNM stage, with a tendency for lower ING3 expression in advanced tumors. Thirty‐nine percent (7/18) of early TNM‐stage tumors demonstrated low ING3 expression, whereas downregulation of ING3 was detected in 60% (30/53) of late TNM‐stage tumors (Table 4; P = 0.19).
However, no significant relationship between ING3 mRNA expression status and any of the applications of chemotherapy or radiotherapy, absence or presence of metastatic lymph nodes, existence or absence of previous cancer history, or p53 mutation status was found (Table 4).
ING3 mRNA expression and survival analysis. Both overall survival (OS) and DFS were determined. All patients were enrolled in a follow‐up program. The follow‐up time was between 2 and 129 months. Valid follow‐up data were available for 67 (94%) out of 71 HNSCC patients. Disease relapses (locoregional recurrence or distant metastases) occurred in 44 patients (62%) and death was confirmed in 39 patients (55%). The median OS was 33 months (range 2–129 months), and the median DFS time was 15 months (range 2–120 months). The mean duration of DFS and OS were 30.3 and 42.7 months, respectively. The 5‐year DFS and OS were 30 and 47%, respectively.
Correlations between ING3 mRNA expression level and the patients’ OS and DFS were analyzed using the univariate Kaplan–Meier method. At 5 years follow up, approximately 60% of the patients with normal to high ING3 expression survived, whereas this was 35% in the patients with low ING3 expression. As shown in Fig. 2a, the mean OS in the low‐expression group was 54 ± 8 months (95% confidence interval [CI] 37–70 months) and that in the normal to high‐expression group was 83 ± 10 months (95% CI 63–102 months). The log rank test showed that patients with low ING3 expression had a significantly shorter OS than those with normal to high expression (P = 0.04; Fig. 2a). However, as shown in Fig. 2b, the mean DFS in the low‐expression group was 43 ± 8 months (95% CI 28–58 months), whereas that in the normal to high‐expression group was 49 ± 9 months (95% CI 32–66 months), with no significant difference between the groups (P = 0.64, log rank test). These data suggest that reduced ING3 mRNA expression results in shorter OS, but no difference in DFS of HNSCC patients.
Figure 2.
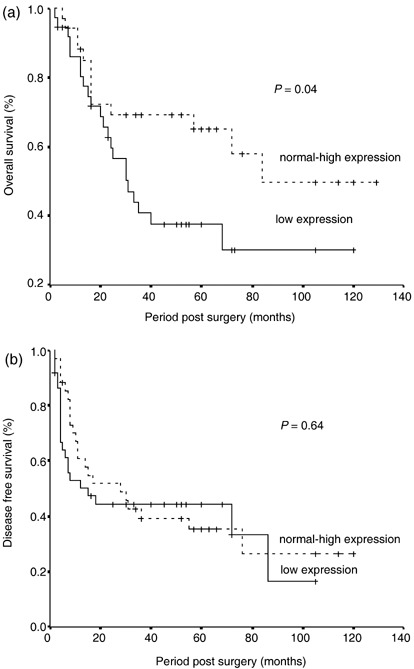
Overall and disease‐free survivals in the groups of head and neck squamous cell carcinoma (HNSCC) patients with decreased and normal to high ING3 expression. Kaplan–Meier survival curves for the total number of cases (n = 71) stratified by ING3 mRNA expression. The cases were divided into the low‐expression group and the normal to high‐expression group. Statistical significance was defined as P < 0.05. (a) The mean overall survival in the low‐expression group was 54 ± 8 months (95% confidence interval [CI] 37–70 months) and that in the normal to high‐expression group was 83 ± 10 months (95% CI 63–102 months) (P‐value for log rank test = 0.04). (b) The mean disease‐free survival in the low‐expression group was 43 ± 8 months (95% CI 28–58 months) and that in the normal to high‐expression group was 49 ± 9 months (95% CI 32–66 months) (P‐value for the log rank test = 0.64).
We also used a multivariate analysis to test the independent value of each parameter predicting OS and DFS. Positive node status (N), advanced tumor status (T), and decreased ING3 expression were independent prognostic factors for poor OS (Table 5). In contrast, positive node status (N), advanced tumor status (T) and TNM stage but decreased ING3 were found to be prognostic factors related with DFS in multivariate analysis (Table 5).
Table 5.
Cox proportional hazard model for survival analysis
| Variable | Disease‐free survival | Overall survival | ||||||
|---|---|---|---|---|---|---|---|---|
| P‐value | 95% CI | P‐value | 95% CI | |||||
| RR | Lower | Upper | RR | Lower | Upper | |||
| Age | 0.084 | 0.963 | 0.922 | 1.005 | 0.069 | 0.956 | 0.912 | 1.003 |
| Smoking | 0.328 | 0.605 | 0.222 | 1.654 | 0.863 | 1.121 | 0.306 | 4.097 |
| Alcohol | 0.613 | 0.800 | 0.337 | 1.901 | 0.746 | 0.839 | 0.290 | 2.427 |
| T stage | 0.009 | 4.927 | 1.500 | 16.185 | 0.020 | 3.757 | 1.231 | 11.470 |
| N stage | 0.012 | 6.078 | 1.480 | 24.962 | 0.002 | 10.492 | 2.320 | 47.440 |
| TNM stage | 0.009 | 0.081 | 0.012 | 0.537 | 0.118 | 0.199 | 0.026 | 1.507 |
| Radiotherapy | 0.911 | 0.952 | 0.405 | 2.239 | 0.111 | 0.449 | 0.167 | 1.203 |
| Chemotherapy | 0.364 | 1.506 | 0.622 | 3.649 | 0.138 | 2.013 | 0.799 | 5.069 |
| Cancer history | 0.315 | 0.574 | 0.195 | 1.695 | 0.190 | 0.441 | 0.130 | 1.500 |
| p53 mutation | 0.472 | 0.765 | 0.369 | 1.588 | 0.947 | 1.028 | 0.458 | 2.308 |
| ING3 expression | 0.551 | 0.800 | 0.384 | 1.665 | 0.034 | 0.395 | 0.168 | 0.931 |
| Localization | 0.658 | 1.099 | 0.723 | 1.672 | 0.887 | 0.966 | 0.598 | 1.561 |
CI, confidence interval; RR, risk ratio. Values in bold are statistically significant.
Relationship between p53 mutation and clinicopathological factors. The frequency of mutations did not correlate with age, sex, smoking, clinical stage, or degree of differentiation. Similarly, mutation status did not display a significant association with OS or DFS (data not shown). However, when we examined the OS and DFS between the combinations of decreased or normal to high ING3 expression and the presence or absence of p53 mutation, the worst OS was detected with the combination of decreased ING3 mRNA expression and the presence of p53 mutation, followed by the groups with low ING3 expression and the absence of p53 mutation (Fig. 3a). Moreover, when the p53 mutation was coassociated with low ING3 expression, a near‐significant relationship was detected compared with the group in which either p53 mutation or downregulation of ING3 was absent (Fig. 3b).
Figure 3.
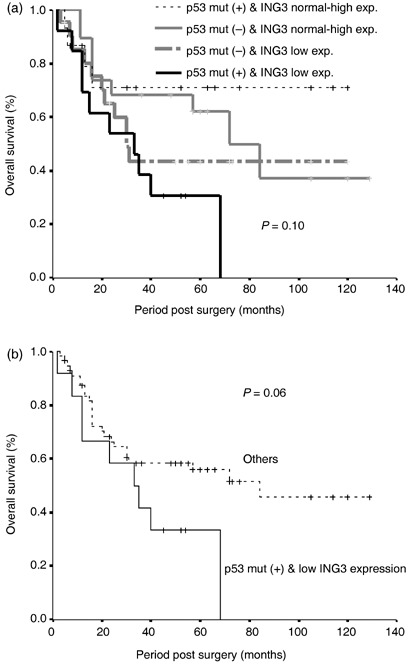
Relationship between p53 mutation status and ING3 mRNA expression. (a) Overall survival for the combinations of decreased or normal to high ING3 expression and the presence or absence of p53 mutation. (b) Overall survival for the combinations of p53 mutation positive, low ING3 expression, and others (absence of one of these variables).
Discussion
Head and neck squamous cell carcinoma is a common cancer worldwide. Despite aggressive and multiple treatment methods including chemotherapy, radiotherapy, and surgery, limited improvement in terms of survival has been obtained. Locoregional recurrence, lymph‐node and distant metastasis, and secondary primary tumors are the major reasons for treatment failure. Moreover, the behavior of the tumors in different locations and even in the same location shows a wide range of clinical features. Some of the tumors may have a better character with good chemoradiotherapy response, and less metastasis and invasion. However, some others may show a highly aggressive phenotype with early metastasis and poor prognosis. So far, the prognostic factors that are considered routinely when deciding on treatment are host and tumor factors such as TNM staging, site of the primary tumor, the presence or absence of nodal and distant metastasis, and pathological grading of differentiation. However, the behavior of tumors is highly variable. Therefore, new predictive markers should be identified both for diagnosis and treatment strategies. Recent developments in human genetics and molecular biology have improved our understanding of the biological basis of tumor development, progression, and metastasis. These studies have included molecules that are altered during development or progression of tumors, such as deletions at TSG loci, amplification of oncogene locations, and various mutations of these and other DNA repair genes.
A novel TSG, ING1, has been cloned recently using a new strategy based on the expression of transforming genetic suppressor elements, which promote transformation presumably by suppressing TSG.( 15 ) We characterized the genomic structure of the human ING1 gene and identified its tumor suppressor function for the first time by showing its chromosomal deletion at the 13q34 region and tumor‐specific mutations in HNSCC.( 5 ) As the next step in our studies on ING families, we identified a novel member of the ING family, ING3.( 6 ) Our efforts and studies from different laboratories led to the identification of new members of this family. Currently, five members of the ING family have been identified and all contain a highly conserved plant homeodomain finger motif in the C‐terminal end of the proteins. Although the exact functions of the ING genes have not been clarified, the gene products are involved in transcriptional regulation, apoptosis, the cell cycle, angiogenesis, and DNA repair through p53‐dependent and ‐independent pathways, and form complexes with histone acetyltransferases and histone deacetylases.( 16 , 17 )
Only a few studies have analyzed the mRNA expression status of ING family genes in various human cancers. Toyama et al. detected 2–10‐fold decreases in ING1 mRNA expression in 44% of breast cancer and in all of 10 breast cancer cell lines examined.( 18 ) Interestingly, the majority of breast cancers showing decreased ING1 expression had metastasized to regional lymph nodes, whereas only a small subset of cancers with elevated ING1 expression compared to adjacent normal tissues were metastatic.( 18 ) Other studies have also demonstrated downregulation of ING1 mRNA in various cancer types, including lymphoid malignancies, brain tumors, lung cancer, and esophagogastric cancer, though no comprehensive clinical correlation was carried out.( 16 , 17 ) Our splice variant‐specific mRNA expression of ING1 showed a prominent decrease in one of the splicing variants (Gunduz et al., unpublished data, 2007).
Regarding the analysis of ING3 mRNA expression, no other study has examined it and compared it with clinicopathological factors, except our previous RT‐PCR study in a limited number of HNSCC( 6 ) and a northern blot analysis reporting the ubiquitous tissue expression of ING3.( 19 ) This latter study also revealed that ING3 activates p53‐transactivated promoters such as p21 and Bax, suggesting its close relationship with a p53‐dependent pathway. In the current study, we analyzed the mRNA expression status of ING3 and compared it with the clinicopathological characteristics of a large population of HNSCC patients. Although no significant relationship was detected between ING3 expression status and most clinical markers, including sex, age, smoking and alcohol consumption, absence or presence of lymph‐node metastasis, application of chemotherapy or radiotherapy, and presence or absence of previous cancer history, downregulation of ING3 was more evident in late‐stage tumors and well‐differentiated cases compared with early stage patients and moderately or poorly differentiated samples, respectively. However, the OS of patients with low ING3 expression was significantly decreased compared with the patients with normal to high ING3 expression. However, no such relationship was detected in terms of DFS. Although we don't know the exact reason for the positive relationship between low ING3 expression and OS but the absence of this relationship for DFS, other genetic and non‐genetic factors could be considered to contribute to the factors related to DFS, such as local recurrences, metastasis, and aggressive phenotype.
The p53 gene, a well‐studied marker, was reported to be a predictive and prognostic factor in HNSCC. In one of these reports, p53 mutation at the margin that was surgically and histopathologically clean revealed much higher recurrence compared to samples without p53 alteration.( 12 ) In another report, detection of p53 mutation status predicted tumor response to chemotherapeutic agents.( 14 ) In the current study, we could not find a relationship between the mutation status of p53 and ING3 mRNA expression. Similarly, p53 mutation status alone did not give significant survival changes either for DFS or OS. However, when we looked at the association between ING3 expression and p53 mutation status, worst survival was obtained in the group with low ING3 expression with p53 mutation. This result is likely to come from the relationship between ING3 downregulation and a weak contribution of p53 mutation status. Because ING3 also had p53‐independent pathways for its action, these pathways could be more important. However, conflicting results have been published for the cellular functional effect of p53 mutation. Though most cases with mutation would show inactivation of p53, it is also possible that some mutation types could promote its tumor suppressive or apoptotic function through gain of function.( 20 ) Thus, characterization of each mutation type could give a better evaluation of its cellular effects and relationship with ING3. Additional genetic changes could also be responsible in HNSCC and a combination of these alterations could serve as better prognostic markers. There have already been reports of prognostic markers in HNSCC. For example, allelic deletion at the 8p21 and 9p21 regions correlates with recurrence and bad prognosis in HNSCC.( 11 , 13 ) Furthermore, the relationship between clinicopathological factors such as stage, differentiation, metastasis, recurrence or survival, and LOH at various loci or alterations of different genes such as p16, p53, fragile histidine triad gene (FHIT), and cyclin D1 have been reported in HNSCC.( 10 , 11 , 12 , 13 , 14 )
In conclusion, our current study has confirmed a reduction of ING3 at the mRNA level in HNSCC and demonstrated an inverse relationship between ING3 mRNA expression and prognostic index. These results indicate ING3 as a potential tumor suppressor molecule and that low levels of ING3 may indicate an aggressive nature of HNSCC. Comparative analysis of other genes, including various ING family members, in subsequent studies would give rise to more valuable and clinically applicable prognostic factors.
Acknowledgments
This work was partially supported by grants‐in‐aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology (19592109 to H. N., 18‐06262 to E. G., and 17406027 to N. N.), Seed Innovation Research from Japan Science and Technology Agency (to M. G.), Sumitomo Trust Haraguchi Memorial Cancer Research Promotion (to M. G.), and Astrazeneca Research Grant (to M. G.).
References
- 1. Ginos MA, Page GP, Michalowicz BS et al . Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res 2004; 64: 55–63. [DOI] [PubMed] [Google Scholar]
- 2. Freier K, Joos S, Flechtenmacher C et al . Tissue microarray analysis reveals site‐specific prevalence of oncogene amplifications in head and neck squamous cell carcinoma. Cancer Res 2003; 63: 1179–82. [PubMed] [Google Scholar]
- 3. Willmore‐Payne C, Holden JA, Layfield LJ. Detection of EGFR‐ and HER2‐activating mutations in squamous cell carcinoma involving the head and neck. Mod Pathol 2006; 19: 634–40. [DOI] [PubMed] [Google Scholar]
- 4. Reed AL, Califano J, Cairns P et al . High frequency of p16 (CDKN2/MTS‐1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res 1996; 56: 3630–3. [PubMed] [Google Scholar]
- 5. Gunduz M, Ouchida M, Fukushima K et al . Genomic structure of the human ING1 gene and tumor‐specific mutations detected in head and neck squamous cell carcinomas. Cancer Res 2000; 60: 3143–6. [PubMed] [Google Scholar]
- 6. Gunduz M, Ouchida M, Fukushima K et al . Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene 2002; 21: 4462–70. [DOI] [PubMed] [Google Scholar]
- 7. Gunduz M, Nagatsuka H, Demircan K et al . Frequent deletion and down‐regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene 2005; 356: 109–17. [DOI] [PubMed] [Google Scholar]
- 8. Lalla RV, Boisoneau DS, Spiro JD, Kreutzer DL. Expression of vascular endothelial growth factor receptors on tumor cells in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 2003; 129: 882–8. [DOI] [PubMed] [Google Scholar]
- 9. Beder LB, Gunduz M, Ouchida M et al . Genome‐wide analyses on loss of heterozygosity in head and neck squamous cell carcinomas. Laboratory Invest 2003; 83: 99–105. [DOI] [PubMed] [Google Scholar]
- 10. Cruz IB, Snijders PJ, Meijer CJ et al . p53 expression above the basal cell layer in oral mucosa is an early event of malignant transformation and has predictive value for developing oral squamous cell carcinoma. J Pathol 1998; 184: 360–8. [DOI] [PubMed] [Google Scholar]
- 11. Lydiatt WM, Davidson BJ, Schantz SP, Caruana S, Chaganti RS. 9p21 deletion correlates with recurrence in head and neck cancer. Head Neck 1998; 20: 113–18. [DOI] [PubMed] [Google Scholar]
- 12. Van Houten VM, Leemans CR, Kummer JA et al . Molecular diagnosis of surgical margins and local recurrence in head and neck cancer patients: a prospective study. Clin Cancer Res 2004; 10: 3614–20. [DOI] [PubMed] [Google Scholar]
- 13. Coon SW, Savera AT, Zarbo RJ et al . Prognostic implications of loss of heterozygosity at 8p21 and 9p21 in head and neck squamous cell carcinoma. Int J Cancer 2004; 111: 206–12. [DOI] [PubMed] [Google Scholar]
- 14. Cabelguenne A, Blons H, De Waziers I et al . p53 alterations predict tumor response to neoadjuvant chemotherapy in head and neck squamous cell carcinoma: a prospective series. J Clin Oncol 2000; 18: 1465–73. [DOI] [PubMed] [Google Scholar]
- 15. Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33 ING1 promotes neoplastic transformation. Nat Genet 1996; 14: 415–20. [DOI] [PubMed] [Google Scholar]
- 16. Campos EI, Chin MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol Life Sci 2004; 61: 2597–613. [DOI] [PubMed] [Google Scholar]
- 17. Gunduz M. Functions of the tumor suppressor ING family genes. J Oral Biosci 2005; 47: 211–20. [Google Scholar]
- 18. Toyama T, Iwase H, Watson P et al . Suppression of ING1 expression in sporadic breast cancer. Oncogene 1999; 18: 5187–93. [DOI] [PubMed] [Google Scholar]
- 19. Nagashima M, Shiseki M, Pedeux RM et al . A novel PHD‐finger motif protein, p47ING3, modulates p53‐mediated transcription, cell cycle control, and apoptosis. Oncogene 2003; 22: 343–50. [DOI] [PubMed] [Google Scholar]
- 20. Koonin EV, Rogozin IB, Glazko GV. p53 gain‐of‐function: tumor biology and bioinformatics come together. Cell Cycle 2005; 4: 686–8. [DOI] [PubMed] [Google Scholar]


