Abstract
Despite long‐term clinical experience with docetaxel, unpredictable severe adverse reactions remain an important determinant for limiting the use of the drug. To identify a genetic factor(s) determining the risk of docetaxel‐induced leukopenia/neutropenia, we selected subjects who received docetaxel chemotherapy from samples recruited at BioBank Japan, and conducted a case–control association study. We genotyped 84 patients, 28 patients with grade 3 or 4 leukopenia/neutropenia, and 56 with no toxicity (patients with grade 1 or 2 were excluded), for a total of 79 single nucleotide polymorphisms (SNPs) in seven genes possibly involved in the metabolism or transport of this drug: CYP3A4, CYP3A5, ABCB1, ABCC2, SLCO1B3, NR1I2, and NR1I3. Since one SNP in ABCB1, four SNPs in ABCC2, four SNPs in SLCO1B3, and one SNP in NR1I2 showed a possible association with the grade 3 leukopenia/neutropenia (P‐value of <0.05), we further examined these 10 SNPs using 29 additionally obtained patients, 11 patients with grade 3/4 leukopenia/neutropenia, and 18 with no toxicity. The combined analysis indicated a significant association of rs12762549 in ABCC2 (P = 0.00022) and rs11045585 in SLCO1B3 (P = 0.00017) with docetaxel‐induced leukopenia/neutropenia. When patients were classified into three groups by the scoring system based on the genotypes of these two SNPs, patients with a score of 1 or 2 were shown to have a significantly higher risk of docetaxel‐induced leukopenia/neutropenia as compared to those with a score of 0 (P = 0.0000057; odds ratio [OR], 7.00; 95% CI [confidence interval], 2.95–16.59). This prediction system correctly classified 69.2% of severe leukopenia/neutropenia and 75.7% of non‐leukopenia/neutropenia into the respective categories, indicating that SNPs in ABCC2 and SLCO1B3 may predict the risk of leukopenia/neutropenia induced by docetaxel chemotherapy. (Cancer Sci 2008; 99: 967–972)
Docetaxel is a semisynthetic taxane derived from an extract of the needles of European yew tree (Taxus baccata) and was approved for treatment of patients with refractory breast cancer, non‐small‐cell lung cancer, ovarian cancer, and head and neck cancer.( 1 , 2 ) Dose‐limiting toxicities of docetaxel are known to be neutropenia, anemia, asthenia, skin toxicity, and nausea. Among these severe adverse reactions, neutropenia is observed most often at a frequency of ~36%.( 3 , 4 ) One of the important factors that causes the limitation of docetaxel use is the unpredictable interindividual variation in toxicity. Potential causes for such variability include the pathogenesis and severity of the disease being treated, the occurrence of unintended drug interactions, and the impairment of hepatic and/or renal functions.( 5 )
Area under the curves (AUCs) of unbound and total docetaxel have been reported to be correlated to the degree of decrements in absolute neutrophil count, as well as development of the worst grade of neutropenia, suggesting that AUC is one of the determinants of docetaxel toxicity.( 6 ) However, AUC also varied among patients even when doses were adjusted by body‐surface area.( 7 ) These results suggest that genetic factors which affect docetaxel metabolism and clearance might change the AUC of docetaxel, and result in the interindividual variation of toxicity.
Docetaxel is predominantly cleared by hepatic disposition in humans; ~5% of docetaxel are excreted in urine and ~75% in bile. CYP3A4 and CYP3A5 are primary enzymes involved in the hepatic oxidation of docetaxel.( 8 ) The metabolites have significantly low cytotoxic activity as compared with the parent drug, making biotransformation by CYP3As a major route of inactivation.( 9 ) The elimination pathway is reported to be mainly mediated by the drug efflux ABC transporter, ABCB1 (also known as P‐glycoprotein or MDR1).( 10 ) Moreover, the expression of these genes is regulated by the nuclear hormone receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR), which are encoded by NR1I2 and NR1I3.( 11 , 12 , 13 , 14 ) It has also been reported that other transporters, ABCC2 (also known as MRP2) and SLCO1B3 (also known as OTAP1B3 or OATP8), were responsible for the efflux and uptake of docetaxel in an in vitro study.( 15 , 16 ) Therefore, these seven genes are considered candidates that may affect docetaxel toxicity. However, there have been few reports on the relationship between the toxicity of docetaxel and genetic variants for CYP3A4, CYP3A5, and ABCB1, and no reports on NR1I2, NR1I3, ABCC2, and SLCO1B3.
In the present study, we selected these seven candidate genes possibly altering docetaxel AUC described above and performed a case–control association study. We found that single nucleotide polymorphisms (SNPs) in two genes, ABCC2 and SLCO1B3, were significantly associated with severe leukopenia/neutropenia observed in patients treated with docetaxel. We also established a scoring system, which may be applicable for prediction of the docetaxel‐induced toxicity, on the basis of the genotypes of these two genes.
Materials and Methods
Subjects. BioBank Japan project (http://biobankjp.org/) was begun in 2003 for the collection of genomic DNA, serum, and clinical information from 270 000 cases diagnosed with either of 47 diseases by a collaboration network of 66 hospitals in Japan. Patients gave written informed consent to participate in this project before information was collected. Clinical information was collected by a questionnaire which also looked at drug use and adverse drug reactions (ADRs). From the registered samples in BioBank Japan, we selected patients who were treated with docetaxel. We defined an ADR group as patients reported to have shown grade 3 or 4 leukopenia/neutropenia and a non‐ADR group as patients reported to have shown no toxicity during docetaxel chemotherapy. The grade of toxicity was classified in accordance with the National Cancer Institute – Common Toxicity Criteria version 2.0.
From June 2003 to March 2006, samples from 253 patients who received the docetaxel treatment were collected in BioBank Japan, and of these, 84 patients classified into the ADR or non‐ADR group were used for a case–control association study (1st set). The remaining 169 patients had shown grade 1/2 leukopenia/neutropenia and/or other toxicities, including nausea, vomiting, anorexia, and diarrhea. During the period between April and December 2006, BioBank Japan additionally obtained samples from 84 patients who were treated with docetaxel, and we selected 29 patients belonging to either group as the 2nd set of samples. In 113 patients in the 1st and 2nd sets, the numbers of patients with lung, breast, esophageal, and other cancers were 32 (28.3%), 38 (33.6%), 11 (9.7%), and 32 (28.3%), respectively. This project was approved by the ethical committees at the SNP Research Center, The Institutes of Physical and Chemical Research (RIKEN), Yokohama, Japan, and The Institute of Medical Science, The University of Tokyo, Tokyo, Japan.
Selection of SNPs and genotyping. We selected the following seven genes as candidates: CYP3A4, CYP3A5, ABCB1, ABCC2, SLCO1B3, NR1I2, and NR1I3, and used haplotype‐tagging SNPs (htSNPs) in ABCB1, ABCC2, SLCO1B3, NR1I2, and NR1I3 that encompass linkage disequilibrium (LD) blocks of these genes. Using genotype data of the Japanese population in the HapMap database, we selected htSNPs by the following criteria: pairwise tagging, r 2 = 0.8, and minor allele frequency (MAF) of >10%. In addition, we selected possible functional polymorphisms reported previously.( 17 , 18 , 19 , 20 ) In the case of CYP3A4 and CYP3A5, we tested only functional SNPs reported in a Japanese population because of the poor information on htSNPs in these gene loci.( 21 , 22 , 23 ) In total, we selected 69 htSNPs that comprehensively captured common variations based on measures of LD (r 2 ≥ 0.8) and 10 functional polymorphisms that would possibly alter the function of either of the gene products. We genotyped these SNPs using multiplex polymerase chain reaction (PCR)‐based Invader assay or direct sequencing as described previously.( 24 )
Statistical analyses and predictive scoring system for leukopenia/neutropenia. We performed association analysis using Fisher's exact test under allelic, dominant‐inheritance, and recessive‐inheritance models. P‐values were adjusted for multiple testing according to strict Bonferronni's correction. The difference in the distribution of age was assessed by the Mann–Whitney U‐test. For the prediction scoring system of leukopenia/neutropenia induced by docetaxel, we assigned scores of 1 to individuals homozygous for the risk allele, and 0 to individuals with other genotypes for ABCC2 rs12762549. For SLCO1B3 rs11045585, individuals were scored as 1 according to homozygous and heterozygous for the risk allele, or as 0 according to homozygous for the low‐risk allele, respectively. On the basis of this system, each patient was classified into either of three groups (group 0, 1, or 2).
Results
Toxicity of docetaxel. Among the 253 cancer patients who received treatment with docetaxel and were enrolled in the BioBank Japan from June 2003 to March 2006, 28 patients experienced grade 3 or 4 leukopenia/neutropenia (ADR group), 56 patients revealed no adverse events (non‐ADR group), and the remaining patients had the mild leukopenia/neutropenia and/or other ADRs, including nausea, vomiting, anorexia, and diarrhea. We selected first two groups as the 1st set of the association study to screen the genetic factors associated with severe leukopenia/neutropenia. Since 11 patients belonging to the ADR group and 18 patients belonging to the non‐ADR group were obtained later, we used these patients as the 2nd set. No significant difference was observed in the types and incidence of ADRs between the 1st and 2nd patient groups treated with docetaxel (P = 0.82). The distributions of gender (male/female) were 17/21 in the ADR group and 29/45 in the non‐ADR group (P = 0.69). There was no significant difference in the age distribution (median, years [range]) between ADRs and non‐ADRs (59 [29–75]versus 59.5 [36–81], P = 0.31). The patients with or without concomitant anticancer drugs were 15/34 or 24/40 in the ADR/non‐ADR groups, suggesting no significant difference (P = 0.55).
Association study with leukopenia/neutropenia by docetaxel. We genotyped 84 patients (the 1st set) for all SNPs in selected candidate genes (Table 1). The MAF of all four SNPs in CYP3A4 were lower than 0.01. CYP3A4*1B (rs2740574), CYP3A4*4 (I118V), and rs2417886 in SLCO1B3 were found to be monomorphic in this population. One SNP (rs2188524) in ABCB1, four SNPs (rs11190291, rs3740065, rs12762549, and rs11190303) in ABCC2, four SNPs (rs7311358, rs11045585, rs2174012, and rs4149117) in SLCO1B3, and one SNP (rs12633127) in NR1I2 revealed a possible association with P‐values of <0.05 with the grade 3 or 4 leukopenia/neutropenia by docetaxel treatment.
Table 1.
Relationship between single nucleotide polymorphisms (SNPs) in candidate genes and leukopenia/neutropenia risk induced by docetaxel
| Gene | SNP ID | ADR, N = 28 | Non‐ADR, N = 56 | P‐value | Odds ratio (95% CI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 † | 12 | 22 | MAF | 11 † | 12 | 22 | MAF | 1 versus 2 | versus 11 | versus 22 | versus 11 | versus 22 | ||
| CYP3A4 | rs2740574 | 27 | 0 | 0 | 0.00 | 56 | 0 | 0 | 0.00 | NA | NA | NA | NA | NA |
| CYP3A4*4 | 27 | 0 | 0 | 0.00 | 55 | 0 | 0 | 0.00 | NA | NA | NA | NA | NA | |
| rs12721627 | 28 | 0 | 0 | 0.00 | 53 | 1 | 0 | 0.01 | 1.0 | 1.0 | 1.0 | NA | NA | |
| rs28371759 | 26 | 1 | 0 | 0.02 | 54 | 1 | 0 | 0.01 | 1.0 | 1.0 | 1.0 | 2.08 [0.12–34.53] | NA | |
| CYP3A5 | rs776746 | 19 | 7 | 1 | 0.17 | 28 | 26 | 2 | 0.27 | 0.18 | 0.10 | 1.0 | 2.38 [0.89–6.32] | 1.04 [0.09–11.98] |
| ABCB1 | rs2157929 | 20 | 6 | 2 | 0.18 | 43 | 12 | 0 | 0.11 | 0.23 | 0.59 | 0.11 | 1.43 [0.50–4.06] | NA |
| rs1978095 | 10 | 16 | 2 | 0.36 | 33 | 17 | 6 | 0.26 | 0.21 | 0.064 | 0.71 | 2.58 [1.01–6.60] | 1.56 [0.29–8.28] | |
| rs13233308 | 11 | 11 | 5 | 0.39 | 19 | 23 | 14 | 0.46 | 0.50 | 0.63 | 0.59 | 1.34 [0.51–3.45] | 1.47 [0.46–4.60] | |
| rs2188524 | 20 | 7 | 0 | 0.13 | 51 | 3 | 1 | 0.05 | 0.062 | 0.035* | 1.0 | 4.46 [1.18–16.92] | NA | |
| rs3789243 | 7 | 18 | 2 | 0.41 | 24 | 19 | 13 | 0.40 | 1.0 | 0.15 | 0.13 | 2.14 [0.78–5.89] | 3.78 [0.78–18.13] | |
| rs1202172 | 21 | 6 | 0 | 0.11 | 50 | 6 | 0 | 0.05 | 0.21 | 0.19 | 1.0 | 2.38 [0.68–8.24] | NA | |
| rs10256836 | 19 | 7 | 1 | 0.17 | 41 | 12 | 3 | 0.16 | 1.0 | 0.80 | 1.0 | 1.15 [0.41–3.18] | 1.47 [0.14–14.85] | |
| rs868755 | 13 | 12 | 3 | 0.32 | 20 | 23 | 13 | 0.44 | 0.18 | 0.48 | 0.24 | 1.56 [0.61–3.92] | 2.52 [0.65–9.71] | |
| rs1922242 | 14 | 9 | 4 | 0.31 | 23 | 27 | 5 | 0.34 | 0.86 | 0.48 | 0.47 | 1.50 [0.59–3.78] | 1.74 [0.42–7.08] | |
| rs3789246 | 19 | 6 | 2 | 0.19 | 41 | 15 | 0 | 0.13 | 0.49 | 0.80 | 0.10 | 1.15 [0.41–3.18] | NA | |
| rs7787082 | 5 | 17 | 5 | 0.50 | 21 | 28 | 6 | 0.36 | 0.13 | 0.083 | 0.49 | 2.72 [0.87–8.27] | 1.86 [0.51–6.74] | |
| rs2373588 | 8 | 14 | 6 | 0.46 | 26 | 23 | 7 | 0.33 | 0.13 | 0.16 | 0.34 | 2.17 [0.81–5.74] | 1.91 [0.57–6.34] | |
| rs6949448 | 14 | 11 | 3 | 0.30 | 18 | 22 | 14 | 0.46 | 0.065 | 0.16 | 0.15 | 2.00 [0.78–5.08] | 2.92 [0.76–11.18] | |
| rs1002205 | 8 | 15 | 5 | 0.45 | 22 | 28 | 6 | 0.36 | 0.31 | 0.47 | 0.49 | 1.62 [0.60–4.31] | 1.81 [0.50–6.55] | |
| rs1882478 | 9 | 17 | 2 | 0.38 | 16 | 27 | 13 | 0.47 | 0.25 | 0.80 | 0.13 | 1.18 [0.44–3.16] | 3.93 [0.82–18.82] | |
| rs2235046 | 12 | 10 | 6 | 0.39 | 20 | 30 | 6 | 0.38 | 0.87 | 0.64 | 0.32 | 1.35 [0.53–3.41] | 2.27 [0.65–7.84] | |
| rs2235067 | 25 | 3 | 0 | 0.05 | 51 | 5 | 0 | 0.05 | 1.0 | 1.0 | 1.0 | 1.22 [0.27–5.54] | NA | |
| rs1045642 | 12 | 13 | 2 | 0.31 | 18 | 25 | 12 | 0.45 | 0.13 | 0.34 | 0.13 | 1.64 [0.63–4.23] | 3.49 [0.72–16.87] | |
| rs1128503 | 12 | 11 | 5 | 0.38 | 21 | 29 | 6 | 0.37 | 1.0 | 0.81 | 0.49 | 1.25 [0.49–3.15] | 1.81 [0.50–6.55] | |
| rs3213619 | 22 | 5 | 0 | 0.09 | 53 | 3 | 0 | 0.03 | 0.11 | 0.11 | 1.0 | 4.02 [0.88–18.27] | NA | |
| ABCC2 | rs12268782 | 18 | 9 | 1 | 0.20 | 45 | 11 | 0 | 0.10 | 0.091 | 0.18 | 0.33 | 2.27 [0.82–6.28] | NA |
| rs2804398 | 24 | 4 | 0 | 0.07 | 43 | 12 | 0 | 0.11 | 0.58 | 0.56 | 1.0 | 1.67 [0.48–5.77] | NA | |
| rs2756109 | 12 | 14 | 2 | 0.32 | 29 | 23 | 3 | 0.26 | 0.47 | 0.49 | 1.0 | 1.49 [0.59–3.72] | 1.33 [0.20–8.48] | |
| rs11190291 | 20 | 7 | 1 | 0.16 | 50 | 6 | 0 | 0.05 | 0.041* | 0.060 | 0.33 | 3.33 [1.03–10.83] | NA | |
| rs2002042 | 11 | 13 | 4 | 0.38 | 31 | 19 | 6 | 0.28 | 0.22 | 0.25 | 0.73 | 1.92 [0.76–4.83] | 1.39 [0.35–5.39] | |
| rs3740065 | 15 | 9 | 4 | 0.30 | 14 | 30 | 11 | 0.47 | 0.046* | 0.015* | 0.57 | 3.38 [1.30–8.82] | 1.50 [0.43–5.22] | |
| rs12762549 | 7 | 12 | 9 | 0.54 | 28 | 23 | 5 | 0.30 | 0.0038* | 0.036* | 0.012* | 3.00 [1.10–8.18] | 4.83 [1.44–16.26] | |
| rs2862691 | 18 | 10 | 0 | 0.18 | 34 | 19 | 3 | 0.22 | 0.55 | 0.82 | 0.55 | 1.16 [0.45–2.98] | NA | |
| rs11598781 | 18 | 10 | 0 | 0.18 | 32 | 22 | 2 | 0.23 | 0.55 | 0.64 | 0.55 | 1.35 [0.52–3.45] | NA | |
| rs11190303 | 17 | 9 | 2 | 0.23 | 15 | 32 | 7 | 0.43 | 0.016* | 0.0047* | 0.49 | 4.02 [1.53–10.54] | 1.94 [0.37–10.01] | |
| rs2273697 | 20 | 7 | 1 | 0.16 | 49 | 7 | 0 | 0.06 | 0.052 | 0.13 | 0.33 | 2.80 [0.89–8.75] | NA | |
| SLCO1B3 | rs7311358 | 17 | 7 | 3 | 0.24 | 21 | 29 | 6 | 0.37 | 0.12 | 0.036* | 1.0 | 2.83 [1.10–7.33] | 1.04 [0.23–4.53] |
| rs4149118 | 7 | 14 | 7 | 0.50 | 20 | 25 | 11 | 0.42 | 0.41 | 0.46 | 0.78 | 1.67 [0.60–4.60] | 1.36 [0.46–4.02] | |
| rs919840 | 18 | 3 | 0 | 0.07 | 35 | 12 | 1 | 0.15 | 0.27 | 0.36 | 1.0 | 2.23 [0.56–8.84] | NA | |
| rs3764006 | 12 | 15 | 0 | 0.28 | 35 | 18 | 2 | 0.20 | 0.32 | 0.15 | 0.55 | 2.19 [0.85–5.58] | NA | |
| rs11045585 | 15 | 13 | 0 | 0.23 | 49 | 7 | 0 | 0.06 | 0.0023* | 0.0010* | 1.0 | 6.07 [2.05–17.97] | NA | |
| rs10841661 | 18 | 8 | 1 | 0.19 | 33 | 15 | 8 | 0.28 | 0.25 | 0.63 | 0.26 | 1.39 [0.53–3.64] | 4.33 [0.51–36.57] | |
| rs3764009 | 20 | 4 | 3 | 0.19 | 29 | 21 | 5 | 0.28 | 0.25 | 0.093 | 1.0 | 2.56 [0.93–7.04] | 1.25 [0.27–5.67] | |
| rs2117032 | 10 | 12 | 5 | 0.41 | 21 | 25 | 10 | 0.40 | 1.0 | 1.0 | 1.0 | 1.02 [0.39–2.64] | 1.05 [0.31–3.43] | |
| rs980084 | 9 | 16 | 2 | 0.37 | 21 | 26 | 8 | 0.38 | 1.0 | 0.81 | 0.49 | 1.24 [0.46–3.25] | 2.13 [0.41–10.79] | |
| rs2417886 | 28 | 0 | 0 | 0.00 | 56 | 0 | 0 | 0.00 | NA | NA | NA | NA | NA | |
| rs7977213 | 17 | 10 | 0 | 0.19 | 34 | 14 | 5 | 0.23 | 0.68 | 1.0 | 0.16 | 1.05 [0.40–2.75] | NA | |
| rs4149154 | 22 | 3 | 2 | 0.13 | 34 | 19 | 3 | 0.22 | 0.21 | 0.080 | 1.0 | 2.85 [0.93–8.63] | 1.41 [0.22–9.00] | |
| rs12228798 | 19 | 8 | 0 | 0.15 | 39 | 15 | 2 | 0.17 | 0.83 | 1.0 | 0.56 | 1.04 [0.37–2.82] | NA | |
| rs7312051 | 22 | 4 | 0 | 0.08 | 41 | 12 | 1 | 0.13 | 0.43 | 0.41 | 1.0 | 1.74 [0.50–5.99] | NA | |
| rs10841714 | 21 | 7 | 0 | 0.13 | 38 | 17 | 1 | 0.17 | 0.51 | 0.62 | 1.0 | 1.42 [0.51–3.95] | NA | |
| rs4424716 | 23 | 4 | 0 | 0.07 | 44 | 10 | 1 | 0.11 | 0.58 | 0.76 | 1.0 | 1.44 [0.41–5.02] | NA | |
| rs10734711 | 13 | 10 | 3 | 0.31 | 31 | 17 | 4 | 0.24 | 0.44 | 0.47 | 0.68 | 1.48 [0.57–3.81] | 1.57 [0.32–7.58] | |
| rs11045639 | 7 | 13 | 7 | 0.50 | 22 | 27 | 7 | 0.37 | 0.13 | 0.33 | 0.21 | 1.85 [0.67–5.10] | 2.45 [0.76–7.89] | |
| rs2174012 | 17 | 10 | 0 | 0.19 | 15 | 33 | 7 | 0.43 | 0.0028* | 0.0034* | 0.089 | 4.53 [1.70–12.09] | NA | |
| rs2900459 | 12 | 11 | 5 | 0.38 | 13 | 32 | 11 | 0.48 | 0.25 | 0.079 | 1.0 | 2.48 [0.93–6.56] | 1.12 [0.34–3.62] | |
| rs4762693 | 7 | 13 | 7 | 0.50 | 20 | 25 | 7 | 0.38 | 0.17 | 0.32 | 0.22 | 1.79 [0.63–4.98] | 2.25 [0.69–7.27] | |
| rs7975631 | 22 | 4 | 0 | 0.08 | 45 | 10 | 1 | 0.11 | 0.59 | 0.77 | 1.0 | 1.34 [0.38–4.71] | NA | |
| rs4149117 | 18 | 7 | 3 | 0.23 | 21 | 29 | 6 | 0.37 | 0.11 | 0.036* | 1.0 | 3.00 [1.17–7.71] | 1.00 [0.23–4.34] | |
| NR1I2 | rs12488820 | 15 | 11 | 1 | 0.24 | 30 | 23 | 3 | 0.26 | 0.85 | 1.0 | 1.0 | 1.08 [0.43–2.73] | 1.47 [0.14–14.85] |
| rs4440154 | 11 | 11 | 6 | 0.41 | 25 | 25 | 5 | 0.32 | 0.30 | 0.65 | 0.17 | 1.29 [0.51–3.25] | 2.73 [0.75–9.89] | |
| rs7643645 | 8 | 14 | 5 | 0.44 | 21 | 24 | 9 | 0.39 | 0.61 | 0.47 | 1.0 | 1.51 [0.56–4.07] | 1.14 [0.34–3.80] | |
| rs2461825 | 11 | 13 | 2 | 0.33 | 23 | 26 | 6 | 0.35 | 0.86 | 1.0 | 0.72 | 1.02 [0.39–2.62] | 1.47 [0.27–7.83] | |
| rs2472682 | 9 | 16 | 2 | 0.37 | 16 | 35 | 4 | 0.39 | 0.87 | 0.80 | 1.0 | 1.22 [0.45–3.28] | 1.02 [0.17–5.95] | |
| rs12633127 | 8 | 16 | 3 | 0.41 | 31 | 22 | 1 | 0.22 | 0.017* | 0.033* | 0.11 | 3.20 [1.19–8.59] | 6.63 [0.65–67.01] | |
| rs6784598 | 3 | 18 | 7 | 0.57 | 12 | 33 | 10 | 0.48 | 0.33 | 0.25 | 0.57 | 2.33 [0.59–9.04] | 1.50 [0.50–4.49] | |
| rs9815093 | 14 | 13 | 1 | 0.27 | 29 | 24 | 3 | 0.27 | 1.0 | 1.0 | 1.0 | 1.07 [0.43–2.66] | 1.53 [0.15–15.40] | |
| rs13070374 | 19 | 9 | 0 | 0.16 | 32 | 22 | 2 | 0.23 | 0.32 | 0.48 | 0.55 | 1.58 [0.61–4.11] | NA | |
| rs11712308 | 15 | 11 | 2 | 0.27 | 25 | 24 | 6 | 0.33 | 0.48 | 0.50 | 0.71 | 1.38 [0.55–3.45] | 1.59 [0.29–8.45] | |
| rs2293214 | 18 | 10 | 0 | 0.18 | 39 | 16 | 1 | 0.16 | 0.83 | 0.81 | 1.0 | 1.27 [0.48–3.33] | NA | |
| NR1I3 | rs11432 | 6 | 13 | 5 | 0.48 | 25 | 19 | 11 | 0.37 | 0.22 | 0.13 | 1.0 | 2.50 [0.86–7.26] | 1.05 [0.32–3.45] |
| rs7528588 | 13 | 13 | 2 | 0.30 | 34 | 15 | 7 | 0.26 | 0.58 | 0.25 | 0.71 | 1.78 [0.71–4.46] | 1.86 [0.35–9.59] | |
| rs5085 | 13 | 12 | 2 | 0.30 | 33 | 15 | 8 | 0.28 | 0.86 | 0.48 | 0.49 | 1.55 [0.61–3.89] | 2.08 [0.41–10.56] | |
| rs4233368 | 9 | 14 | 5 | 0.43 | 20 | 26 | 10 | 0.41 | 0.87 | 0.81 | 1.0 | 1.17 [0.44–3.07] | 1.00 [0.30–3.27] | |
| rs2307418 | 27 | 1 | 0 | 0.02 | 45 | 11 | 0 | 0.10 | 0.11 | 0.094 | 1.0 | 6.60 [0.80–54.01] | NA | |
| rs7417867 | 9 | 13 | 6 | 0.45 | 28 | 21 | 7 | 0.31 | 0.12 | 0.16 | 0.34 | 2.11 [0.81–5.46] | 1.91 [0.57–6.34] | |
| rs11584174 | 15 | 11 | 1 | 0.24 | 24 | 24 | 7 | 0.35 | 0.21 | 0.35 | 0.26 | 1.61 [0.63–4.08] | 3.79 [0.44–32.52] | |
| rs2501870 | 16 | 9 | 2 | 0.24 | 28 | 20 | 7 | 0.31 | 0.46 | 0.49 | 0.71 | 1.40 [0.55–3.56] | 1.82 [0.35–9.44] | |
| rs11265572 | 24 | 3 | 0 | 0.06 | 44 | 11 | 0 | 0.10 | 0.39 | 0.37 | 1.0 | 2.00 [0.50–7.87] | NA | |
Major allele in non‐ADRs was defined as allele 1.
P < 0.05.
ADR, adverse drug reaction; CI, confidence interval; NA, not available.
We further genotyped 29 additional samples of the 2nd set using these nine SNPs, except rs4149117 in SLCO1B3 because it revealed absolute LD with rs7311358 (r 2 = 1.0) (Table 2). Only rs12762549 in ABCC2 showed P‐values of less than 0.05 (P = 0.048) in the 2nd set. Because the sample size of the 2nd set was very small, we combined samples of the 1st and 2nd sets (Table 2). Two SNPs were significantly associated with docetaxel‐induced leukopenia/neutropenia after the correction of multiple testing. We observed the most significant association for SNP rs11045585 in SLCO1B3 (P = 0.00017, adjusted P = 0.013; odds ratio [OR], 5.44; 95% CI [confidence interval], 2.22–13.34). rs12762549 in ABCC2 was also showed a significant P‐value of 0.00022 (adjusted P = 0.017; OR, 3.12; 95% CI, 1.27–7.69). For these two SNPs, the OR and direction of risk alleles were maintained in both the 1st and 2nd screenings. In 68 patients with grade 1/2 leukopenia/neutropenia, rs11045585 and rs12762549 showed lower effects than in patients with grade 3/4. The ORs (95% CIs) of rs11045585 and rs12762549 in grade 1/2 patients were 1.35 (0.56–3.27, P = 0.51) and 1.93 (0.83–5.48, P = 0.16), respectively, compared with non‐ADR (data not shown).
Table 2.
Replication study for single nucleotide polymorphisms (SNPs) showing P ‐values of <0.05 in 1st screening
| Gene | SNP ID | Sample set | ADR † | Non‐ADR † | P‐value | Odds ratio (95% CI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 ‡ | 12 | 22 | MAF | 11 ‡ | 12 | 22 | MAF | 1 versus 2 | versus 11 | versus 22 | versus 11 | versus 22 | |||
| ABCB1 | rs2188524 | 1st | 20 | 7 | 0 | 0.13 | 51 | 3 | 1 | 0.05 | 0.062 | 0.035 | 1.0 | 4.46 [1.18–16.92] | NA |
| 2nd | 9 | 1 | 0 | 0.05 | 14 | 3 | 0 | 0.09 | 1.0 | 1.0 | 1.0 | 1.93 [0.17–21.54] | NA | ||
| 1st + 2nd | 29 | 8 | 0 | 0.11 | 65 | 6 | 1 | 0.06 | 0.18 | 0.14 | 1.0 | 2.56 [0.84–7.73] | NA | ||
| ABCC2 | rs11190291 | 1st | 20 | 7 | 1 | 0.16 | 50 | 6 | 0 | 0.05 | 0.041 | 0.060 | 0.33 | 3.33 [1.03–10.83] | NA |
| 2nd | 4 | 7 | 0 | 0.32 | 13 | 5 | 0 | 0.14 | 0.18 | 0.12 | 1.0 | 4.55 [0.91–22.63] | NA | ||
| 1st + 2nd | 24 | 14 | 1 | 0.21 | 63 | 11 | 0 | 0.07 | 0.0053 | 0.0058 | 0.35 | 3.58 [1.44–8.88] | NA | ||
| rs3740065 | 1st | 15 | 9 | 4 | 0.30 | 14 | 30 | 11 | 0.47 | 0.046 | 0.015 | 0.57 | 3.38 [1.30–8.82] | 1.50 [0.43–5.22] | |
| 2nd | 8 | 3 | 0 | 0.14 | 9 | 6 | 2 | 0.29 | 0.21 | 0.44 | 0.51 | 2.37 [0.46–12.14] | NA | ||
| 1st + 2nd | 23 | 12 | 4 | 0.26 | 23 | 36 | 13 | 0.43 | 0.013 | 0.0085 | 0.41 | 3.06 [1.37–6.87] | 1.93 [0.58–6.38] | ||
| rs12762549 | 1st | 7 | 12 | 9 | 0.54 | 28 | 23 | 5 | 0.30 | 0.0038 | 0.036 | 0.012 | 3.00 [1.10–8.18] | 4.83 [1.44–16.26] | |
| 2nd | 1 | 4 | 6 | 0.73 | 5 | 10 | 3 | 0.44 | 0.056 | 0.36 | 0.048 | 3.85 [0.38–38.36] | 6.00 [1.08–33.38] | ||
| 1st + 2nd | 8 | 16 | 15 | 0.59 | 33 | 33 | 8 | 0.33 | 0.00022 § | 0.014 | 0.00080 | 3.12 [1.27–7.69] | 5.16 [1.94–13.70] | ||
| rs11190303 | 1st | 17 | 9 | 2 | 0.23 | 15 | 32 | 7 | 0.43 | 0.016 | 0.0047 | 0.49 | 4.02 [1.53–10.54] | 1.94 [0.37–10.01] | |
| 2nd | 8 | 3 | 0 | 0.14 | 9 | 7 | 2 | 0.31 | 0.21 | 0.27 | 0.51 | 2.67 [0.52–13.43] | NA | ||
| 1st + 2nd | 25 | 12 | 2 | 0.21 | 24 | 39 | 9 | 0.40 | 0.0044 | 0.0026 | 0.32 | 3.57 [1.58–8.09] | 2.64 [0.54–12.90] | ||
| SLCO1B3 | rs7311358 | 1st | 17 | 7 | 3 | 0.24 | 21 | 29 | 6 | 0.37 | 0.12 | 0.036 | 1.0 | 2.83 [1.10–7.33] | 1.04 [0.23–4.53] |
| 2nd | 5 | 6 | 0 | 0.27 | 11 | 7 | 0 | 0.19 | 0.53 | 0.47 | 1.0 | 1.89 [0.41–8.61] | NA | ||
| 1st + 2nd | 22 | 13 | 3 | 0.25 | 32 | 36 | 6 | 0.32 | 0.28 | 0.17 | 1.0 | 1.80 [0.81–3.98] | 1.03 [0.24–4.37] | ||
| rs11045585 | 1st | 15 | 13 | 0 | 0.23 | 49 | 7 | 0 | 0.06 | 0.0023 | 0.0010 | 1.0 | 6.07 [2.05–17.97] | NA | |
| 2nd | 5 | 6 | 0 | 0.27 | 14 | 4 | 0 | 0.11 | 0.16 | 0.11 | 1.0 | 4.20 [0.82–21.35] | NA | ||
| 1st + 2nd | 20 | 19 | 0 | 0.23 | 63 | 11 | 0 | 0.07 | 0.00054 § | 0.00017 § | 1.0 | 5.44 [2.22–14.12] | NA | ||
| rs2174012 | 1st | 17 | 10 | 0 | 0.19 | 15 | 33 | 7 | 0.43 | 0.0028 | 0.0034 | 0.089 | 4.53 [1.70–13.34] | NA | |
| 2nd | 5 | 4 | 2 | 0.36 | 6 | 9 | 3 | 0.42 | 0.79 | 0.67 | 1.0 | 1.67 [0.35–7.77] | 1.11 [0.15–7.97] | ||
| 1st + 2nd | 22 | 14 | 2 | 0.24 | 21 | 42 | 10 | 0.42 | 0.0078 | 0.0040 | 0.21 | 3.40 [1.50–7.73] | 2.86 [0.59–13.77] | ||
| NR1I2 | rs12633127 | 1st | 8 | 16 | 3 | 0.41 | 31 | 22 | 1 | 0.22 | 0.017 | 0.033 | 0.11 | 3.20 [1.19–8.59] | 6.63 [0.65–67.01] |
| 2nd | 7 | 3 | 0 | 0.15 | 10 | 7 | 0 | 0.21 | 0.73 | 0.69 | 1.0 | 1.63 [0.30–8.61] | NA | ||
| 1st + 2nd | 15 | 19 | 3 | 0.34 | 41 | 29 | 1 | 0.22 | 0.072 | 0.11 | 0.12 | 2.00 [0.89–4.50] | 6.18 [0.61–61.61] | ||
1st: ADR, N = 28; non‐ADR, N = 56.
2nd: ADR, N = 11; non‐ADR, N = 18.
1st + 2nd: ADR, N = 39; non‐ADR, N = 74.
Major allele in non‐ADRs was defined as allele 1.
Statistically significant after Bonferroni correction, based on 79 independent effective tests (P < 0.00063).
ADR, adverse drug reaction; CI, confidence interval; NA, not available.
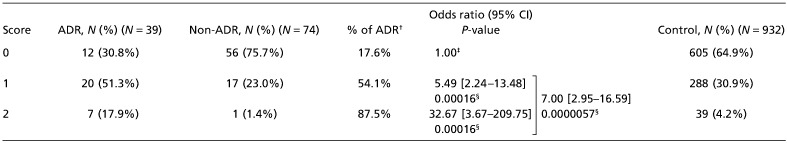
Establishment of predictive scoring system for leukopenia/neutropenia. Based on the results of case–control association studies, we investigated a combined effect of these two genes on the risks for grade 3/4 leukopenia/neutropenia using a scoring system. For the prediction scoring system, we gave a score of 1 to individuals homozygous for the risk allele, and 0 to those with other genotypes at ABCC2 rs12762549. For SLCO1B3 rs11045585, we gave a score of 1 to individuals for homozygous or heterozygous of the risk allele, and a score of 0 to those for homozygous of the low‐risk allele. As shown in Table 3, a proportion of patients with docetaxel‐induced leukopenia/neutropenia were significantly increased in a group with a higher prediction score (P = 0.0000042 by χ2‐test; 2 degrees of freedom); the incidence of grade 3/4 leukopenia/neutropenia was 17.6% (12/68) in the score 0 group, 54.1% (20/37) in the score 1 group, and 87.5% (7/8) in the score 2 group. Correspondingly, the OR of the ADR in the score 2 group was as high as 32.67 (95% CI, 3.67–209.75; P = 0.00016) and that of the score 1 group was 5.49 (95% CI, 2.24–13.48; P = 0.00016), compared with that of the score 0 group. When ‘risk’ and ‘non‐risk’ groups were defined as patients with scores of 1/2 and 0, respectively, the prediction‐scoring system using the data from all 113 subjects with two SNPs (rs11045585 and rs12762549) clearly separated the two patient groups (P = 0.0000057; OR, 7.00; 95% CI, 2.95–16.59). The sensitivity and specificity were calculated to be 69.2% and 75.7%, respectively. To confirm this scoring system, the genotype data of the 932 Japanese samples were analyzed as a control Japanese population. The proportion of subjects in this control population classified as belonging to the higher‐risk group was 35.1%.
Table 3.
Prediction scores of docetaxel‐induced leukopenia/neutropenia using ABCC2 rs12762549 and SLCO1B3 rs11045585
Discussion
Although there have been several reports concerning the relationship between AUC and the toxicity of docetaxel, the genetic determinant(s) of docetaxel toxicity has not been clarified yet.( 6 ) To identify the gene(s) related to docetaxel‐induced leukopenia/neutropenia, we genotyped 79 polymorphisms located on or near to the seven candidate genes, which are considered to be involved in the metabolism (CYP3A4 and CYP3A5) and transport (ABCB1, ABCC2, and SLCO1B3) of docetaxel,( 8 , 10 , 15 , 16 ) and in the regulation of these genes (NR1I2 and NR1I3).( 11 , 12 , 13 , 14 ) We found that rs12762549 in ABCC2 and rs11045585 in SLCO1B3 were significantly associated with severe leukopenia/neutropenia, indicating that ABCC2 and SLCO1B3 might be involved in the transport of docetaxel.
ABCC2 is a member of the ABC transporters, which is expressed in the bile canalicular membrane, in the apical membranes of pharmacologically important epithelia such as the small intestine, brain endothelial cells, and placental trophoblasts, as well as in a variety of human tumor cells.( 25 , 26 ) Involvement of ABCC2 in the efflux of docetaxel has been reported in an in vitro study using epithelial cells over‐expressing ABCC2.( 15 ) SLCO1B3, one of the solute carrier organic anion transporter members, is expressed specifically in the basolateral membrane of hepatocytes.( 27 ) This protein is shown to be involved in the docetaxel uptake from blood into hepatocytes.( 16 ) Both ABCC2 and SLCO1B3 are considered to be of particular importance for hepatic docetaxel elimination. Recently, it is reported that vectorial transport of substrates in the double‐transfected cells of ABCC2 and SLCO1B3 was at least six‐fold faster than in the single‐transfected cells of ABCC2 or SLCO1B3, suggesting that docetaxel may also be cooperatively transported by these two transporters.( 28 ) Therefore, quantitative or qualitative reduction of ABCC2 and/or SLCO1B3 activity by functional SNPs may result in an increase of docetaxel AUC because of a decrease of hepatic elimination. In the predictive scoring system, on the basis of the combined SNP information on ABCC2 rs12762549 and SLCO1B3 rs11045585, the OR of 5.49 in patients possessing one risk genotype for either the former or later SNP (score 1 group) was increased about six‐fold to 32.67 in patients carrying the risk genotypes for both SNPs (score 2 group), indicating the synergistic effect of these two proteins.
There was additional evidence supporting our hypothesis. Rougier et al. demonstrated that docetaxel clearance was reduced in patients with elevated concentration of bilirubin in serum.( 3 ) ABCC2 is well known as a gene responsible for Dubin–Johnson syndrome, characterized by conjugated hyperbilirubinemia.( 29 ) SLCO1B3 is also reported to be responsible for the hepatic uptake of bilirubin.( 30 ) These lines of evidence suggest that both reduction of docetaxel clearance and elevation of serum bilirubin concentration could be caused by similar mechanisms, namely the dysfunction (or reduced function) of ABCC2 and/or SLCO1B3.
In the association study, SNPs which showed a significant association with docetaxel‐induced leukopenia/neutropenia were tag SNPs in the non‐coding region. We also genotyped three non‐synonymous SNPs found in these genes: one in ABCC2 (rs2273697, I417V) and two in SLCO1B3 (rs4149117, S112A and rs7311358, M233I). However, these non‐synonymous SNPs showed no association with docetaxel‐induced toxicity. Thus, these tag SNPs may influence the expression levels of these genes, or some other genetic variations which linked to these tag SNPs in ABCC2 and SLCO1B3 may alter these gene products and affect the risk of leukopenia/neutropenia induced by docetaxel. Further investigations will be needed to clarify the biological mechanisms associated with the ADRs for docetaxel chemotherapy.
The US Food and Drug Administration recommended genotyping prior to drug administration for avoidance of severe ADRs for two anticancer drugs, 6‐mercaptopurine and irinotecan. The application of pharmacogenomic/pharmacogenetic information to clinical treatment is expected to help when predicting individual efficacy and/or toxicity of drugs. In this study, we established a predictive scoring system for docetaxel toxicity. A combination of ABCC2 SNP rs12762549 and SLCO1B3 SNP rs11045585 resulted in the higher sensitivity of 69.2% compared with those calculated by only genotype for rs12762549 (38.5%) or rs11045585 (48.7%). Considering the incidence (~36%) of docetaxel‐induced leukopenia/neutropenia,( 4 ) the positive predictive value was calculated to be 61.6%, while the negative predictive value was 81.4%. These results demonstrate the feasibility of using our scoring system as a means of predictive genetic testing to distinguish between ADR and non‐ADR patients before administering docetaxel.
In conclusion, through the candidate gene approach, we identified SNPs which might be related to the grade 3/4 leukopenia/neutropenia in patients treated with docetaxel. Although further validation and improvement of the scoring system described here will be required using a larger number of patients, our results open the possibility that prediction of docetaxel toxicity can lead to better prognosis and quality of life for patients with cancer.
Acknowledgments
We thank all the patients who participated in the BioBank Japan project. We thank all members of BioBank Japan, Institute of Medical Science, The University of Tokyo, and the SNP Research Center, The Institute of Physical and Chemical Research, for their contribution to the completion of our study.
References
- 1. Montero A, Fossella F, Hortobagyi G, Valero V. Docetaxel for treatment of solid tumours: a systematic review of clinical data. Lancet Oncol 2005; 6: 229–39. [DOI] [PubMed] [Google Scholar]
- 2. Tannock IF, De Wit R, Berry WR et al . Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 2004; 351: 1502–12. [DOI] [PubMed] [Google Scholar]
- 3. Rougier P, Adenis A, Ducreux M et al . A phase II study: docetaxel as first‐line chemotherapy for advanced pancreatic adenocarcinoma. Eur J Cancer 2000; 36: 1016–25. [DOI] [PubMed] [Google Scholar]
- 4. Ishimoto O, Sugawara S, Inoue A et al . Phase II study of carboplatin combined with biweekly docetaxel for advanced non‐small cell lung cancer. J Thorac Oncol 2006; 1: 979–83. [PubMed] [Google Scholar]
- 5. Evans WE, McLeod HL. Pharmacogenomics drug disposition, drug targets, and side effects. N Engl J Med 2003; 348: 538–49. [DOI] [PubMed] [Google Scholar]
- 6. Baker SD, Li J, Ten Tije AJ et al . Relationship of systemic exposure to unbound docetaxel and neutropenia. Clin Pharmacol Ther 2005; 77: 43–53. [DOI] [PubMed] [Google Scholar]
- 7. Yamamoto N, Tamura T, Murakami H et al . Randomized pharmacokinetic and pharmacodynamic study of docetaxel: dosing based on body‐surface area compared with individualized dosing based on cytochrome P450 activity estimated using a urinary metabolite of exogenous cortisol. J Clin Oncol 2005; 23: 1061–9. [DOI] [PubMed] [Google Scholar]
- 8. Marre F, Sanderink GJ, De Sousa G et al . Hepatic biotransformation of docetaxel (Taxotere) in vitro: involvement of the CYP3A subfamily in humans. Cancer Res 1996; 56: 1296–302. [PubMed] [Google Scholar]
- 9. Sparreboom A, Van Tellingen O, Scherrenburg EJ et al . Isolation, purification and biological activity of major docetaxel metabolites from human feces. Drug Metab Dispos 1996; 24: 655–8. [PubMed] [Google Scholar]
- 10. Van Zuylen L, Verweij J, Nooter K, Brouwer E, Stoter G, Sparreboom A. Role of intestinal P‐glycoprotein in the plasma and fecal disposition of docetaxel in humans. Clin Cancer Res 2000; 6: 2598–603. [PubMed] [Google Scholar]
- 11. Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 1998; 102: 1016–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 1999; 274: 6043–6. [DOI] [PubMed] [Google Scholar]
- 13. Burk O, Koch I, Raucy J et al . The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR). J Biol Chem 2004; 279: 38379–85. [DOI] [PubMed] [Google Scholar]
- 14. Geick A, Eichelbaum M, Burk O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem 2001; 276: 14581–7. [DOI] [PubMed] [Google Scholar]
- 15. Huisman MT, Chhatta AA, Van Tellingen O, Beijnen JH, Schinkel AH. MRP2 (ABCC2) transports taxanes and confers paclitaxel resistance and both processes are stimulated by probenecid. Int J Cancer 2005; 116: 824–9. [DOI] [PubMed] [Google Scholar]
- 16. Smith NF, Acharya MR, Desai N, Figg WD, Sparreboom A. Identification of OATP1B3 as a high‐affinity hepatocellular transporter of paclitaxel. Cancer Biol Ther 2005; 4: 815–8. [DOI] [PubMed] [Google Scholar]
- 17. Tanabe M, Ieiri I, Nagata N et al . Expression of P‐glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)‐1 gene. J Pharmacol Exp Ther 2001; 297: 1137–43. [PubMed] [Google Scholar]
- 18. Kim RB, Leake BF, Choo EF et al . Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther 2001; 70: 189–99. [DOI] [PubMed] [Google Scholar]
- 19. Itoda M, Saito Y, Soyama A et al . Polymorphisms in the ABCC2 (cMOAT/MRP2) gene found in 72 established cell lines derived from Japanese individuals: an association between single nucleotide polymorphisms in the 5′‐untranslated region and exon 28. Drug Metab Dispos 2002; 30: 363–4. [DOI] [PubMed] [Google Scholar]
- 20. Letschert K, Keppler D, König J. Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8). Pharmacogenetics 2004; 14: 441–52. [DOI] [PubMed] [Google Scholar]
- 21. Hsieh KP, Lin YY, Cheng CL et al . Novel mutations of CYP3A4 in Chinese. Drug Metab Dispos 2001; 29: 268–73. [PubMed] [Google Scholar]
- 22. Fukushima‐Uesaka H, Saito Y, Watanabe H et al . Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat 2004; 23: 100. [DOI] [PubMed] [Google Scholar]
- 23. Kuehl P, Zhang J, Lin Y et al . Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 2001; 27: 383–91. [DOI] [PubMed] [Google Scholar]
- 24. Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, Nakamura Y. A high‐throughput SNP typing system for genome–wide association studies. J Hum Genet 2001; 46: 471–7. [DOI] [PubMed] [Google Scholar]
- 25. Sandusky GE, Mintze KS, Pratt SE, Dantzig AH. Expression of multidrug resistance‐associated protein 2 (MRP2) in normal human tissues and carcinomas using tissue microarrays. Histopathology 2002; 41: 65–74. [DOI] [PubMed] [Google Scholar]
- 26. König J, Nies AT, Cui Y, Leier I, Keppler D. Conjugate export pumps of the multidrug resistance protein (MRP) family. Localization, substrate specificity, and MRP2‐mediated drug resistance. Biochim Biophys Acta 1999; 1461: 377–94. [DOI] [PubMed] [Google Scholar]
- 27. König J, Cui Y, Nies AT, Keppler D. Localization and genomic organization of a new hepatocellular organic anion transporting polypeptide. J Biol Chem 2000; 275: 23161–8. [DOI] [PubMed] [Google Scholar]
- 28. Cui Y, König J, Keppler D. Vectorial transport by double‐transfected cells expressing the human uptake transporter SLC21A8 and the apical export pump ABCC2. Mol Pharmacol 2001; 60: 934–43. [DOI] [PubMed] [Google Scholar]
- 29. Wada M, Toh S, Taniguchi K et al . Mutations in the canilicular multispecific organic anion transporter (cMOAT) gene, a novel ABC transporter, in patients with hyperbilirubinemia II/Dubin–Johnson syndrome. Hum Mol Genet 1998; 7: 203–7. [DOI] [PubMed] [Google Scholar]
- 30. Cui Y, König J, Leier I, Buchholz U, Keppler D. Hepatic uptake of bilirubin and its conjugates by the human organic anion transporter SLC21A6. J Biol Chem 2001; 276: 9626–30. [DOI] [PubMed] [Google Scholar]


