Abstract
(Cancer Sci 2010; 101: 631–638)
Stem cells of acute myeloid leukemia (AML) have been identified as immunodeficient mouse‐repopulating cells with a Lin−CD34+38− phenotype similar to normal hematopoietic stem cells. To identify the leukemia‐propagating stem cell fraction of Philadelphia chromosome‐positive (Ph+) leukemia, we serially transplanted human leukemia cells from patients with chronic myeloid leukemia blast crisis (n = 3) or Ph+ acute lymphoblastic leukemia (n = 3) into NOD/SCID/IL‐2Rγc−/− mice. Engrafted cells were almost identical to the original leukemia cells as to phenotypes, IGH rearrangements, and karyotypes. CD34+CD38−CD19+, CD34+38+CD19+, and CD34−CD38+CD19+ fractions could self‐renew and transfer the leukemia, whereas the CD34−CD38+CD19+ fraction did not stably propagate in NOD/SCID mice. These findings suggest that leukemia‐repopulating cells in transformed Ph+ leukemia are included in a lineage‐committed but multilayered fraction, and that CD34+ leukemia cells potentially emerge from CD34− populations.
The current concept of leukemia stem cells (LSCs) has significantly influenced the view of the development and treatment of leukemia.( 1 , 2 , 3 , 4 , 5 ) Intravenous injection of human leukemia cells into NOD/SCID mice, which is now a common and reliable method to identify human hematopoietic stem cells (HSCs) in vivo, shows that leukemia cells expressing primitive cell surface makers such as Lin−CD34+CD38− can repopulate NOD/SCID mice regardless of the type of acute myeloid leukemia (AML).( 1 , 6 , 7 , 8 , 9 ) Serial transplantation of only this fraction can propagate AML from mouse to mouse. AML LSCs have a hierarchy similar to normal HSCs and also show other characteristics of HSCs such as quiescence, chemotherapy‐resistance, and osteoblastic niche‐dependence.( 9 , 10 ) However, there are limited but conflicting data about LSCs in acute lymphoblastic leukemia (ALL).( 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 ) For example, it remains unclear whether ALL LSCs have a stem cell phenotype. Chronic myelogenous leukemia (CML) has been thought to be a stem cell disease, because the Philadelphia chromosome (Ph) can be detected in not only all myeloid cells but also in some lymphoid cells.( 20 ) In addition, BCR‐ABL mRNA is detected in the patients’ HSC fraction (Lin−CD34+CD38−CD90+).( 21 ) In CML blast crisis (CML‐BC), progenitor cells have been shown to acquire self‐renewal capacity through additional gene alterations,( 21 , 22 ) however, it remains unclear whether LSCs in CML‐BC are phenotypically different from AML or ALL, and how leukemia cells are hierarchically controlled. Furthermore, the difference between leukemia stem cells in Ph+ ALL and CML‐BC remains unknown. To address these questions, we used NOD/SCID/IL‐2Rγc−/− (NOG) mice, which are more severely immunocompromised than NOD/SCID mice, and therefore permit increased engraftment of human cells and present a more sensitive mouse model for detecting human HSCs and LSCs.( 10 , 23 ) In this system, we found that engrafted cells were lineage‐committed and clonal in the IGH gene rearrangements, irrespective of stem/progenitor cell markers. Lineage‐committed fractions, including CD34+CD38+ and CD34−CD38+ fractions, could self‐renew and transfer leukemia in mice, indicating that leukemia‐repopulating cells in Ph+ leukemia are multilayered but phenotypically different from normal HSCs. Ex vivo culture of Ph+ leukemia cells showed a similar maturation course based on surface makers. However, transplantation of the CD34−CD38+ fraction gave rise to immature populations, such as the CD34+CD38+ and CD34+CD38− fractions, in NOG mice, suggesting that the CD34 expression of human leukemia cells is potentially reversible in vivo.
Materials and Methods
Patients. The patients’ characteristics are shown in Table 1. CML‐BC or Ph+ ALL was diagnosed according to hematological findings, the clinical course, chromosomal analyses, and cell phenotypes. Samples from unique patient number (UPN)2, UPN3, and UPN4 and those from UPN1, UPN5, and UPN6 were obtained at diagnosis and at relapse after imatinib‐containing therapy, respectively. Primary leukemia cells from patient bone marrow (BM) were collected after obtaining written informed consent. Each sample contained more than 80% of leukemia cells.
Table 1.
Comparison of cell markers and karyotypes between original and engrafted leukemia cells
| Patient (age/gender, type of leukemia, ABL mutation) | UPN1 (31/F, CML‐BC, T315I) | UPN2 (67/M, CML‐BC, F311I) | UPN3 (52/F, CML‐BC, wild) | UPN4 (77/M, Ph+ ALL, wild) | UPN5 (68/F, Ph+ ALL, wild) | UPN6 (77/F, Ph+ ALL, Y253H) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Original | INH (engrafted) | Original | MKS (engrafted) | Original | MIZ (engrafted) | Original | SGR (engrafted) | Original | OMR (engrafted) | Original | KWI (engrafted) | |
| Engraftment efficiency | 10 × 106 | 5/5 | 4.0 × 106 | 3/3 | 4.0 × 106 | 3/3 | 10 × 106 | 3/3 | 2.5 × 106 | 4/4 | 1.5 × 106 | 5/5 |
| CD2 | – | – | – | – | – | – | – | – | – | – | – | ND |
| CD3 | – | – | – | – | – | – | – | – | +/− | Low | – | – |
| CD4 | – | – | – | – | – | – | – | – | – | – | – | – |
| CD5 | – | – | – | – | – | – | – | – | – | – | – | – |
| CD7 | – | – | Low | +/− | – | – | – | – | – | – | – | – |
| CD10 | + | +/− | – | +/− | + | + | + | + | + | +/− | + | + |
| CD13 | – | – | + | + | – | +/− | Low | – | +/− | Low | Low | – |
| CD14 | Low | – | – | – | +/− | – | – | – | – | – | – | – |
| CD19 | + | + | – | – | + | + | + | + | + | + | + | + |
| cyCD22 | ND | + | ND | – | ND | +/− | ND | Low | Low | Low | ND | ND |
| CD33 | – | +/− | +/− | Low | Low | Low | – | +/− | +/− | +/− | Low | – |
| CD34 | +/− | +/− | + | + | +/− | +/− | + | +/− | + | + | +/− | – |
| CD79a | ND | + | ND | +/− | ND | +/− | ND | + | + | + | ND | ND |
| MPO | ND | Low | ND | +/− | ND | Low | ND | – | – | – | ND | ND |
| Karyotype | 47,XX,t(9;22)(q34;q11),+mar | ND | 46,XY,t(9;22)(q34;q11) | 46,XY,del(1)(p13p22),add(5),t(9,22)(q34;q11) | 46,XX,t(9;22)(q34;q11) | 44,X,‐X or‐Y, ‐6.‐10.‐12,‐18,‐21,‐22,+4mar | 45,XY,der(7:12)(q10;q10),t(9:22)(q34;q11) | 45,XY,der(7;12)(q10;q10),t(9;22)(q34+q11.2),‐10,mar | 46,XX,t(9;22)(q34;q11) | 46,XX,t(9;22)(q34;q11.2) | 46,XX‐2,‐7,t(9;22)(q34;q11.2),+2mar | 46,XX,t(2;7)(p11.2;p13),t(9;22)(q34;q11.2) |
| 46,X,‐X or ‐Y,‐9,‐10,‐17,;3mar | 45,XX,idem,dic(8;12)(p11;p11) | |||||||||||
Original and engrafted leukemia cells were analyzed for surface markers, gene mutations at the kinase domain of the BCR‐ABL, and karyotypes. The number of original cells transplanted into NOD/SCID/IL‐2Rγc−/− mice and engraft‐efficiency are described. +, positive; −, negative, +/−, positive/negative. ALL, acute lymphoblastic leukemia; CML‐BC, Chronic myelogenous leukemia blast crisis; cy, cytoplasmic; MPO, myeloperoxidase; ND, not done; Ph+, Philadelphia chromosome‐positive.
Flow cytometry analysis and cell sorting. Bone marrow mononuclear cells were separated by Ficoll‐Paque PLUS (Amersham Biosciences, Uppsala, Sweden) density‐gradient centrifugation and suspended in PBS with 2% FBS (Gibco BRL, Carlsbad, CA, USA). Mononuclear cells were stained with lineage‐associated phycoerythrin (PE)‐Cy5.5‐conjugated antibodies, including CD2, CD3, CD4, CD8, CD14, CD19, CD20, and CD56 from Caltag Laboratories (Burlingame, CA, USA), except the markers positive in leukemia cells. Cells with lineage cocktail antibodies were further incubated with HSC‐associated antibodies consisting of allophycocyanin (APC)‐conjugated anti‐CD34 (HPCA‐2; Becton Dickinson PharMingen, Franklin Lakes, NJ, USA), PE‐Cy7‐conjugated anti‐CD38 (Becton Dickinson PharMingen), FITC‐labeled CD47 and PE‐conjugated anti‐CD90, or with progenitor‐associated antibodies consisting of APC‐conjugated anti‐CD34, PE‐Cy7‐conjugated anti‐CD38, PE‐conjugated anti‐interleukin‐3 receptor (IL‐3R) (9F5; Becton Dickinson PharMingen), and FITC‐conjugated anti‐CD45RA (MEM56; Caltag Laboratories).
Flow cytometry analysis and cell sorting were carried out as previously reported.( 21 , 22 , 24 ) After staining, cells were analyzed and sorted using a modified FACSAria (Becton Dickinson Immunocytometry Systems, Franklin Lakes, NJ, USA) equipped with a 488‐nm argon laser and a 633‐nm He‐Ne laser and FlowJo software (Tree Star, San Carlos, CA, USA). Lin−CD34+ cells were divided into HSCs (Lin−CD34+CD38−) and progenitors (Lin−CD34+CD38+). HSCs were further separated into long‐term HSCs (Lin−CD34+CD47+CD90+) and short‐term HSCs (Lin−CD34+CD47+CD90−). Progenitors were further separated into common myeloid progenitors (Lin−CD34+CD38+IL‐3R+CD45RA−) and their progeny, including granulocyte/monocyte progenitor (GMP; Lin−CD34+CD38+IL‐3R+CD45RA+), and megakaryocyte/erythroid progenitors (Lin−CD34+CD38+IL‐3R−CD45RA−).
Xenotransplantation into NOG mice. NOG and NOD/SCID mice were obtained from the Central Institute for Experimental Animals (Kawasaki, Japan) and Clea Japan (Tokyo, Japan), respectively. Human leukemia cells (1.5–10 × 106 cells) were injected into the tail vein of non‐irradiated 8‐week‐old male NOG mice. To prevent human T cell expansion, 0.1 mg anti‐CD3 antibody (Janssen Pharmaceutical, Tokyo, Japan) was injected on the same day, as described previously.( 10 ) For the in vivo passage of leukemia cells, BM cells or spleen cells (1–5 × 106 cells) from a NOG mouse were injected into another NOG mouse with an 8‐ to 16‐week interval. To analyze the repopulating activity in mice, 5–10 × 103 fractionated leukemia cells were serially transplanted together with 0.1 × 106 mouse BM cells. Pathological examination was done as described.( 10 ) The animal experiments were approved by the institutional ethics committee for Laboratory Animal Research, Nagoya University School of Medicine (Nagoya, Japan) and carried out according to the guidelines of the institute.
Mutation analysis of the ABL kinase domain. RNA was isolated from BM mononuclear cells by RNA STAT‐60TM (Tel‐Test, Friendswood, TX, USA) and cDNA synthesized by reverse transcriptase (TaqMan Gold RT‐PCR Kit; Applied Biosystems, Foster City, CA, USA). The kinase domain of BCR‐ABL was sequenced following the nested PCR method. BCR‐ABL was first amplified for 35 cycles (94°C for 30 sec, 60°C for 30 sec, 72°C for 3 min) followed by two primers, BCR‐2682F (5′‐TCCGCTGACCATCAATAAGGA‐3′) and ABL‐1745R (5′‐CTCCGGGGGACCTCGCTCTTT‐3′). The amplified fragment was subjected to nested PCR for 30 cycles (94°C for 30 s, 60°C for 30 s, 72°C for 2 min) with primers ABL‐772F (5′‐GAGGGCGTGTGGAAGAAATA‐3′) and ABL‐1616R (5′‐GCAGCTCTCCTGGAGGTCCTCG‐3′). After purification of PCR products using the GeneClean III Kit (Qbiogene, Vista, CA, USA), standard dideoxy chain‐termination DNA sequencing was carried out using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) on an automated ABI3700 analyzer and analyzed with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). All mutations were confirmed by sequencing forward and reverse strands.
Southern blot analysis and cloning of IGH complementarity‐determining region‐3 (CDR‐3). High molecular weight DNA was extracted from samples. Southern blot analysis using the JH probe was carried out as previously described.( 25 )
Ex vivo culture of human leukemia cells. Cells were resuspended and cultured in QBSF‐60 serum‐free media (Quality Biological, Gaithersburg, MD, USA) containing 10 ng/mL recombinant human thrombopoietin (rhTPO), recombinant human stem cell factor (rhSCF), recombinant human FLT3 ligand (rhFLT3L), and 10 ng/mL recombinant human interleukin‐7 (rhIL‐7). rhTPO and rhSCF were kindly provided by Kirin (Tokyo, Japan). rhFLT3L and rhIL‐7 were purchased from PeproTech (Rocky Hill, NJ, USA).
Results
Xenotransplantation of human leukemia cells into NOG mice. We injected unsorted human leukemia cells (1.5–10 × 106 cells/mouse) from six patients with CML‐BC (n = 3) or Ph+ ALL (n = 3) into three to five NOG mice per patient, and observed their engraftment in all mice injected (Table 1). The engraftment of human leukemia cells was confirmed by more than 1% of human CD45+ peripheral blood cells at week 8, and the BM or spleen cells were analyzed and passed intravenously until week 16. Cell markers, karyotypes, and gene mutations at the kinase domain of BCR‐ABL were almost the same as the original samples (Table 1). In the MIZ line, the karyotype became complex whereas BCR‐ABL was detected by RT‐PCR (data not shown). These six leukemia lines, serially passed in vivo, have been propagated for more than 1 year, but were unable to stably proliferate ex vivo in culture.
Analysis of HSC and progenitor phenotypes of transplanted leukemia cells. To study the differentiation hierarchy, lineage and stem/progenitor markers were compared between the original and engrafted cells. As shown in Figure 1(a), the CD34/CD38 patterns were almost the same in each experiment. In CML‐BC, the major fraction was CD34+CD38+IL‐3R+CD45RA+, indicative of the GMP phenotype. There were minor fractions of CD34+CD38− or CD34−CD38+. In Ph+ ALL, the phenotype of the OMR line was similar to those of CML‐BC lines. The SGR line showed a relatively homogeneous phenotype with regards to CD34/CD38 expression. The KWI line phenotype was shifted to CD34−CD38+ compared with the original leukemia cells. All leukemia cells, irrespective of CD34 and CD38 expression, expressed lineage markers; CD19 in the INH, MIZ, SGR, OMR, and KWI lines, and CD13 in the MKS line. These data indicate that NOG mice transplanted with leukemia cells can recapture a similar differentiation pattern.
Figure 1.
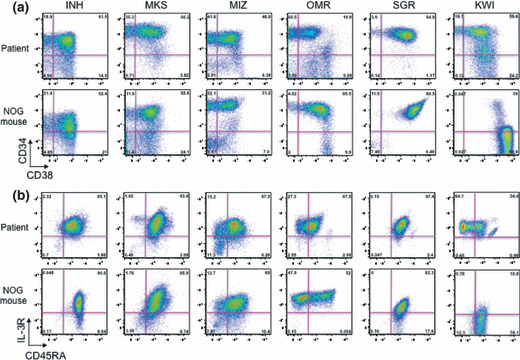
Bone marrow cells of leukemia‐transplanted NOD/SCID/IL‐2Rγc−/− (NOG) mice showed a similar human hematopoietic stem cell/progenitor profile to the original patient’s bone marrow. (a) CD34/CD38 pattern. (b) Interleukin‐3R (IL‐3R)/CD45RA pattern in a CD34+CD38+ fraction. Human hematopoietic stem cell/progenitor profiles, which were analyzed by FACSAria and FlowJo software, showed that the percentage of granulocyte/monocyte progenitor fraction is increased.
BCR‐ABL fusion transcripts and mutations at the ABL kinase domain. The major type of BCR‐ABL fusion transcripts was detected in the original cells and the established INH, MKS, MIZ, and KWI lines. The minor type of transcripts was detected in SGR and OMR. ABL kinase domain mutations T315I, F311I, and Y253H were detected in the INH, MKS, and KWI lines and their original cells, respectively (Table 1).
Serial passages of INH and OMR leukemia cells sorted according to CD34 expression. Both CD34+ and CD34− populations were maintained in the INH and OMR lines. To study which fraction could repopulate leukemia into NOG mice, we transplanted 5 × 103 or 104 leukemia cells sorted based on CD34/CD38 expression. The purity of sorted cells was over 98% (Fig. S1). In the INH line, CD34+CD38−, CD34+CD38+, and CD34−CD38+ cells repopulated human leukemia 8 weeks after transplantation (n = 3; Fig. 2a). Serial transplantation of 1 × 104 CD34−CD38+ cells mainly generated CD34−CD38+ leukemia in the BM; however, CD34+CD38+ cells faintly emerged in the spleen and BM (Fig. 2a).
Figure 2.
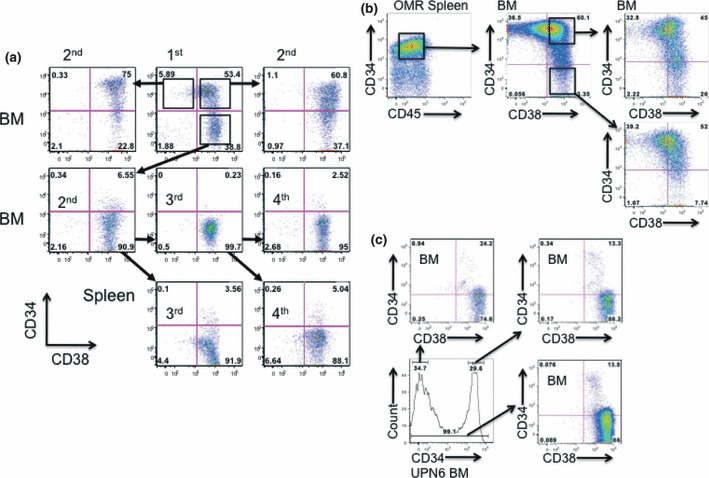
Both the CD34+ and CD34− populations of chronic myelogenous leukemia blast crisis (INH) and Philadelphia chromosome‐positive acute lymphoblastic leukemia (OMR) cells from primary recipient mouse bone marrow (BM) developed disease. (a) INH cells from a primary recipient mouse of UPN1 were sorted into CD34+CD38−, CD34+CD38+, and CD34−CD38+ fractions. Sorted cells (5 × 103 to 1 × 104) were transplanted into the secondary NOD/SCID/IL‐2Rγc−/− (NOG) mice. Tertiary and quaternary transplantations using the CD34−CD38+ population were carried out and CD34/CD38 profiles of leukemia cells from the BM and spleen are shown. (b) OMR cells from the primary recipient mouse spleen of UPN5 were sorted into CD34+CD38+ and CD34−CD38+ fractions and 5 × 104 sorted cells were transplanted into NOG mice. Tertiary transplantation was also carried out. The CD34/CD38 pattern was almost the same between mice transplanted with CD34+CD38+ and CD34−CD38+ BM cells. (c) CD34+ (n = 3) or CD34− (n = 2) BM cells, or total BM cells (n = 2) (all at 2.5 × 105) from UPN6 were transplanted into NOG mice.
In the OMR line, injection of CD34−CD38+ cells generated all the three factions: CD34+CD38−, CD34+CD38+ and CD34−CD38+ (n = 3; Fig. 2b). To further confirm whether CD34− fresh leukemia cells generated CD34+ cells, we transplanted 2.5 × 104 of CD34+ or CD34− cells of UPN6 sample into NOG mice (n = 3 and n = 3, respectively). BM cells showed almost the same pattern of CD34/CD38 expression. Although the main population was CD34−CD38+, a small population of CD34+CD38+ was also detected (Fig. 2c). These findings suggest that either CD34+CD38−, CD34+38+, or CD34−CD38+ fractions self‐renewed and transferred leukemia in NOG mice, which does not correspond with previous reports in AML.
To study the possibility that the difference came from using NOG mice, we transplanted CD34/CD38‐sorted fractions into NOD/SCID mice. CD34+CD38+ and CD34−CD38+ fractions passed from the CD34+CD38+ fraction of the INH line [(1) and (2) in Fig. 3a] engrafted in NOD/SCID mice as efficiently as in NOG mice (Fig. 2b). The CD34/CD38 pattern was the same as in NOG mice (Fig. 2b). However, the CD34+CD38+ and CD34−CD38+ fractions of INH cells passed from the CD34−CD38+ fraction [(3) and (4) in Fig. 3a] had reduced engraftment in NOD/SCID mice [(3) and (4) in Fig. 3b]. Neither fraction of 1 × 105 OMR cells from NOG mouse BM was transplantable into NOD/SCID mice (Fig. 3c), although it was stably transferred into NOG mice (Fig. 2b). These findings suggest that mouse immunocompetence significantly affects the repopulating activity of leukemia cells, and that the expression of CD34 might be reversible in vivo.
Figure 3.
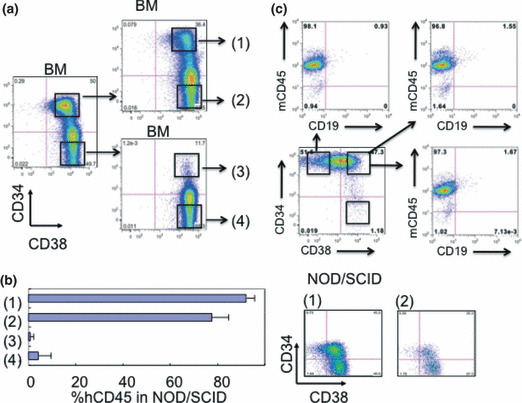
Human leukemia cells transplanted into NOD/SCID/IL‐2Rγc−/− (NOG) mice were not efficiently transferred into NOD/SCID mice. (a) Chronic myelogenous leukemia blast crisis (INH) cells repopulated in NOG mouse bone marrow (BM) were sorted into CD34+CD38+ and CD34−CD38+ fractions and these sorted cells were secondarily transplanted into NOG mice. Cells from the secondary recipient mice BM were further separated according to CD34 expression and 104 cells were transplanted into NOD/SCID mice. (b) Chimerism of human CD45+ (hCD45) leukemia cells in NOD/SCID BM was measured by flow cytometry 8 weeks after transplantation (n = 3; left panel). CD34/CD38 expression profiles of fractions (1) and (2)‐transplanted NOD/SCID mouse BM are shown (right panel). (c) Fractionated 104 OMR cells from NOG mouse BM were transplanted into NOD/SCID mice (n = 3). mCD45, murine CD45.
Ex vivo cell culture. Each leukemia line was cultured in medium supplemented with FCS and/or an appropriate cytokine cocktail; however, no lines were able to be maintained for more than 2 weeks in culture. INH cells were cultured for 8 days and injected into NOG mice. Although more than 30% of cells were positive for CD34+ after 8 days, the cultured INH cells did not engraft in NOG mice (Fig. 4). Similar to normal hematopoietic cells, cultured INH CD34+ cells showed a decrease in CD34 expression (Fig. 4a), and INH CD34− cells did not gain CD34 expression during ex vivo culture (Fig. 4b). These data suggest that lines passed in mice were different from cell lines cultured in medium, and that the maintenance of self‐renewal activity and the reversibility of CD34 expression need an in vivo environment.
Figure 4.
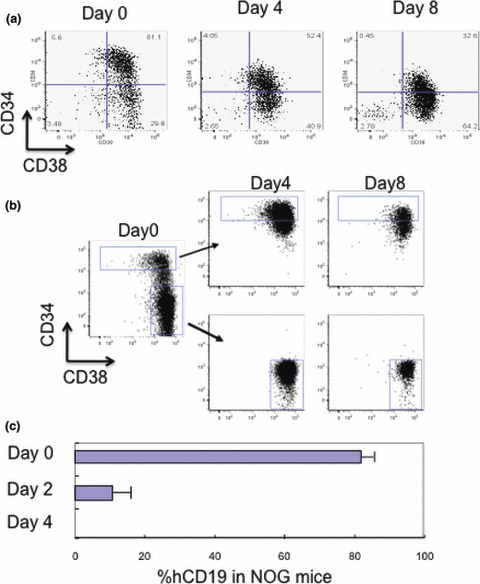
In vitro cultured chronic myelogenous leukemia blast crisis (INH) cells were not transplantable into NOD/SCID mice. Unsorted (a) and sorted (b) INH cells from the bone marrow of primary recipient mice were cultured in serum‐free medium containing rhSCF, rhTPO, rhFLT3L, and rhIL‐7. Immunophenotypic analysis was carried out at the designated time points. (c) Engrafted INH cells were detected by anti‐human CD19 (hCD19) antibody. Cells cultured in vitro for more than 2 days were not transplantable into NOD/SCID mice.
Lineage commitment. Southern blot analysis of IGH shows rearranged bands (Fig. S2a), which were identical to those of the original samples (data not shown). To analyze the existence of leukemia clones in HSC and progenitors, we determined the CDR‐3 sequence and used the leukemia‐CDR‐3‐specific primer to detect leukemia cells in the HSC and progenitor populations (Fig. S2b,c). The expression of MIZ‐specific CDR‐3 was detected in all fractions (Fig. S2d). Moreover, the expression level was similar in these fractions, indicating the general existence of leukemia cells even in the HSC compartment.
Immunostaining analysis of leukemic mouse BM. To examine where CD34+ leukemia cells localize in vivo, sterna of leukemic mice (INH, OMR, MKS, and MIZ) were immunohistochemically examined for human CD34 expression (Fig. 5a). Similar to flow cytometric results, leukemic cells of MKS and MIZ mice were almost positive for CD34. However, INH and OMR BM consisted of both positive and negative cells for CD34 and showed a mosaic‐like structure. A sternum of an INH leukemic mouse transplanted with the CD34− INH cells was stained for human CD45 and CD34 (Fig. 5b). CD34 re‐expression from the CD34− INH cells was also confirmed by immunohistochemical staining. CD34+ cells were scattered and there was no preference of CD34+ cells to the endosteal region.
Figure 5.
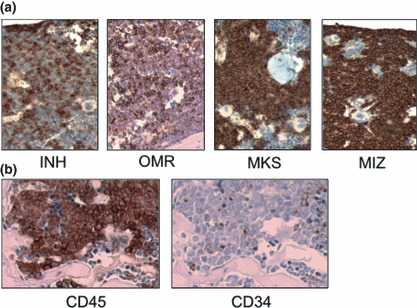
Immunostaining analysis of leukemic mouse bone marrow. Each leukemic mouse was dissected 8 weeks after transplantation. (a) Sterna of leukemic mice transplanted with unfractionated original leukemia cells (Table 1) were immunohistochemically stained for human CD34. (b) A sternum obtained from a mouse transplanted with CD34− chronic myelogenous leukemia blast crisis (INH) cells was stained for human CD45 (left panel) and CD34 (right panel).
Discussion
In this study, we transplanted Ph+ leukemia samples into NOG mice, passed leukemia serially, and established six Ph+ leukemia lines in vivo in NOG mice. The transplanted and passed leukemia lines reproduced an apparently hierarchical control of CD34/CD38 expression.
Chronic myelogenous leukemia is a myeloproliferative neoplasm that originates from a pluripotent BM HSC with the BCR‐ABL fusion gene.( 20 ) It is evident that CML originates from an HSC based on the following findings: Ph+ cells are detected in all lineages of peripheral blood; and the BCR‐ABL fusion gene is detected in the stem cell fraction, even in CML patients in molecular complete remission (CR) due to imatinib treatment.( 26 , 27 ) However, CML‐BC appears to also harbor progenitor cells capable of initiating leukemia.( 21 , 22 ) The acquisition of additional gene abnormalities is believed to block differentiation and accelerate the self‐renewal capacity of these cells.( 28 , 29 ) Our previous studies and those of others have shown that the GMP‐like population is expanded in the CML‐BC population,( 21 , 24 ) and it also expressed CD19/CD10 or CD13. In the current study, we confirmed this finding and showed that CD34+CD38−, CD34+CD34+, or CD34−CD38+ fractions were able to repopulate CML‐BC in NOG mice. Transplantation of the CD34−CD38+ fraction resulted in the expansion of a CD34−CD38+ leukemia in the early phase, but later a CD34+CD38+ fraction emerged in the BM and spleen in the INH line. In the OMR line and UPN6 sample, injection of either CD34+ or CD34− reconstituted the same pattern of CD34/CD38 expression. Similar findings were recently reported, in which the CD34−CD19+ ALL population can self‐renew and serially transfer ALL in NOD/SCID mice.( 17 )
There have been conflicting data published regarding the LSC fraction of ALL. It was reported that the stem cell fraction had either CD34 or CD19 expression. CD133+/CD19− cells were also shown to have stem cell activity in precursor‐B cell ALL using a NOD/SCID mouse xenotransplantation method.( 16 ) Furthermore, serial transplantation studies indicated that CD133+/CD19− ALL cells were capable of self‐renewal and expansion of the progenitor cell pool. However, it was also reported that the CD34+/CD38−/CD19+ but not the CD34+/CD38+/CD19+ fraction repopulated ALL in NOD/SCID mice.( 30 ) Thus, how the expression of CD34, CD38, or CD19 relates to the ALL LSCs remains unclear. It is possible that there is heterogeneity at the stem cell level among ALL cases and that serial transplantation might promote or select for xeno‐adaptation. Using the intrafemoral NOD/SCID serial transplantation assay, le Viseur et al. described that the leukemia‐initiating ability of standard‐risk ALL was found in both the CD34+CD19− and CD34+CD19+ populations, and that the CD34−CD19+ population was able to serially transplant leukemia in high‐risk ALL, exclusive of Ph+ ALL.( 17 ) In Ph+ ALL, there is a report that only the CD34+CD38− fraction can repopulate in NOD/SCID mice and this is in line with the report that a primitive hematopoietic cell is the target for leukemic transformation.( 1 , 11 ) Our result regarding Ph+ ALL LSCs is different from the report by Cobaleda et al. ( 11 ) In our system, not only the CD34+CD38− but also CD34+CD38+ and CD34−CD38+ fractions can repopulate by transplantation into mice.
This discrepancy is probably due to the mouse strain we used. The more severely immunocompromised NOG mice, which not only have impaired T cells, B cells and natural killer cells but also impaired complements, macrophages, and dendritic cells, likely permit higher levels of engraftment of human cells than NOD/SCID mice.( 25 ) In our study, the engraftment of the INH CD34+ fraction into NOD/SCID was similar to that seen in NOG mice, whereas the engraftment of CD34− was unsuccessful. In the OMR line, however, neither CD34+ nor CD34− fractions were transferred into NOD/SCID mice if 1 × 104 cells were injected. When 1 × 106 OMR cells were transplanted into irradiated NOD/SCID mice, half of the mice received the OMR line (data not shown). These findings suggest that the mouse strain significantly contributes to engraftment of human leukemia cells. Recently, a report by Kong et al. has also shown that the CD34+/CD38−/CD19+ as well as CD34+/CD38+/CD19+ cells are leukemia‐initiating cells in human B‐precursor ALL using NOG mice.( 15 )Their results are quite compatible with our data, and to develop and use better humanized mice is preferable, in order to properly evaluate engraftments and reconstitutions.( 31 )
We then asked whether these Ph+ leukemia lines were able to be maintained in ex vivo culture. Although several culture media were tested with or without murine feeder cells, cell culture was unsuccessful. In general, lymphoid cells are considerably different from myeloid cells whose fate is normally determined only in BM and toward differentiation and death.( 32 ) In contrast, lymphoid cells migrate from BM into lymphoid tissues and mature there. Only a small portion of them can survive from selection and most of them are induced into apoptosis, suggesting the mechanism to control their survival is more complicated.
Although AML LSCs are thought to express CD34 on their surface, it is controversial whether ALL LSCs exist within the CD34+ fraction. In the report by le Viseur et al., both CD34+ and CD34− fractions of high risk ALL can serially transplant leukemia in immunodeficient mice.( 17 ) Furthermore, both the CD34+ and CD34− fractions reproduced the original phenotype of CD34 expression in those mice. One explanation for the different CD34 expression in LSCs among leukemias is their lineage. Interestingly, our serial transplantation of CD34− cells from Ph+ ALL (OMR) produced both CD34+ and CD34− cells, and showed a similar CD34 expression pattern to the parental leukemia, which is compatible with le Visueur’s result. In contrast, CD34− cells from CML‐BC (INH) produced a limited portion of CD34+ cells and showed a different CD34 expression pattern than the original leukemia. Unlike myeloid cells whose self‐renewal is lost during differentiation, mature lymphoid cells survive for a long period and even restart proliferation upon stimulation. This self‐renewal capability of lymphoid cells might account for the higher reproducibility of CD34 expression in ALL leukemia cells and, to a lesser extent, in CML‐BC, but not in AML. Recent studies suggest that LSCs are responsible for leukemia relapse following conventional or targeted therapies and that eradication of LSCs might be necessary to cure the disease. This Ph+ leukemia NOG mice model will be useful not only to characterize LSCs but also to develop anti‐LSC therapy.
Finally, our study suggested that the leukemia‐repopulating capacity of transformed Ph+ leukemia cells is not restricted by the fractionation of CD34 and CD38, as is the case in AML, but rather that the repopulating ability is associated with the host mouse species, and that CD34 expression is reversible in vivo from negative to positive (Fig. 6).
Figure 6.
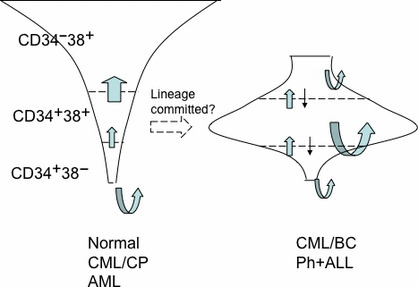
Summary of Philadelphia chromosome‐positive (Ph+) leukemia stem cells. Differential distribution of the self‐renewal populations in normal hematopoiesis (Normal), chronic myelogenous leukemia chronic phase (CML‐CP), and acute myeloid leukemia (AML), and CML blast crisis (CML‐BC) and Ph+ acute lymphoblastic leukemia (ALL). In CML‐CP and AML, only the CD34+CD38− population possesses self‐renewal capability but proliferative potential is increased along with shifting from CD34+CD38− and CD34+CD38+ to CD34−CD38+. In contrast, in CML‐BC and Ph+ ALL, the CD34+CD38− population is dominant and has greater self‐renewal capability but all compartments can self‐renew. Furthermore, leukemia cells of CML‐BC and Ph+ ALL reversibly shift between CD34+ and CD34−. Curved arrow, self‐renewal capability; straight arrow, shift between compartments.
Disclosure Statement
None.
Supporting information
Fig. S1. Purity analyses of sorted cells. Chronic myelogenous leukemia blast crisis (INH) cells were sorted by FACSAria according to CD34/CD38 expression. The purities of sorted cells were more than 98%.
Fig. S2. Analysis of IGH rearrangements in leukemia lines and detection of (complementarity‐determining region‐3) CDR‐3 sequence in hematopoietic stem cells (HSCs)/progenitors from MIZ (Table 1). (a) Southern blot analysis of genomic DNA from INH, MKS, and MIZ. Red arrows indicate rearranged bands. (b) Nucleotide sequence of IGH CDR‐3 of MIZ. Location of the oligonucleotide primers is shown. (c) HSC/progenitor profiles of MIZ. Granulocyte/monocyte progenitor (GMP)‐like population was prominent. CMP, common myeloid progenitor; IL, interleukin; MEP, megakaryocyte/erythroid progenitor; Thy1, CD90. (d) Detection of CDR‐3 in HSC/progenitors from MIZ. Expression of leukemia‐specific CDR‐3 was detected in all fractions at a similar level. The amounts of cDNA for PCR amplification were standardized by the value of real‐time quantitative PCR data of GAPDH.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Acknowledgments
This study was supported by Grants‐in‐Aid from the National Institute of Biomedical Innovation and from the Ministry of Education, Culture, Sports, Science and Technology on Scientific Research, and a Grant‐in‐Aid for Scientific Research (B) from the Japan Society for the Promotion of Science.
References
- 1. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–737. [DOI] [PubMed] [Google Scholar]
- 2. Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem‐cell biology to cancer. Nat Rev Cancer 2003; 3: 895–902. [DOI] [PubMed] [Google Scholar]
- 3. Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol 2005; 15: 494–501. [DOI] [PubMed] [Google Scholar]
- 4. Jordan CT. The leukemic stem cell. Best Pract Res Clin Haematol 2007; 20: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krause DS, Van Etten RA. Right on target: eradicating leukemic stem cells. Trends Mol Med 2007; 13: 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jordan CT, Upchurch D, Szilvassy SJ et al. The interleukin‐3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 2000; 14: 1777–1784. [DOI] [PubMed] [Google Scholar]
- 7. Lumkul R, Gorin NC, Malehorn MT et al. Human AML cells in NOD/SCID mice: engraftment potential and gene expression. Leukemia 2002; 16: 1818–1826. [DOI] [PubMed] [Google Scholar]
- 8. Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self‐renewal capacity. Nat Immunol 2004; 5: 738–743. [DOI] [PubMed] [Google Scholar]
- 9. Ishikawa F, Yoshida S, Saito Y et al. Chemotherapy‐resistant human AML stem cells home to and engraft within the bone‐marrow endosteal region. Nat Biotechnol 2007; 25: 1315–1321. [DOI] [PubMed] [Google Scholar]
- 10. Ninomiya M, Abe A, Katsumi A et al. Homing, proliferation and survival sites of human leukemia cells in vivo in immunodeficient mice. Leukemia 2007; 21: 136–142. [DOI] [PubMed] [Google Scholar]
- 11. Cobaleda C, Gutierrez‐Cianca N, Perez‐Losada J et al. A primitive hematopoietic cell is the target for the leukemic transformation in human philadelphia‐positive acute lymphoblastic leukemia. Blood 2000; 95: 1007–1013. [PubMed] [Google Scholar]
- 12. George AA, Franklin J, Kerkof K et al. Detection of leukemic cells in the CD34(+)CD38(−) bone marrow progenitor population in children with acute lymphoblastic leukemia. Blood 2001; 97: 3925–3930. [DOI] [PubMed] [Google Scholar]
- 13. Cox CV, Evely RS, Oakhill A, Pamphilon DH, Goulden NJ, Blair A. Characterization of acute lymphoblastic leukemia progenitor cells. Blood 2004; 104: 2919–2925. [DOI] [PubMed] [Google Scholar]
- 14. Castor A, Nilsson L, Astrand‐Grundstrom I et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med 2005; 11: 630–637. [DOI] [PubMed] [Google Scholar]
- 15. Kong Y, Yoshida S, Saito Y et al. CD34+ CD38+ CD19+ as well as CD34+ CD38− CD19+ cells are leukemia‐initiating cells with self‐renewal capacity in human B‐precursor ALL. Leukemia 2008; 22: 1207–1213. [DOI] [PubMed] [Google Scholar]
- 16. Cox CV, Diamanti P, Evely RS, Kearns PR, Blair A. Expression of CD133 on leukemia‐initiating cells in childhood ALL. Blood 2009; 113: 3287–3296. [DOI] [PubMed] [Google Scholar]
- 17. Le Viseur C, Hotfilder M, Bomken S et al. In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell 2008; 14: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bernt KM, Armstrong SA. Leukemia stem cells and human acute lymphoblastic leukemia. Semin Hematol 2009; 46: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heidenreich O, Vormoor J. Malignant stem cells in childhood ALL: the debate continues! Blood 2009; 113: 4476–4477; author reply 4477. [DOI] [PubMed] [Google Scholar]
- 20. Ren R. Mechanisms of BCR‐ABL in the pathogenesis of chronic myelogenous leukemia. Nat Rev Cancer 2005; 5: 172–183. [DOI] [PubMed] [Google Scholar]
- 21. Jamieson CH, Ailles LE, Dylla SJ et al. Granulocyte‐macrophage progenitors as candidate leukemic stem cells in blast‐crisis CML. N Engl J Med 2004; 351: 657–667. [DOI] [PubMed] [Google Scholar]
- 22. Abrahamsson AE, Geron I, Gotlib J et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci U S A 2009; 106: 3925–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ito M, Hiramatsu H, Kobayashi K et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 2002; 100: 3175–3182. [DOI] [PubMed] [Google Scholar]
- 24. Abe A, Minami Y, Hayakawa F et al. Retention but significant reduction of BCR‐ABL transcript in hematopoietic stem cells in chronic myelogenous leukemia after imatinib therapy. Int J Hematol 2008; 88: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kiyoi H, Naoe T, Horibe K, Ohno R. Characterization of the immunoglobulin heavy chain complementarity determining region (CDR)‐III sequences from human B cell precursor acute lymphoblastic leukemia cells. J Clin Invest 1992; 89: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elrick LJ, Jorgensen HG, Mountford JC, Holyoake TL. Punish the parent not the progeny. Blood 2005; 105: 1862–1866. [DOI] [PubMed] [Google Scholar]
- 27. O’Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr Opin Genet Dev 2006; 16: 92–99. [DOI] [PubMed] [Google Scholar]
- 28. Mullighan CG, Miller CB, Radtke I et al. BCR‐ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008; 453: 110–114. [DOI] [PubMed] [Google Scholar]
- 29. Calabretta B, Perrotti D. The biology of CML blast crisis. Blood 2004; 103: 4010–4022. [DOI] [PubMed] [Google Scholar]
- 30. Hong D, Gupta R, Ancliff P et al. Initiating and cancer‐propagating cells in TEL‐AML1‐associated childhood leukemia. Science 2008; 319: 336–339. [DOI] [PubMed] [Google Scholar]
- 31. Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol 2007; 7: 118–130. [DOI] [PubMed] [Google Scholar]
- 32. Metcalf D. Hematopoietic cytokines. Blood 2008; 111: 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Purity analyses of sorted cells. Chronic myelogenous leukemia blast crisis (INH) cells were sorted by FACSAria according to CD34/CD38 expression. The purities of sorted cells were more than 98%.
Fig. S2. Analysis of IGH rearrangements in leukemia lines and detection of (complementarity‐determining region‐3) CDR‐3 sequence in hematopoietic stem cells (HSCs)/progenitors from MIZ (Table 1). (a) Southern blot analysis of genomic DNA from INH, MKS, and MIZ. Red arrows indicate rearranged bands. (b) Nucleotide sequence of IGH CDR‐3 of MIZ. Location of the oligonucleotide primers is shown. (c) HSC/progenitor profiles of MIZ. Granulocyte/monocyte progenitor (GMP)‐like population was prominent. CMP, common myeloid progenitor; IL, interleukin; MEP, megakaryocyte/erythroid progenitor; Thy1, CD90. (d) Detection of CDR‐3 in HSC/progenitors from MIZ. Expression of leukemia‐specific CDR‐3 was detected in all fractions at a similar level. The amounts of cDNA for PCR amplification were standardized by the value of real‐time quantitative PCR data of GAPDH.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


