Abstract
Metastasis‐associated gene C4.4A is a glycolipid‐anchored membrane protein expressed in several human malignancies. The aim of this study was to explore the expression and clinical relevance of C4.4A in colorectal cancer. By quantitative RT‐PCR, 154 colorectal cancer tissues were examined for C4.4A mRNA. We examined 132 colorectal cancer tissues by immunohistochemistry using a new polyclonal antibody that recognizes the C4.4A protein C‐terminus containing the glycosylphosphatidyl‐inositol anchor signaling sequence. A significant difference in 5‐year overall survival was found between samples with high and low expression of C4.4A mRNA (P = 0.0005). Immunohistochemistry showed strong membranous staining of C4.4A at the invasive front of colorectal cancer tumors and at the frontier of metastatic lesions to lymph node and lung. The membranous staining with enhanced intensity at the invasive front of the primary colorectal cancer (Type A: 34/132, 25.6%) was associated with depth of invasion (P = 0.033) and venous invasion (P = 0.003), and was a significant independent prognostic factor (5‐year overall survival in the entire series [n = 132; P = 0.004] and disease‐free survival in stage II and III colorectal cancers [n = 82; P = 0.003]). Moreover, Type A C4.4A expression was linked to shorter liver metastasis‐free survival rate, lung metastasis‐free survival rate, or hematogenous metastasis‐free survival (P = 0.0279, P = 0.0061, and P = 0.0006, respectively). Our data indicate that expression of the C4.4A protein at the invasive front acts as a novel prognostic marker in colorectal cancer, possibly through invasion‐related mechanisms. (Cancer Sci 2010)
Colorectal cancer is one of the world’s major human malignancies. According to a report from the National Cancer Institute (USA), estimated new cases of colon and rectal cancer were 106 100 and 40 870, respectively, in the USA in 2009.( 1 ) Despite recent advances, especially in the field of chemotherapy using 5‐fluorouracil, folinic acid, and oxaliplain or irinotecan,( 2 ) total deaths from colon and rectal cancers combined was expected to reach approximately 49 920 in 2009.( 1 ) Molecularly targeted agents such as bevacizumab, a mAb against vascular endothelial growth factor,( 3 ) or cetuximab, a monoclonal antibody against epithelial growth factor receptor,( 4 ) are current therapeutic options to improve outcomes. Thus, intensive analysis and comprehension of CRC at a molecular level is of great importance.
The C4.4A protein was identified in a highly metastasizing rat pancreatic adenocarcinoma cell line, but not in locally growing rat tumors.( 5 , 6 ) Rat C4.4A cDNA was cloned in 1998 and the molecular structure indicates GPI‐anchored membrane proteins with 30% homology to the urokinase‐type plasminogen activator receptor.( 7 ) Subsequently, the human homologue of rat C4.4A, located on chromosome 19q13.1–q13.2, was cloned in 2001.( 8 ) In normal human tissues, C4.4A mRNA is present in placental tissue, skin, esophagus, and peripheral blood leukocytes, but not in other tissues, by Northern blot analysis.( 8 ) Although the physiological function of the C4.4A protein is largely unknown, upregulation of C4.4A expression is observed during the wound healing process of migrating keratinocytes or urothelium.( 9 , 10 )
Recent studies showed that C4.4A expression is also present in subsets of human malignancies. Human C4.4A mRNA was detected in cancer cell lines, including melanoma, breast, bladder, and renal cell carcinoma as well as in tumor tissue samples from malignant melanoma, CRC, breast cancer, lung carcinoma, and urothelial tumors.( 9 , 11 , 12 , 13 , 14 ) It is reported that C4.4A expression increases in metastatic lymph nodes and metastasized skin lesions compared with primary malignant melanoma.( 11 ) There is also evidence that C4.4A expression is associated with poor prognosis of non‐small cell lung cancer patients.( 14 )
In the present study, we first examined the C4.4A mRNA level in CRC tissue samples to assess the prognostic value of metastasis‐associated gene C4.4A. To further elucidate how C4.4A is involved in the progression of CRC, we generated two polyclonal antibodies that recognize the N‐terminus or C‐terminus of the C4.4A protein. Using the antibody that reacts with the amino acid sequence at the C‐terminal GPI anchor, we found that C4.4A was specifically induced at the tumor invasive front by immunohistochemistry. These findings suggest a crucial role for C4.4A in the progression of CRC, especially in tumor invasion.
Materials and Methods
Tissue samples and cell lines. Colorectal tissue samples were collected during surgery at the Department of Surgery, Osaka University (Osaka, Japan) (n = 132; 1995–2007) and the Department of Surgery and Molecular Oncology, Kyushu University (Beppu, Japan) (n = 108; 1993–1999). In this study, none of the patients had preoperative chemotherapy or irradiation. After surgery, patients with stage III/IV tumor were basically treated with 5‐fluorouracil‐based chemotherapy. The samples were fixed in buffered formalin at 4°C overnight, processed through graded ethanol solutions, and embedded in paraffin. A piece of tissue sample was frozen in a tube containing RNA preservation solution and stored in −20°C until RNA extraction. The samples were appropriately used under approval by the ethics committee at Graduate School of Medicine, Osaka University.
The human colon cancer cell line, HCT116 was obtained from American Type Culture Collection (Manassas, VA, USA). The KM12SM colon cancer cell line( 15 ) was a kind gift from Prof. T. Minamoto (Cancer Research Institute, Kanazawa, Japan). These cells were grown in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin, and grown at 37°C in a humidified incubator, under 5% CO2 in air.
Antibodies. Anti‐human C4.4A polyclonal antibodies were generated for this study. Rabbits were immunized with the target peptides bound to thyroglobulin. Among the full amino acid sequence of human C4.4A,( 9 ) two amino acid sequences were selected as immunogens: CGSGLPGKNDRGLDL near the N‐terminus; and AGHQDRSNSGQYPAKG at the C‐terminus containing a portion of the GPI anchor (Fig. 1). The C4.4A‐specific IgG was purified by passage of the antisera over a peptide column where the peptide had been coupled to the beads. The resultant N‐terminal and C‐terminal antibodies were named anti‐C4.4A‐1 and anti‐C4.4A‐2, respectively. The rabbit anti‐human actin antibody was purchased from Sigma‐Aldrich (St. Louis, MO, USA).
Figure 1.

Amino acid sequences of C4.4A protein and epitopes used as immunogens. (i) CGSGLPGKNDRGLDL near the N‐terminus, and (ii) AGHQDRSNSGQYPAKG at the C‐terminus. The boxed area indicates the glycosylphosphatidyl‐inositol (GPI) anchor signaling sequence.
Immunohistochemistry. Tissue sections (4‐μm thick) were prepared from paraffin‐embedded blocks. After antigen retrieval treatment in 10 mM citrate buffer (pH 6.0) at 95°C for 40 min, immunostaining was carried out using the Vectastain ABC peroxidase kit (Vector Laboratories, Burlingame, CA, USA) as described previously.( 16 , 17 ) The slides were incubated with anti‐C4.4A‐1 or anti‐C4.4A‐2 antibody at a 1:200 dilution overnight at 4°C. Non‐immunized rabbit IgG (Vector Laboratories) was used as a negative control and substituted for the primary antibody to exclude possible false‐positive responses from the secondary antibody or from non‐specific binding of IgG. The stained slides were carefully evaluated by H.Y., K.K., and N.M. (Department of Pathology, Faculty of Medicine, Osaka University) and final agreement was achieved under the microscope.
Immunocytochemistry. Cells were fixed in 10% formalin for 10 min and ethanol for 30 min. Immunostaining was carried out as described above. Slides were incubated with anti‐C4.4A‐2 antibody at a 1:1000 dilution overnight at 4°C.
Collagen gel culture. Collagen gel culture was carried out using a collagen gel kit (Nitta Gelatin, Osaka, Japan), according to the manufacturer’s protocol. One milliliter of collagen solution containing 5 × 105 HCT116 cells was overlaid on a pre‐prepared basal collagen layer in a 12‐well dish, and incubated at 37°C for 30 min. DMEM supplemented with 10% FBS was added after gelatinization. The culture medium was changed every day and the gels were fixed on day 10 in 10% buffered formalin overnight, then embedded in paraffin.
Western blot analysis. Western blot analysis was carried out as described previously.( 18 , 19 ) Briefly, the protein samples (20 μg) were separated by 12.5% PAGE followed by electroblotting onto a PVDF membrane. The membrane was incubated with the primary antibodies at the appropriate concentrations (1:500 for anti‐C4.4A‐1 and anti‐C4.4A‐2, and 1:1000 for actin) for 1 h. The protein bands were detected using the Amersham ECL Detection System (Amersham Biosciences, Piscataway, NJ, USA).
Quantitative real‐time PCR. Total RNA was extracted using TRIzol reagent (Life Technologies, Rockville, MD, USA). cDNA was generated from 1 μg total RNA with avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI, USA). Quantitative real‐time PCR was carried out using LightCycler (Idaho Technology, ID, USA) as described previously.( 20 ) Quantification data from each sample were analyzed using the LightCycler analysis software. The transcription value of C4.4A was determined by plotting against a standard curve constructed using MDA‐MB‐468( 21 ) breast cancer cells. The primer sequences were:( 12 ) C4.4A sense, 5′‐ AAGAATGACCG‐CGGCCTGGATC‐3′; and C4.4A antisense, 5′‐ GACATG‐ATCGCTGGCGTTGTAG‐3′. The amount of each transcript was normalized against the expression of the housekeeping gene, GAPDH, from the same sample: sense, 5′‐ CAACTACA‐TGGTTTACATGTTC‐3′; antisense, 5′‐ GCCAGTGGACTCCACGAC‐3′.
Statistical analysis. Statistical analysis was carried out using the StatView J‐5.0 program (Abacus Concepts, Berkeley, CA, USA). The Kaplan–Meier method was used to estimate tumor recurrence from CRC, and the log–rank test was used to determine the statistical significance. Associations between discrete variables were assessed using the χ2‐test. Mean values were compared using the Mann–Whitney U‐test. Spearman’s rank correlation test was used to analyze the correlation between the two factors. The data were expressed as the median value. P‐values <0.05 were considered statistically significant.
Results
Relationship between C4.4A mRNA expression and CRC patient survival. The relationship between C4.4A mRNA level in tumor tissues (n = 154; stage I–IV) and patients’ prognosis was analyzed. When high expression was defined as the top one‐third group (33.3%), significant differences in the 5‐year OS were found between the high expression and low expression groups (P = 0.0005; Fig. 2). When the cut‐off point was set as the mean value of C4.4A mRNA expression, a similar result showing a worse prognosis was noted in the high expression group (P = 0.024, data not shown).
Figure 2.
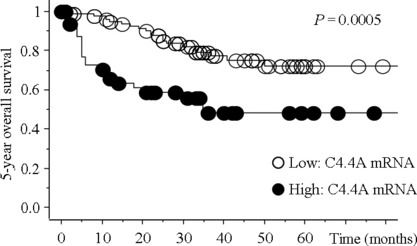
Kaplan–Meier plots showing survival curves stratified by C4.4A mRNA expression in colorectal cancer tissues. A total of 154 colorectal cancer tissues were examined for C4.4A mRNA expression using quantitative RT‐PCR. A significant difference in 5‐year overall survival rate was noted between high and low C4.4A expression (P = 0.0005).
Reactivity of anti‐human C4.4A antibodies. We produced two antibodies against the C4.4A protein (for details, see “Materials and Methods”). By Western blot analysis, the positive control sample of esophageal epithelium( 22 ) displayed an intense band around 67 kDa with both C4.4A antibodies (Fig. 3). However, in colon cancer cell lines and colon tissue samples, anti‐C4.4A‐2 antibody alone displayed a prominent band at approximately 40 kDa, whereas anti‐C4.4A‐1 antibody produced no bands (Fig. 3).
Figure 3.
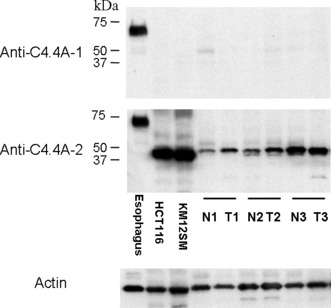
Western blot analysis of the C4.4A protein in normal esophageal tissue, colorectal cancer cell lines (HCT116 and KM12SM), and in three samples from patients taken from normal (N) and cancerous colorectal tissues (T). Esophageal epithelium displayed an intense band of approximately 67 kDa with both the anti‐C4.4A‐1 and anti‐C4.4A‐2 antibodies. However, in both colon cancer cell lines and colon tissue samples, anti‐C4.4A‐2 antibody alone displayed a prominent band at approximately 40 kDa. Anti‐C4.4A‐1 antibody did not produce any bands. The blot for actin served as a loading control.
Concordant results were confirmed by immunohistochemistry. Thus, membranous staining was observed with both antibodies at the suprabasal squamous epithelium (Fig. 4A,B). In CRC tissues, although anti‐C4.4A‐1 antibody gave no signal, staining was obtained with anti‐C4.4A‐2 antibody (Fig. 4C,D). Negative control tests showed that neither non‐immunized rabbit IgG nor the antibody pre‐absorbed with an excess amount of the C4.4A‐2 immunogen peptide stained esophageal epithelium (data not shown). The anti‐C4.4A‐2 antibody detected the C4.4A protein at the bottom of the normal colonic epithelium (Fig. 4E).
Figure 4.
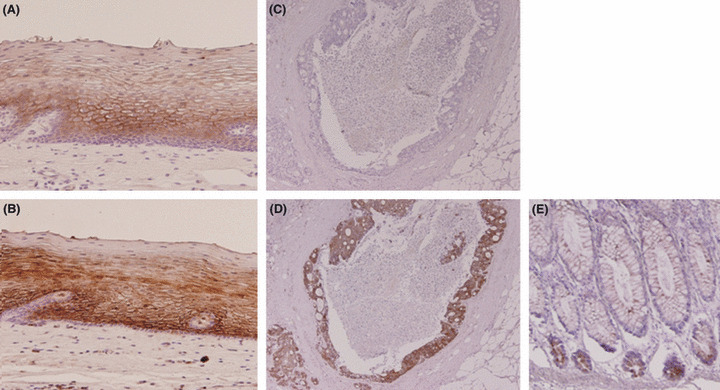
Tissue sections of normal esophageal mucosa (A,B) and colorectal cancer tissue (C,D) were immunostained with anti‐C4.4A‐1 (A,C) and anti‐C4.4A‐2 (B,D) antibodies. (A,B) Membranous staining was observed in the suprabasal squamous epithelium. (C) No signal was observed with the C4.4A‐1 antibody in a colorectal cancer tissue sample. (D) Strong staining was observed with anti‐C4.4A‐2 antibody on the same sample. (E) Tissue sections of normal colonic mucosa. In normal colonic mucosa, staining was observed with anti‐C4.4A‐2 antibody at the bottom of the gland. Magnification, (A,B) ×100; (C–E) ×40.
Expression pattern of C4.4A in CRC tissue and metastatic lesions. In cancer tissues, C4.4A expression was observed in the majority of CRC cases (105 of 132, 79.5%). The superficial or intermediate portion of the cancer body displayed weak C4.4A expression that was present mostly in the cytoplasm (Fig. 5A‐I), whereas C4.4A expression at the invasive front was often found on the plasma membrane (Fig. 5A‐II). It is noteworthy that among the expansive population of cancer specimens, only those cells that faced toward virgin stroma expressed the C4.4A protein (Fig. 5A‐III). Metastasis to lymph nodes or lung displayed a similar expression pattern with C4.4A at the invasive margin (Fig. 5B,C).
Figure 5.
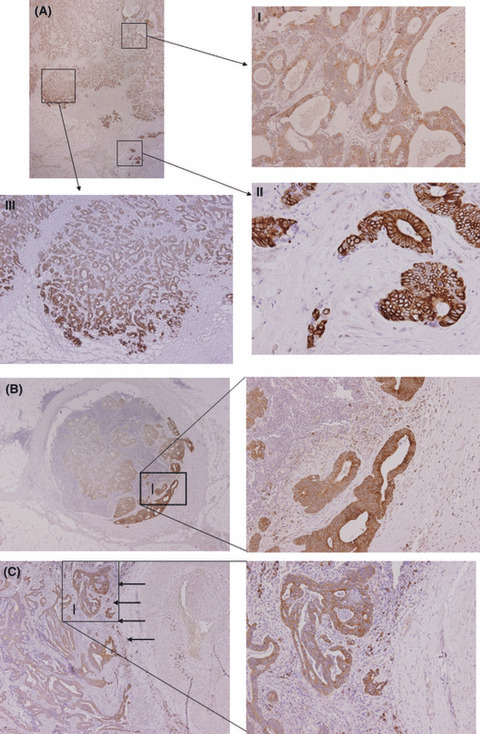
Immunohistochemistry of C4.4A expression in colorectal cancer. (A) Unique expression pattern of C4.4A at the invasive front of specimens from colorectal cancer. (A‐I) In the superficial or intermediate portion, weak C4.4A expression was observed mostly in the cytoplasm. (A‐II) C4.4A expression was present on the plasma membrane at the invasive front. (A‐III) Among the expansive population of cancer specimens, only those cells that faced virgin stroma expressed the C4.4A protein. (B) Metastatic lesion to lymph node. Metastatic tumor cells invading into the fibrous capsule exclusively expressed intense, membranous C4.4A. (C) Metastatic lesion from the lung. Metastatic tumor cells displayed a similar expression pattern of C4.4A at the invasive margin (indicated by arrows). Magnification, (A) upper left, ×12.5; (A‐I,II) ×100; (A‐III) ×40; (B) left, ×12.5; right, ×100; (C) left, ×40; right, ×100.
Based on the staining pattern at the invasive front compared with that in the superficial or intermediate portion of the cancer body, we divided 132 CRC cases into the following four categories: (A) translocation from cytoplasm to membrane and with enhanced intensity (n = 34); (B1) translocation but without enhanced intensity (n = 12); (B2) no translocation but with enhanced intensity (n = 15); and (C) no translocation and without enhanced intensity (n = 71) (Table 1).
Table 1.
Classification of changes in C4.4A staining patterns at the invasive front of colorectal cancer
| Translocation to membrane | Increase in intensity | ||
|---|---|---|---|
| + | − | Total | |
| + | A (34) | B1 (12) | 46 |
| − | B2 (15) | C (71) | 86 |
| Total | 49 | 83 | 132 |
When Type A and the other types (B1 + B2 + C) were compared for the various clinical and pathological parameters listed in Table 2, Type A was associated with deeper tumor invasion and venous invasion (P = 0.033 and P = 0.003, respectively). When stage II and III patients (n = 82) were analyzed with regard to sex, age, tumor size, differentiation, depth of invasion, lymph node metastasis, and venous invasion, Type A was solely associated with venous invasion (P = 0.0027; Table 3).
Table 2.
Expression pattern of C4.4A and clinicopathological parameters in colorectal cancer patients with stage I–IV tumors (n = 132)
| A (n = 34) | B1, B2, C (n = 98) | P‐value | |
|---|---|---|---|
| Age in years, mean (range) | 61.5 (39–85) | 62 (40–93) | NS |
| Sex | |||
| Male | 22 | 59 | NS |
| Female | 12 | 39 | |
| Tumor size (cm), mean (range) | 5 (1.8–9) | 4.5 (1.1–17) | NS |
| Differentiation | |||
| Well, mod | 34 | 88 | NS |
| Por, muc | 0 | 10 | |
| Depth of invasion | |||
| ∼mp | 3 | 26 | 0.033 |
| ss∼ | 31 | 72 | |
| Lymph node metastasis | |||
| Negative | 14 | 55 | NS |
| Positive | 20 | 43 | |
| Venous invasion | |||
| Negative | 16 | 73 | 0.003 |
| Positive | 18 | 25 | |
| Distant metastasis | |||
| Negative | 27 | 82 | NS |
| Positive | 7 | 16 | |
| Liver metastasis | |||
| Negative | 27 | 85 | NS |
| Positive | 7 | 13 | |
| Lung metastasis | |||
| Negative | 32 | 94 | NS |
| Positive | 2 | 4 | |
| Peritoneal dissemination | |||
| Negative | 34 | 90 | NS |
| Positive | 0 | 8 | |
| Stage | |||
| I, II | 12 | 52 | NS |
| III, IV | 22 | 46 | |
Colorectal cancer cases (n = 132) were categorized as: (A) translocation from cytoplasm to membrane and with enhanced intensity (n = 34); (B1) translocation but without enhanced intensity (n = 12); (B2) no translocation but with enhanced intensity (n = 15); or (C) no translocation and without enhanced intensity (n = 71). mp, muscularis propria; NS, not significant; Por, muc, poorly differentiated adenocarcinoma or mucinous carcinoma; ss, subserosa; Well, mod, well or moderately differentiated adenocarcinoma.
Table 3.
Expression pattern of C4.4A and clinicopathological parameters in colorectal cancer patients with stage II and III tumors (n = 82)
| A (n = 34) | B1, B2, C (n = 98) | P‐value | |
|---|---|---|---|
| Age in years, mean (range) | 64 (39–85) | 62 (47–79) | NS |
| Sex | |||
| Male | 14 | 36 | NS |
| Female | 11 | 21 | |
| Tumor size (cm), mean (range) | 5 (1.8–9) | 5 (1.4–17) | NS |
| Differentiation | |||
| Well, mod | 25 | 52 | NS |
| Por, muc | 0 | 5 | |
| Depth of invasion | |||
| ∼mp | 2 | 6 | NS |
| ss∼ | 23 | 51 | |
| Lymph node metastasis | |||
| Negative | 11 | 33 | NS |
| Positive | 14 | 24 | |
| Venous invasion | |||
| Negative | 12 | 46 | 0.003 |
| Positive | 13 | 11 | |
Colorectal cancer cases were categorized as: (A) translocation from cytoplasm to membrane and with enhanced intensity; (B1) translocation but without enhanced intensity; (B2) no translocation but with enhanced intensity; or (C) no translocation and without enhanced intensity. mp, muscularis propria; NS, not significant; Por, muc, poorly differentiated adenocarcinoma or mucinous carcinoma; ss, subserosa; Well, mod, well or moderately differentiated adenocarcinoma.
Survival analysis. Survival analyses indicated that either change of localization (A + B1) or increase of intensity (A + B2) was associated with shorter 5‐year OS (P = 0.023 and P = 0.0031, respectively; Fig. 6A). A more striking difference was noted between Type A and the other types (B1 + B2 + C) (P = 0.0013; Fig. 6A). When stage II and III patients were analyzed for 5‐year DFS after curative surgery, similar results were obtained (Fig. 6B).
Figure 6.

Staining pattern of C4.4A and prognosis of patients with colorectal cancer. Kaplan–Meier 5‐year overall survival (OS) curves for patients with stage I–IV tumors (n = 132). The cases were categorized as: (A) translocation from cytoplasm to membrane and with enhanced intensity (n = 34); (B1) translocation but without enhanced intensity (n = 12); (B2) no translocation but with enhanced intensity (n = 15); or (C) no translocation and without enhanced intensity (n = 71). Change of localization from cytoplasm to the plasma membrane (A + B1) or increase in intensity (A + B2) at the invasive front was associated with shorter 5‐year OS (P = 0.023 and P = 0.0031, respectively). More striking differences were noted between Type A and other expression and localization types (B1 + B2 + C) (P = 0.0013). (B) The 5‐year disease‐free survival in patients with stage II and III tumors (n = 82). When these patients were analyzed for 5‐year disease‐free survival after curative surgery, similar results to those in 5‐year OS were obtained (P = 0.011, P = 0.0016, and P = 0.0004, respectively).
The univariate analysis indicated that larger tumor size, the presence of lymph node metastasis, distant metastasis, liver metastasis, lung metastasis, and peritoneal dissemination were predictors of poor 5‐year OS in addition to the Type A expression pattern of C4.4A. We then carried out a multivariate analysis to further determine the most significant prognostic factors. The Type A expression pattern of C4.4A, larger tumor size, lymph node metastasis, and peritoneal metastasis were identified as independent prognostic factors (Table 4). In the stage II and III subgroup (n = 82), the Type A C4.4A expression pattern and the presence of lymph node metastasis were both predictors of shorter DFS by multivariate analysis (P = 0.003 and 0.043, respectively; Table 5). We then analyzed site‐specific relapse‐free survival according to the recurrent mode, that is, recurrence in liver, lung, bone, lymph node, and local site, and we found that Type A C4.4A expression was linked to shorter liver metastasis‐free survival rate, lung metastasis‐free survival rate, or hematogenous metastasis‐free survival (Fig. 7; P = 0.0279, P = 0.0061, and P = 0.0006, respectively). No difference was observed in other site‐specific disease‐free survival curves (data not shown).
Table 4.
Overall survival analysis in colorectal cancer patients with stage I–IV tumors (n = 132)
| Univariate analysis | P‐value |
|---|---|
| Sex (male vs female) | NS |
| Age (≥62 years vs <62 years) | NS |
| Tumor size (≥4.5 cm vs <4.5 cm) median | 0.004 |
| Differentiation (well,mod vs por,muc) | NS |
| Depth of invasion (∼mp vs ss∼) | NS |
| Lymph node metastasis (positive vs negative) | <0.001 |
| Venous invasion (positive vs negative) | NS |
| Distant metastasis (positive vs negative) | <0.001 |
| Liver metastasis (positive vs negative) | 0.002 |
| Lung metastasis (positive vs negative) | 0.048 |
| Peritoneal dissemination (positive vs pegative) | 0.007 |
| Expression of C4.4A (A vs B and C) | 0.001 |
| Multivariate analysis | Risk ratio | Confidence interval | P‐value |
|---|---|---|---|
| Expression of C4.4A (A vs B and C) | 3.536 | 1.497–8.354 | 0.004 |
| Tumor size (≥4.5 cm vs <4.5 cm) | 3.155 | 1.134–8.780 | 0.028 |
| Lymph node metastasis (positive vs negative) | 6.218 | 1.998–19.352 | 0.002 |
| Distant metastasis (positive vs negative) | 1.771 | 0.739–4.247 | NS |
| Peritoneal dissemination (positive vs negative) | 5.689 | 1.641–19.717 | 0.004 |
Colorectal cancer cases were categorized as: (A) translocation from cytoplasm to membrane and with enhanced intensity; (B1) translocation but without enhanced intensity; (B2) no translocation but with enhanced intensity; or (C) no translocation and without enhanced intensity. mp, muscularis propria; NS, not significant; por,muc, poorly differentiated adenocarcinoma or mucinous carcinoma; ss, subserosa; well,mod, well or moderately differentiated adenocarcinoma.
Table 5.
Disease‐free survival analysis in colorectal cancer patients with stage II and III tumors (n = 82)
| Univariate analysis | P‐value |
|---|---|
| Sex (male vs female) | NS |
| Age (≥62 years vs <62 years) | NS |
| Tumor size (≥5.0 cm vs <5.0 cm) | NS |
| Differentiation (well,mod vs por,muc) | NS |
| Depth of invasion (∼mp vs ss∼) | NS |
| Tumor site (colon vs rectum) | NS |
| Lymph node metastasis (positive vs negative) | 0.017 |
| Venous invasion (positive vs negative) | NS |
| Expression of C4.4A (A vs B and C) | <0.001 |
| Multivariate analysis | Risk ratio | Confidence interval | P‐value |
|---|---|---|---|
| Expression of C4.4A (A vs B and C) | 4.261 | 1.664–10.907 | 0.003 |
| Lymph node metastasis (positive vs negative) | 2.732 | 1.033–7.426 | 0.043 |
Colorectal cancer cases were categorized as: (A) translocation from cytoplasm to membrane and with enhanced intensity; (B1) translocation but without enhanced intensity; (B2) no translocation but with enhanced intensity; or (C) no translocation and without enhanced intensity. mp, muscularis propria; NS, not significant; por,muc, poorly differentiated adenocarcinoma or mucinous carcinoma; ss, subserosa; well,mod, well or moderately differentiated adenocarcinoma.
Figure 7.
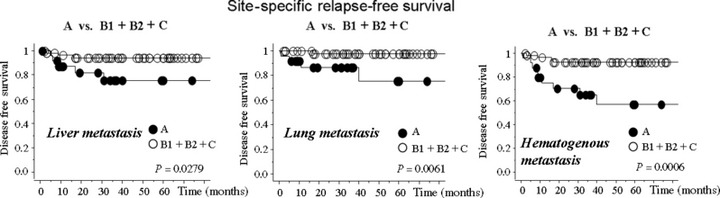
Site‐specific relapse‐free survival in colorectal cancer patients with stage II and III tumors (n = 82). The patients in stage II and III developed disease recurrence: liver metastasis (n = 8); lung metastasis (n = 5); bone metastasis (n = 1); local recurrence (n = 3); and lymph node metastasis (n = 3). Among them, one patient had both liver metastasis and lung metastasis, and another had both liver metastasis and local recurrence. The cases were categorized as: (A) translocation from cytoplasm to membrane and with enhanced intensity; (B1) translocation but without enhanced intensity; (B2) no translocation but with enhanced intensity; or (C) no translocation and without enhanced intensity. Type A C4.4A expression was linked to shorter liver metastasis‐free survival rate, lung metastasis‐free survival rate, or hematogenous metastasis‐free survival (P = 0.0279, P = 0.0061, and P = 0.0006, respectively). No difference was observed in other site‐specific disease‐free survival curves (data not shown).
In vitro collagen culture of HCT116 colon cancer cells. C4.4A protein expression was found in the nucleus of HCT116 in 2D culture (Fig. 8A). As C4.4A expression on the plasma membrane appears to be tightly linked to cell invasion into the stroma, we examined the localization of the C4.4A protein in the 3D collagen culture system. HCT116 cells on the collagen gels expressed the C4.4A protein in the cytoplasm, whereas the C4.4A expression moved to the plasma membrane when cells were surrounded by a collagen matrix (Fig. 8B).
Figure 8.

In vitro cultures of HCT116 colon cancer cells. (A) The C4.4A protein expression was observed in the nucleus of HCT116 in 2D culture. (B) The 3D cell cultures in collagen gels. The HCT116 cells on the gels expressed C4.4A protein in the cytoplasm, whereas the protein was often localized to the plasma membrane when the cells were surrounded by a collagen matrix. Note that the collagen matrix tended to become degraded when the cells had membranous C4.4A expression. The arrows indicate intense membranous staining. Magnification, ×200.
Discussion
A proteomics study began 5 years ago in collaboration with Prof. T. Takao (a co‐author of this manuscript) to identify certain peptide fragments in the urine of cancer patients. One of the peptide fragments was actually identical to part of C4.4A in patients with pancreatic or colorectal cancer. As C4.4A is a GPI‐anchored membrane protein,( 7 , 9 ) it is likely that the soluble form of the C4.4A molecule or the related fragment may readily circulate throughout the body and eventually be excreted in the urine. The amount of the C4.4A fragment in urine was small, requiring a laborious mass spectrometry, so we gave up on an extended large‐scale study. However, we considered this finding attractive because C4.4A has been shown to be a metastasis‐associated molecule by Zöller and the colleagues.( 6 , 7 )
To address the clinical value of C4.4A, we first examined C4.4A mRNA levels in CRC samples using RT‐PCR. To permit a power analysis for a survival survey, we collected CRC tissue samples from two independent institutes, Osaka University (n = 46) and Kyushu University (n = 108). As a result, we found a significant association between the high expression of C4.4A mRNA and shorter OS (P = 0.0005; Fig. 2), suggesting that C4.4A is indeed associated with poor prognosis of CRC. This finding is in agreement with a recent study by Hansen et al. ( 14 ) showing that C4.4A expression correlates with poor prognosis in non‐small cell lung cancer by an unknown mechanism.
To explore how C4.4A affects the survival of CRC patients, we developed two antibodies, anti‐C4.4A‐1 and anti‐C4.4A‐2, which recognize the N‐terminus and C‐terminus of C4.4A, respectively. As other investigators previously reported,( 22 ) both antibodies clearly reacted with a 67 kDa protein from esophageal squamous epithelium (Fig. 3), which was greater than the estimated molecular weight of 32 kDa, possibly due to modification by N‐ and O‐glycosylation.( 9 , 13 ) However, in colonic epithelium, CRC cell lines, or CRC tissues, the anti‐C4.4A‐1 antibody did not react with the C4.4A protein at all, whereas the anti‐C4.4A‐2 antibody displayed a prominent band of approximately 40 kDa for the C4.4A protein. One monogenetic interpretation of this result is that the C4.4A protein may be routinely subjected to proteolytic cleavage at a certain amino acid site in colon tissue and CRC cell lines. Another possibility is that the target epitope recognized by anti‐C4.4A‐1 might become hidden by glycosylation, as the gycosylation site may differ greatly among cell types.( 13 ) Based on these results, we used the anti‐C4.4A‐2 antibody for subsequent experiments.
We found a unique staining pattern of C4.4A in CRC tissues by immunostaining. Thus, the superficial to intermediate cancer body displayed no or weak cytoplasmic C4.4A expression, but intense C4.4A expression was noted on the plasma membrane at the invasive front in subsets of CRC tissue. It is of interest that either translocation to the cell membrane, an increase in intensity, or both was associated with 5‐year OS and 5‐year DFS, suggesting that C4.4A may play a pivotal role at the invasive front of CRC tumors that is tightly linked to disease recurrence or metastasis. When we examined the recurrent mode of stage II and III CRC patients, we found that the Type A staining pattern was significantly associated with shorter liver metastasis‐free survival (P = 0.0279), lung metastasis‐free survival (P = 0.0061), and hematogenous metastasis‐free survival (P = 0.0006). Taken together, these findings suggest that C4.4A expression at the invasive front may be a sensitive marker for metachronous metastasis, which could be due to invasion of tumor cells into the tumor vessels.
Prior to our current report, Paret et al. ( 13 ) showed that C4.4A was a candidate marker in the diagnosis of colorectal cancer. However, they did not find a positive impact of C4.4A expression on patients’ prognosis. The difference could be due to distinct antibodies. In particular, our anti‐C4.4A‐2 antibody was raised against the C‐terminus of C4.4A that contains the GPI anchor signaling sequence, which may contribute to the detection of actively invasive CRC cells.
During experiments and the preparation of this manuscript, other investigators showed that C4.4A predicted poor prognosis in non‐small cell lung cancer patients,( 14 ) and that squamous esophageal carcinoma expressed C4.4A at the invasive front.( 22 ) These findings are in line with our results in CRC. The current unique expression pattern of C4.4A (e.g. intense membranous expression at the invasive front) was highly specific to the C4.4A molecule and totally different from those we previously reported as the biomarkers for poor prognosis or malignant features of colorectal tumor.( 18 , 19 , 23 , 24 , 25 , 26 )
A clinicopathologic survey indicated that intense membranous staining of C4.4A was associated with depth of invasion, and venous invasion (Table 2), so it is postulated that C4.4A expressed at the invasive front may advance the frontier line into cancer‐associated stroma and facilitate cancer cell invasion into the tumor vessels (2, 3). It is of interest that similar C4.4A expression patterns at the invasive front were noted in lymph node metastasis and lung metastasis. It may supposed that invasive cells are likely to be related to dedifferentiation of the tumor cells, but our data indicated that C4.4A expression was not associated with histological differentiation grade. Moreover, we also examined the relationship between C4.4A expression and focal histology at the invasive front, that is, interferon α, β, and γ (defined by the General Rules for Clinical and Pathological Studies on Cancer of the Colon, Rectum and Anus, The 7th Edition, Japanese Society for Cancer of the Colon and Rectum), as the focal histology at the invasive front is not always identical to the representative histological type. However, there was no significant relation (data not shown). The findings suggest that C4.4A may facilitate cell invasion independent of histological feature. In this regard we assume that tumor budding might be one mechanism for C4.4A‐associated invasion as the budding number correlated well with C4.4A expression at the invasive front (our unpublished observation, 2010).
As reported in other human malignancies,( 11 ) it appears that C4.4A is overexpressed in CRC compared to the normal mucosa. Würfel et al. ( 8 ) also showed that C4.4A mRNA was scarcely detected by Northern blot analysis, but esophageal squamous epithelium expressed it. We found by immunohistochemistry that C4.4A expression was slightly noted in the bottom of the normal colonic epithelium (Fig. 4E). When we compared C4.4A mRNA levels between normal and cancer tissues we found that tumor tissues expressed approximately 1.34‐fold higher C4.4A mRNA than did the normal tissue (n = 15 for each, data not shown). When comparing C4.4A expression in CRC tissues at the RNA level and at the protein level, we found that high C4.4A RNA expression was not associated with a Type A staining pattern (data not shown), suggesting the existence of other regulatory mechanisms for the intense membranous C4.4A staining at the invasive front.
The precise mechanism of how C4.4A is induced at the invasive front is not known. Paret et al. ( 27 ) previously showed that C4.4A bound LN5 and supported cell migration. As LN5 binds to collagen VII, 28 one possibility is that C4.4A may also be involved in forming an anchor between the cell surface and the collagen matrix. In support of this hypothesis, we found that translocation to the plasma membrane of C4.4A was induced in a 3D collagen culture, whereas such translocation of C4.4A was not observed in 2D cultures (Fig. 8A,B). It was also reported that cleavage of LN5 by MT1‐MMP( 29 ) or MMP19( 30 ) facilitates cell migration. In this regard, it is suggested that these molecules help cancer cells migrate and invade the cancer‐associated stroma in a coordinated manner.
Alternatively, at the microenvironment of the tumor invasive front, where tumor‐associated vessels have not yet developed, the cancer cells may be hypoxic or malnourished. In fact, under low‐serum culture conditions, C4.4A and MT1‐MMP were highly induced, at least in subsets of CRC cell lines (our unpublished data, 2010). A recent study showed that C4.4A transcription is activated by C/EBPβ transcription factor, 31 which fibroblasts produce in response to anoxic conditions.( 32 ) Investigation of this possibility is currently underway in our laboratory.
In conclusion, we found that overexpression of C4.4A mRNA or intense membranous expression of C4.4A at the tumor front was associated with poor prognosis in CRC. These findings also suggest that a soluble form or certain fragments of C4.4A in the serum of CRC patients may be a useful marker of risk of disease recurrence, especially metachronous hematogenous metastasis. Therefore, further investigation should be of clinical value.
Abbreviations
- CRC
colorectal cancer
- DFS
disease free survival rate
- ESCC
esophageal squamous cell carcinoma
- GAPDH
glyceraldehydes‐3‐phosphate dehydrogenase
- GPI
glycosylphosphatidyl‐inositol
- LN
laminin
- MT1‐MMP
membrane‐type matrix metalloproteinase‐1
- OS
overall survival
- uPAR
urokinase‐type plasminogen activator receptor
Acknowledgments
We thank Prof. T. Minamoto (Cancer Research Institute, Kanazawa, Japan) for providing the KM12SM colon cancer cell line. We are also grateful to Y. Nakamura for valuable discussions and technical support. The work was supported by a Grant‐in‐Aid for Cancer Research from the Ministry of Education, Science, Sports, and Culture Technology, Japan, to H.Y.
References
- 1. National Cancer Institute . Colon and rectal cancers. [Cited July 1, 2009.] Available from URL: http://www.cancer.gov/cancertopics/types/colon‐and‐rectal.
- 2. Wadhawan A, Stephens R, Adams R. Intermittent therapy in the palliative treatment of metastatic colorectal cancer. Expert Rev Anticancer Ther 2009; 9: 125–34. [DOI] [PubMed] [Google Scholar]
- 3. Hurwitz H, Fehrenbacher L, Novotny W et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004; 350: 2335–42. [DOI] [PubMed] [Google Scholar]
- 4. Van Cutsem E, Köhne CH, Hitre E et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009; 360: 1408–17. [DOI] [PubMed] [Google Scholar]
- 5. Matzku S, Wenzel A, Liu S, Zöller M. Antigenic differences between metastatic and nonmetastatic BSp73 rat tumor variants characterized by monoclonal antibodies. Cancer Res 1989; 49: 1294–9. [PubMed] [Google Scholar]
- 6. Claas C, Herrmann K, Matzku S, Möller P, Zöller M. Developmentally regulated expression of metastasisassociated antigens in the rat. Cell Growth Differ 1996; 7: 663–78. [PubMed] [Google Scholar]
- 7. Rösel M, Claas C, Seiter S, Herlevsen M, Zöller M. Cloning and functional characterization of a new phosphatidyl‐inositol anchored molecule of a metastasizing rat pancreatic tumor. Oncogene 1998; 17: 1989–2002. [DOI] [PubMed] [Google Scholar]
- 8. Würfel J, Seiter S, Stassar M et al. Cloning of the human homologue of the metastasis‐associated rat C4.4A. Gene 2001; 262: 35–41. [DOI] [PubMed] [Google Scholar]
- 9. Hansen LV, Gårdsvoll H, Nielsen BS et al. Structural analysis and tissue localization of human C4.4A: a protein homologue of the urokinase receptor. Biochem J 2004; 380: 845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith BA, Kennedy WJ, Harnden P, Selby PJ, Trejdosiewicz LK, Southgate J. Identification of genes involved in human urothelial cell –matrix interactions: implications for the progression pathways of malignant urothelium. Cancer Res 2001; 61: 1678–85. [PubMed] [Google Scholar]
- 11. Seiter S, Stassar M, Rappl G, Reinhold U, Tilgen W, Zöller M. Upregulation of C4.4A expression during progression of melanoma. J Invest Dermatol 2001; 116: 344–7. [DOI] [PubMed] [Google Scholar]
- 12. Fletcher GC, Patel S, Tyson K et al. hAG‐2 and hAG‐3, human homologues of genes involved in differentiation, are associated with oestrogen receptor‐positive breast tumors and interact with metastasis gene C4.4a and dystroglycan. Br J Cancer 2003; 88: 579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paret C, Hildebrand D, Weitz J et al. C4.4A as a candidate marker in the diagnosis of colorectal cancer. Br J Cancer 2007; 97: 1146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hansen LV, Skov BG, Ploug M, Pappot H. Tumor cell expression of C4.4A, a structural homologue of the urokinase receptor, correlates with poor prognosis in non‐small cell lung cancer. Lung Cancer 2007; 58: 260–6. [DOI] [PubMed] [Google Scholar]
- 15. Morikawa K, Walker SM, Nakajima M, Pathak S, Jessup JM, Fidler IJ. Influence of organ environment on the growth, selection, and metastasis of human colon carcinoma cells in nude mice. Cancer Res 1988; 48: 6863–71. [PubMed] [Google Scholar]
- 16. Hayashi N, Yamamoto H, Hiraoka N et al. Differential expression of cyclooxygenase‐2 (COX‐2) in human bile duct epithelial cells and bile duct neoplasm. Hepatology 2001; 34: 638–50. [DOI] [PubMed] [Google Scholar]
- 17. Noura S, Yamamoto H, Ohnishi T et al. Comparative detection of lymph node micrometastases of stage II colorectal cancer by reverse transcriptase polymerase chain reaction and immunohistochemistry. J Clin Oncol 2002; 20: 4232–41. [DOI] [PubMed] [Google Scholar]
- 18. Takemasa I, Yamamoto H, Sekimoto M et al. Overexpression of CDC25B phosphatase as a novel marker of poor prognosis of human colorectal carcinoma. Cancer Res 2000; 60: 3043–50. [PubMed] [Google Scholar]
- 19. Yamamoto H, Soh JW, Shirin H et al. Comparative effects of overexpression of p27Kip1 and p21Cip1/Waf1 on growth and differentiation in human colon carcinoma cells. Oncogene 1999; 18: 103–15. [DOI] [PubMed] [Google Scholar]
- 20. Yamamoto H, Kondo M, Nakamori S et al. JTE‐522, a cyclooxygenase‐2 inhibitor, is an effective chemopreventive agent against rat experimental liver fibrosis. Gastroenterology 2003; 125: 556–71. [DOI] [PubMed] [Google Scholar]
- 21. Adam PJ, Boyd R, Tyson KL et al. Comprehensive proteomic analysis of breast cancer cell membranes reveals unique proteins with potential roles in clinical cancer. J Biol Chem 2003; 278: 6482–9. [DOI] [PubMed] [Google Scholar]
- 22. Hansen LV, Laerum OD, Illemann M, Nielsen BS, Ploug M. Altered expression of the urokinase receptor homologue, C4.4A, in invasive areas of human esophageal squamous cell carcinoma. Int J Cancer 2008; 122: 734–41. [DOI] [PubMed] [Google Scholar]
- 23. Takayama O, Yamamoto H, Damdinsuren B et al. Expression of PPARδ in multistage carcinogenesis of the colorectum: implications of malignant cancer morphology. Br J Cancer 2006; 95: 889–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ngan CY, Yamamoto H, Seshimo I et al. A multivariate analysis of adhesion molecules expression in assessment of colorectal cancer. J Surg Oncol 2007; 95: 652–62. [DOI] [PubMed] [Google Scholar]
- 25. Ezumi K, Yamamoto H, Murata K et al. Aberrant expression of connexin 26 is associated with lung metastasis of colorectal cancer. Clin Cancer Res 2008; 14: 677–84. [DOI] [PubMed] [Google Scholar]
- 26. Tsujino T, Seshimo I, Yamamoto H et al. Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin Cancer Res 2007; 13: 2082–90. [DOI] [PubMed] [Google Scholar]
- 27. Paret C, Bourouba M, Beer A et al. Ly6 family member C4.4A binds laminins 1 and 5, associates with Galectin‐3 and supports cell migration. Int J Cancer 2005; 115: 724–33. [DOI] [PubMed] [Google Scholar]
- 28. Marinkovich MP. Tumor microenvironment: laminin 332 in squamous‐cell carcinoma. Nat Rev Cancer 2007; 7: 370–80. [DOI] [PubMed] [Google Scholar]
- 29. Udayakumar TS, Chen ML, Bair EL et al. Membrane type‐1‐matrix metalloproteinase expressed by prostate carcinoma cells cleaves human laminin‐5 beta3 chain and induces cell migration. Cancer Res 2003; 63: 2292–9. [PubMed] [Google Scholar]
- 30. Sadowski T, Dietrich S, Koschinsky F et al. Matrix metalloproteinase 19 processes the laminin 5 gamma 2 chain and induces epithelial cell migration. Cell Mol Life Sci 2005; 62: 870–80. [DOI] [PubMed] [Google Scholar]
- 31. Fries F, Nazarenko I, Hess J, Claas A, Angel P, Zöller M. CEBPbeta, JunD and c‐Jun contribute to the transcriptional activation of the metastasis‐associated C4.4A gene. Int J Cancer 2007; 120: 2135–47. [DOI] [PubMed] [Google Scholar]
- 32. Estes SD, Stoler DL, Anderson GR. Normal fibroblasts induce the C/EBP beta and ATF‐4 bZIP transcription factors in response to anoxia. Exp Cell Res 1995; 220: 47–54. [DOI] [PubMed] [Google Scholar]


