Abstract
The Y‐box binding protein 1 (YB‐1) is a multifunctional protein that affects transcription, splicing, and translation. Overexpression of YB‐1 in breast cancers causes cisplatin resistance. The exact mechanism by which YB‐1 confers cisplatin resistance is unknown. The aim of the present study was to identify, using mass spectrometry, proteins that interact with YB‐1 that are important for cisplatin resistance in two breast cancer cell lines, namely MCF7 and MDA‐MB‐231. A tagged YB‐1 construct was used to identify proteins interacting directly with YB‐1 in breast cancer cells. We then focused on proteins that are potentially involved in breast cancer progression based on the ONCOMINE public microarray database. Genes encoding for these YB‐1‐interacting proteins were examined in the public NCBI comparative genomic hybridization database to determine whether they are localized to regions of chromosomes that are rearranged in breast cancer tissues. From these analyses, we generated a list of proteins potentially involved in cisplatin resistance. Cisplatin dose–response curves were constructed in MCF7 and MDA‐MB‐231 transfected with four siRNA corresponding to each of these YB‐1 interactors to identify proteins significantly affecting cisplatin sensitivity upon gene silencing. Depletion of only the X‐linked ribosomal protein S4 (RPS4X) resulted in consistent resistance to cisplatin in both cell lines with at least three different siRNA sequences against RPS4X. Further analyses indicated that the knock down of RPS4X decreased DNA synthesis, induced cisplatin resistance, and is equivalent to the overexpression of YB‐1 in both MCF7 and MDA‐MB‐231 cells. These results suggest that the RPS4X/YB‐1 complex is a significant potential target to counteract cisplatin resistance in breast cancer. (Cancer Sci 2011; 102: 1410–1417)
Cisplatin is an effective first‐line therapy for many types of cancer. It creates intrastrand cross‐links onto the DNA, which, in turn, leads to replication fork collapse during replication and cell death. However, a major problem with the cisplatin regimen is the appearance of resistant tumor cells during the course of the treatment.( 1 ) Importantly, overexpression of the Y box‐binding protein 1 (YB‐1) has been correlated with the appearance of cisplatin‐resistant tumor cells in several cancer patients.( 2 ) The YB‐1 protein is a multifunctional protein that affects the transcription, splicing, and translation of specific mRNAs.( 3 , 4 , 5 , 6 ) In recent years, several laboratories have demonstrated that YB‐1 is directly involved in the cellular response to genotoxic stress. Depletion of YB‐1 expression with anti‐sense RNA results in increased sensitivity to cisplatin.( 7 ) In fact, several studies have indicated that the level of nuclear expression of YB‐1 is predictive of drug resistance and patient outcome in breast tumors, ovarian cancers, and synovial sarcomas.( 2 , 8 , 9 , 10 , 11 ) Upon ultraviolet (UV) irradiation, YB‐1 translocates from the cytoplasm to the nucleus( 12 ) and is known to bind to modified nucleic acid.( 13 ) Moreover, YB‐1 preferentially binds to cisplatin‐modified DNA.( 14 ) Further analyses have indicated that YB‐1 actively promotes strand separation of duplex DNA containing either mismatches or cisplatin modifications independent of the nucleotide sequence( 15 ) and that it has exonuclease activity.( 16 ) Finally, YB‐1 stimulates an endonuclease (human eNdonuclease THree like protein 1 [hNTH1]) involved in base excision repair.( 17 , 18 ) Interestingly, knock down of hNTH1‐specific mRNA increases the cisplatin sensitivity of YB‐1‐overexpressing cells.( 18 ) However, there is no evidence that overexpression of hNTH1 in breast cancer could be used as a marker of cisplatin resistance. Conversely, overexpression of nuclear YB‐1 is important in conferring drug resistance in tumor cells. This indicates that YB‐1 is a potential target for chemotherapeutic intervention. In order to design appropriate molecules to reverse the cisplatin resistance conferred by YB‐1, we need to find additional proteins that participate with YB‐1 in this resistance and may be potential markers of cisplatin resistance. In the present study, we used large‐scale mass spectrometry analysis of a tandem affinity purification (TAP)‐tagged–YB‐1 construct in MCF7 mammary tumor cells to identify the cellular partners of YB‐1 that exhibit deregulated expression in breast cancer tissues. Knock down analyses of several of these proteins provided additional potential targets for the enhancement of chemotherapeutic interventions in two breast cancer cell lines. We specifically focused on antisense technology because it has been described as a new potential anticancer therapy for the treatment of several clinical cancers.( 19 )
Materials and Methods
Cell lines. Human MCF7 breast cancer cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were maintained in RPMI 1640 medium supplemented with 2 mM l‐glutamine, 10% FBS, and 1% antibiotic–antimycotic solution (Invitrogen, Carlsbad, CA, USA). Human MDA‐MB‐231 breast cancer cells were obtained from ATCC and were maintained in Eagle’s Minimum Essential Medium (EMEM) supplemented with 2 mM l‐glutamine, 10% FBS, and 1% antibiotic–antimytotic solution (Invitrogen). All cell lines were routinely maintained at 37°C in a humidified 5% CO2 atmosphere. Cisplatin was obtained from the Hôtel‐Dieu de Québec Hospital (Quebec City, Québec, Canada) and prepared as a stock solution (100 mM) in DMSO. Drug solutions were prepared using RPMI 1640 or EMEM supplemented with 2 mM l‐glutamine and 5% FBS. A detailed description of the methodology is available online as supplementary information for this paper (Data S1).
Results
Identification of YB‐1‐interacting proteins in MCF7 breast cancer cells. To identify new proteins that interact with YB‐1 and may affect cisplatin resistance, YB‐1 cDNA was cloned in frame with a TAP tag containing a calmodulin and streptavidin binding peptide for TAP. This construct was transfected into the MCF7 breast cancer cell line and several stable, viable clones were obtained. However, subsequent experiments were performed using one clone only that expressed the TAP‐YB‐1 protein bait at a lower level than endogenous YB‐1 (clone A in Fig. 1a) to avoid the identification of artifacts due to overexpression of TAP‐YB‐1 in the cells. Importantly, TAP‐YB‐1 clone A was more resistant to cisplatin than the TAP clone used in the present study (Fig. S1). Copurification of proteins was achieved on exponentially growing MCF7 cells expressing either the TAP alone or the TAP‐YB‐1 construct. Because nuclear YB‐1 is important for cisplatin resistance, we isolated nuclei using a standard procedure before the chromatographic step to remove as many cytoplasmic proteins as possible. We knew that our nuclear fraction was contaminated with mitochondria. Because YB‐1 is also present in the mitochondria and this localization may be important for cisplatin‐induced mitochondrial DNA damage,( 20 ) we did not perform an additional fractionation step before the chromatography procedure. To identify mainly YB‐1–protein interactions, RNAse A (100 μg/mL) was added to all buffers. Unbound proteins at each step of the chromatography process were removed by extensive washing, thus yielding proteins stringently bound to the TAP‐YB‐1 construct in cell lysates. Bound proteins were identified by liquid chromatography–tandem mass spectrometry (LC‐MS/MS). The experiment was repeated six times with the same stable TAP‐YB‐1 clone (clone A in Fig. 1a). Proteins identified from both MCF7 TAP and TAP‐YB‐1‐expressing cells were considered artifacts and removed from the final list of potential YB‐1‐interacting proteins. Table 1 lists 36 potential proteins (in addition to YB‐1) that were identified by more than two unique peptides in all six experiments as interacting with the TAP‐YB‐1 construct in MCF7 cells. Descriptive details for each protein (excluding YB‐1, which was used as bait) are given in Table S1. Cisplatin treatment did not increase or decrease the number of identified peptides specific to a protein interacting with the TAP‐YB‐1 construct (data not shown).
Figure 1.

Validation of an interaction between tandem affinity purification (TAP)‐tagged Y box‐binding protein 1 (YB‐1) and 11 proteins identified from liquid chromatography–tandem mass spectrometry (LC‐MS/MS). (a) MCF7 clones expressing the construct. Clone A was used for LC‐MS/MS analyses. (b) Interaction between TAP‐YB‐1 and the C1QBP, DAP3, HNRPK, HNRPL, PABPC1, RALYL, RIBC2, RPL31, RPS7, RPS4X, and SARG proteins. Proteins from TAP‐ and TAP‐YB‐1‐expressing MCF7 cells were eluted from the streptavidin beads and analyzed by SDS‐PAGE with antibodies against the proteins indicated. Proteins were revealed using an ECL Plus kit (GE Healthcare Bio‐Sciences Corp., Piscataway, NJ, USA). WCE, whole‐cell lysate; TAP eluate, protein eluted from the streptavidin beads. (c) Nuclear expression of C1QBP, DAP3, HNRPK, HNRPL, PABPC1, RALYL, RIBC2, RPL31, RPS7, RPS4X, SARG, and lamin B1 (protein control) in TAP cells and the TAP‐YB‐1 clone A.
Table 1.
List of proteins identified by liquid chromatography–tandem mass spectrometry in the tandem affinity purification–Y‐box binding protein 1 eluate
| Protein | Accession no. | Localization | No. unique peptides identified |
|---|---|---|---|
| YB‐1 | P67809 | N, M, C | 79 |
| C1QBP | Q07021 | N, M | 48 |
| FBL | P22087 | N | 29 |
| RPS8 | P62241 | C | 9 |
| RPL5 | B3KTM6 | N, C | 9 |
| RPS7 | P62081 | N, C | 6 |
| RAVER1 | UPI0000E042A4 | N, C | 5 |
| EEF1A1 | B4DNE0 | C | 4 |
| RPS20 | B4DW28 | C | 4 |
| HSPD1 | B2R5M6 | M | 4 |
| APLF | A8K476 | N, C | 3 |
| LOC646864 | UPI0000EE61E0 | Unknown | 3 |
| HADHBE | B4DDC9 | M | 3 |
| CHCH9 | Q5T1J5 | M | 2 |
| KLC1 | B4DGB3 | C | 2 |
| VPS39 | B3KY10 | C | 2 |
| RPL31 | B2R4C1 | C | 2 |
| MRPL9 | Q9BYD2 | M | 2 |
| RPS4X | P62701 | C | 2 |
| SARG | Q9BW04 | N, C | 2 |
| CLUL1 | Q15846 | C | 2 |
| RPL23A | A8MUS3 | C | 2 |
| MRPS22 | A8K9Y7 | M | 2 |
| DDX5 | B4DLW8 | N | 2 |
| NDUFS3 | B4DFM8 | M | 2 |
| RB1CC1 | A8K6N4 | N, C | 2 |
| RIBC2 | Q9H4K1 | N, C | 2 |
| HNRNPK | P61978 | N, C | 2 |
| HNRNPR | O43390 | N, C | 2 |
| DAP3 | P51398 | M | 2 |
| MRPL19 | P49406 | M | 2 |
| PABPC1 | P11940 | N, C | 2 |
| MRPS28 | Q53G62 | M | 2 |
| HNRNPL | A6ND69 | N, C | 2 |
| MRPL13 | Q9BYD1 | M | 2 |
| MRPS7 | Q9Y2R9 | M | 2 |
| RALYL | A6NNK2 | N, C | 2 |
N, nuclear; M, mitochondrial; C, cytoplasmic.
We next confirmed the interaction of 11 of these proteins, selected at random from our list, using western blot analysis (Fig. 1b). Total cell lysates from TAP‐YB‐1 and TAP MCF7 clones were incubated with streptavidin binding resin overnight in a cold room and proteins were eluted the next day for immunoblot analysis with antibodies against complement component 1, q subcomponent binding protein (C1QBP), death associated protein 3 (DAP3), heterogeneous nuclear ribonucleoprotein K (HNRPK), heterogeneous nuclear ribonucleoprotein L (HNRPL), poly(A) binding protein, cytoplasmic 1 (PABPC1), RALY RNA binding protein‐like (RALYL), RIB43A domain with coiled‐coils 2 (RIBC2), ribosomal protein L31 (RPL31), ribosomal protein S7 (RPS7), RPS4X, and specifically androgen‐regulated gene protein (SARG). As indicated in Fig. 1b, these proteins bound the TAP‐YB‐1 construct but not the TAP construct alone. Cisplatin had no effect on the amount of each protein interacting with TAP‐YB‐1 in MCF7 cells (data not shown). Although all these proteins were expressed at the same level in the whole‐cell lysate of both TAP and TAP‐YB‐1 clones (Fig. 1b), nuclear levels of the C1QBP, DAP3, RIBC2, RPS4X, and SARG proteins were increased in TAP‐YB‐1 clone A compared with levels in the control TAP clone (Fig. 1c).
Identification of the differential expression of YB‐1‐interacting proteins in breast cancer. To determine whether YB‐1‐interacting proteins were potentially relevant in breast cancer (for prognosis or diagnosis), we searched for their presence in the ONCOMINE public database.( 21 ) ONCOMINE is a cancer microarray database and web‐based data‐mining platform that aims to facilitate discoveries from genome‐wide expression analyses. Data can be queried and visualized for a selected gene across all microarray analyses available to the public. Thus, we queried the expression levels of genes encoding each YB‐1‐interacting protein in breast cancer compared with normal breast tissues. In addition, we determined whether each gene was localized to a region of the genome that was rearranged in breast cancer using the comparative genomic hybridization (CGH) database from the NCBI web site (http://www.ncbi.nlm.nih.gov/projects/sky/, accessed Jan 5, 2011). As of January 2011, this database contained 144 breast cancer cases analyzed by CGH methods. Table 2 gives details regarding the localization of the genes encoding each of the YB‐1 interacting proteins and the number of cases with either amplification or a deletion of the genomic regions containing these genes in breast cancer tissues. The localization of the genes encoding the proteins was obtained from the University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/, accessed Jan 5, 2011). Overall, 25 genes coding for a potential YB‐1‐interacting protein (of 36 potential candidates) were amplified and overexpressed, or deleted and underexpressed, in breast cancer tissues (Table 2). There were no significant changes in the expression of the other genes in breast cancer tissues based on the ONCOMINE microarray database (Table S1) and so they were not analyzed further in the present study. Importantly, based on the information contained in the public databases, nothing is known about the expression of the proteins we identified with regard to cisplatin resistance in breast cancer.
Table 2.
List of potential Y‐box binding protein 1‐interacting proteins, their chromosomal location and expression levels in breast cancer
| ONCOMINE database | CGH database (144 breast cancer cases) | Comments | ||||
|---|---|---|---|---|---|---|
| Protein name | Accession no. | Chromosome | No. cases | Amplification (n) | Deletion (n) | |
| CIQBP | Q07021 | 17p13.3 | 47 | 0 | 47 | Down in tumors versus normal |
| FBL | P22087 | 19q13.2 | 29 | 3 | 26 | Down in tumors versus normal |
| RPS8 | P62241 | 1p34.1 | 26 | 2 | 24 | Down in relapsed tumors |
| RPS7 | P62081 | 2p25.3 | 9 | 7 | 2 | Up with grades |
| RPS4X | P62701 | Xq13.1 | 46 | 41 | 5 | Up in tumors versus normal |
| HADHB | B4DDC9 | 2p23 | 10 | 8 | 2 | Up with grades |
| VPS39 | B3KY10 | 15q15.1 | 10 | 1 | 9 | Down in poor differentiated tumors |
| RPL31 | B2R4C1 | 2q11.2 | 13 | 12 | 1 | Up with grades |
| MRPL9 | Q9BYD2 | 1q21.3 | 55 | 55 | 0 | Up in tumors versus normal |
| CLUL1 | Q15846 | 2q11.2 | 13 | 12 | 1 | Up in tumors versus normal |
| RPL23A | A8MUS3 | 17p11.2 | 32 | 3 | 29 | Down in tumors versus normal |
| MRPS22 | A8K9Y7 | 3q23 | 12 | 11 | 1 | Up with grades |
| NDUFS3 | B4DFM8 | 11p11.2 | 10 | 3 | 7 | Down in tumors versus normal |
| RB1CC1 | A8K6N4 | 8q11.23 | 34 | 30 | 4 | Up with grades |
| RIBC2 | Q9H4K1 | 22q13.31 | 33 | 32 | 1 | Up with grades |
| HNRNPK | P61978 | 9q21.32 | 7 | 2 | 5 | Down in tumors versus normal |
| HNRNPR | O43390 | 1p36.12 | 25 | 2 | 23 | Down with survival (death) |
| DAP3 | P51398 | 1q22 | 57 | 57 | 0 | Up in tumors versus normal |
| MRPL19 | P49406 | 2p12 | 18 | 16 | 2 | Up in tumors versus normal |
| PABPC1 | P11940 | 8q22.3 | 47 | 45 | 2 | Up in tumors versus normal |
| MRPS28 | Q53G62 | 8q21.3 | 49 | 46 | 3 | Up with bad prognosis |
| HNRNPL | A6ND69 | 19q13.2 | 29 | 3 | 26 | Down in tumors versus normal |
| RALYL | O60812 | 8q21.2 | 63 | 59 | 4 | Up in tumors versus normal |
| MRPL13 | Q9BYD1 | 8q24.12 | 47 | 46 | 1 | Up in tumors versus normal |
| MRPS7 | Q9Y2R9 | 17q25.1 | 37 | 19 | 18 | Up in tumors versus normal |
CGH, comparative genomic hybridization.
Identification of YB‐1‐interacting proteins that affect cisplatin resistance in MCF7 and MDA‐MB‐231 cells. To determine whether YB‐1‐interacting proteins affect cisplatin resistance or sensitivity, MCF7 cells were subjected to high‐throughput screening analysis using four different siRNA for each target gene. We first evaluated the sensitivity of the MCF7 cells to cisplatin by determining its EC50. MCF7 cells were then transfected with each unique siRNA sequence. An siRNA against GFP was used as a control. A concentration–response curve was constructed for cisplatin using eight different concentrations (0 to 100 M for MCF7 cells and 0 to 500 M for MDA‐MB‐231 cells) for each of the siRNA‐transfected cell lines. To eliminate the risk of off‐target effects of a specific siRNA sequence, we prioritized targets that had at least two different siRNA sequences specific to their corresponding mRNA demonstrating the same effect on cell viability in the presence of cisplatin as determined using the CellTiter‐Glo (Promega, Madison, Wisconsin) luminescent assay. From this preliminary screen, six proteins of interest were identified that had an impact on the viability of MCF7 cells treated with cisplatin, namely PABPC1, MRPS7, RB1CC1, RPL31, RPS4X, and RPS7. We next determined the efficiency of the knock down of each siRNA sequence by western blotting. The sequences of the siRNAs for the six genes tested are given in Table S2. We observed significant decreases in the protein levels of PABPC1, MRPS7, RB1CC1 (Fig. S2), and RPS4X, but not PRL31 and RPS7 (data not shown). Thus, RPL31 and RPS7 were not considered further in the present study. The drug sensitivity experiments were repeated with biological duplicates to obtain an EC50 value from the cisplatin concentration–response curves for PABPC1, MRPS7, RB1CC1, and RPS4X in MCF7 cells. The effects of each siRNA on the EC50 value of cisplatin compared with that in control transfected MCF7 cells are summarized in Fig. 2a, whereas Fig. S3 shows examples of normalized responses of transfected MCF7 cells to specific concentrations of cisplatin and the EC50 values calculated for each siRNA. Although two siRNA molecules against PABPC1 (siRNAs b and d) tended to sensitize MCF7 cells to cisplatin (by 25%; Fig. 2a) and two siRNA molecules against RB1CC1 (siRNAs a and b) increased cisplatin resistance by more than 48%, only the effect of siRNA b reached statistical significance. Two siRNA molecules against MRPS7 (siRNAs b and d) tended to increase cisplatin resistance by more than 34%. Strikingly, three siRNA sequences against RPS4X (siRNAs b, c, and d) increased cisplatin resistance in MCF7 cells by more than 58%, with the effects of siRNAs c and d being significant (P < 0.05). To conclude, several siRNAs against RPS4X increased cisplatin resistance in MCF7 cells by more than 50%.
Figure 2.
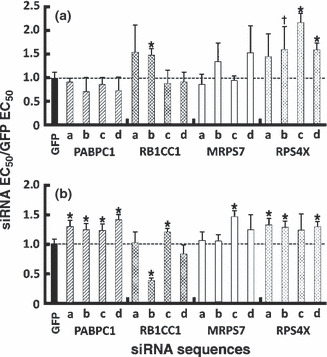
Histograms showing changes in cisplatin EC50 values for each siRNA sequence as a ratio of the EC50 in cells transfected with an siRNA against GFP (control) in (a) MCF7 and (b) MDA‐MB‐231 transfected cells. The EC50 values were calculated from cisplatin concentration–response curves obtained after transfection of individual siRNA sequences a, b, c, and d targeting the proteins indicated. Data are the mean ± SEM (n = 4). *P < 0.05, † P = 0.0719 compared with control (unpaired Student’s t‐test).
Next, to validate our findings, we tested the same siRNA molecules against cisplatin in another breast cancer cell line, namely MDA‐MB‐231. The MDA‐MB‐231 cells were more resistant to cisplatin than MCF7 cells because they had a higher EC50 in the GFP siRNA control (284 vs 84 μM; Figs S3 and S4). The effects of each siRNA on the response to cisplatin in transfected MDA‐MB‐231 cells are shown in Fig. 2b and examples of normalized response of transfected MDA‐MB‐231 cells to specific concentrations of cisplatin and the EC50 calculated for each siRNA are shown in Fig. S4. All siRNA molecules against PABPC1 significantly increased cisplatin resistance in MDA‐MB‐231 cells by more than 24% (Fig. 2b). This is in contrast with the findings in MCF7 cells (cf. Fig. 2a,b). We observed one siRNA molecule against RB1CC1 (siRNA b) that sensitized MDA‐MB‐231 cells to cisplatin by 61% and yet another siRNA sequence (siRNAs c) that increased resistance by 21%. These result differ from those in MCF7 cells (Fig. 2a,b). One siRNA molecule against MRPS7 (siRNA c) increased cisplatin resistance by 47%, which also differs from the findings in MCF7 cells (Fig. 2a). Three siRNA sequences against RPS4X significantly increased cisplatin resistance in MDA‐MB‐231 cells by more than 29% (Fig. 2b). To conclude, knocking down PABPC1, MRPS7, or RB1CC1 resulted in inconsistent survival outcomes when comparing MCF7 and MDA‐MB‐231 cells and these genes were not pursued further. Finally, several siRNAs against RPS4X increased cisplatin resistance in both MCF7 and MDA‐MB‐231 cells.
Knocking down RPS4X or overexpressing YB‐1 decreases bromodeoxyuridine incorporation and cell growth in MCF7 and MDA‐MB‐231 cells. Cisplatin causes DNA cross‐links that can lead to DNA breaks upon replication and eventually cell death. Cells that grow slowly or that are cell arrested would be more resistant to these effects. Thus, we investigated the effects of RPS4X protein on cell growth using a bromodeoxyuridine (BrdU) incorporation assay, which provides an estimate of the level of DNA synthesis in cells. We first determined the efficiency of knock down for each siRNA sequence. As shown in Fig. 3a,b siRNAs a, c, and d specific to RPS4X mRNA significantly depleted RPS4X protein levels in both MCF7 and MDA‐MB‐231 cells, but siRNA b was less efficient. Because β‐actin levels were also decreased by siRNAs against RPS4X in both cell lines, we examined levels of the DNA repair protein KU86 in transfected cells as a second loading control. As indicated in the bottom panels of Fig. 3a,b the siRNAs against RPS4X had a less negative impact on KU86 protein levels compared with β‐actin levels in transfected cells. Therefore, we concluded that the protein levels of RPS4X were significant decreased after siRNA transfection in both breast cancer cell lines. The incorporation of BrdU was significantly less in RPS4X‐knockdown cells using siRNAs a, c, and d compared with control siRNA‐transfected MCF7 cells (Fig. 3c). Interestingly, siRNA b, which was less efficient in depleting RPS4X in MCF7 cells, had little impact on BrdU incorporation compared with control siRNA. The same results were observed in MDA‐MB‐231 cells (Fig. 3d).
Figure 3.
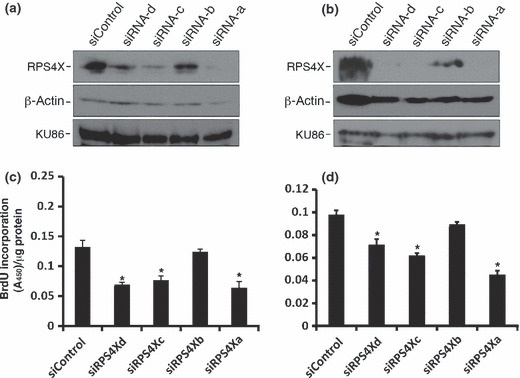
Knock down of RPS4X with specific siRNA molecules in (a,c) MCF7 and (b,d) MDA‐MB‐231 cells. Cells were transfected with the indicated siRNA molecules and, 96 h later, proteins were extracted for western blot analyses. (a,b) Representative blots following transfection of MCF7 (a) and MDA‐MB‐231 (b) cells with siRNAs against RPS4X. siControl, non‐specific siRNA molecules. In every blot, β‐actin and KU86 were used as loading controls. (c,d) Effect of RPS4X depletions on bromodeoxyuridine (BrdU) incorporation in MCF7 (c) and MDA‐MB‐231 (d) cells transfected with the indicated siRNA sequences, as determined by ELISA. Cells were transfected with the indicated siRNA sequences for 72 h and then incubated with medium containing BrdU for an additional 24 h before the ELISA. Data are the mean ± SEM (n = 4). *P < 0.03 (unpaired Student’s t‐test).
We next examined the impact of YB‐1 overexpression on BrdU incorporation in both MCF7 and MDA‐MB‐231 cells. As indicated in Fig. 4a, overexpression of YB‐1 in MCF7 and MDA‐MB‐231 cells resulted in a significant 26% and 55% decrease in BrdU incorporation, respectively. Figure 4bb shows examples of YB‐1 overexpression in MCF7 and MDA‐MB‐231 transfected cells. Scan analyses of the blots revealed a 1.5‐ and a 3‐fold increase in total YB‐1 protein levels compared with control MCF7 and MDA‐MB‐231 transfected cells, respectively. Finally, FACS analyses indicated a 3% and 11% decrease in the number of YB‐1‐overexpressing cells in the S phase of the cell cycle compared with control MCF7 and MDA‐MB‐231 transfected cells, respectively (Fig. 4c). Examples of cell cycle analyses for MDA‐MB‐231‐transfected cells are shown in Fig. S5. Overall, the results indicate that overexpression of YB‐1 decreases DNA synthesis in both MCF7 and MDA‐MB‐231 cells.
Figure 4.
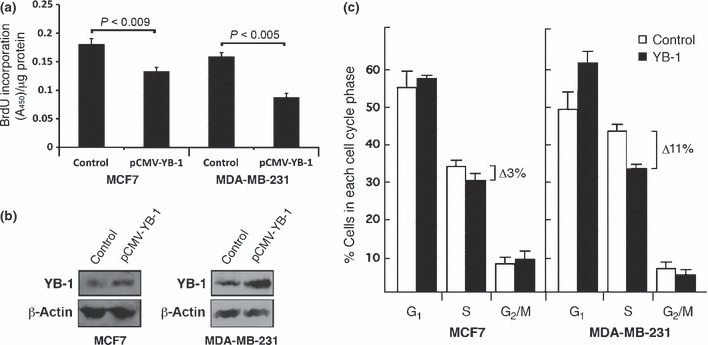
Effect of Y box‐binding protein 1 (YB‐1) overexpression on bromodeoxyuridine (BrdU) incorporation in MCF7 and MDA‐MB‐231 cells. (a) Incorporation of BrdU, as determined by ELISA, in MCF7 and MDA‐MB‐231 cells. Cells were transfected with a YB‐1 expression vector or a control empty vector and 24 h later were incubated with medium containing BrdU for an additional 24 h before the ELISA. Indicated P‐values were determined using an unpaired Student’s t‐test. (b) Representative western blots showing increased expression of YB‐1 in MCF7 and MDA‐MB‐231 cells transfected with a YB‐1 expression vector compared with an empty control vector. Cells were transfected and 24 h later proteins were extracted for western blot analysis. (c) Percentage of MCF7 and MDA‐MB‐231 transfected cells in each phase of the cell cycle. Cells were transfected with the indicated constructs and subjected to FACS analysis 24 h later. Data are the mean ± SEM (n = 4).
Finally, we examined the growth rate of TAP‐YB‐1‐expressing MCF7 cells, which are more resistant to cisplatin than TAP‐expressing cells (Fig. S1). We found that the growth rate of TAP‐YB‐1‐expressing MCF7 cells was slower than that of TAP‐expressing MCF7 cells (Fig. 5a). Similarly, the growth rate of YB‐1‐transfected MDA‐MB‐231 was slower than that of MDA‐MB‐231 cells transfected with a control vector (Fig. 5b).
Figure 5.
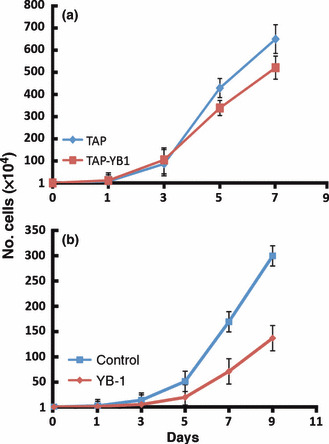
Cell growth of Y box‐binding protein 1 (YB‐1)‐overexpressing cells. (a) Growth curves for tandem affinity purification (TAP) and TAP‐YB‐1‐expressing MCF7 clones. (b) Growth curves for MDA‐MB‐231 cells transfected with control or YB‐1 expression vectors. Fifty thousand cells were plated on 60‐mm Petri dishes 24 h after transfection in duplicate. Cells were counted every other day with a hemocytometer. Data are the mean ± SEM. MCF7 and MDA‐MB‐231 cells reached confluence on Days 9 and 11, respectively.
Depletion of RPS4X in YB‐1‐overexpressing cells maintains cisplatin resistance. We next examined the impact of knocking down RPS4X expression in cisplatin‐resistant YB‐1‐overexpressing breast cancer cells. Knocking down the expression of RPS4X in MDA‐MB‐231 cells increased cisplatin resistance (Fig. 6a), but transfection of YB‐1 in RPS4X‐depleted cells did not increase cisplatin resistance further. Similar results were obtained in MCF7 cells expressing the TAP‐YB‐1 construct (Fig. 6b).
Figure 6.

Resistance of Y box‐binding protein 1 (YB‐1)‐overexpressing cells to cisplatin after RPS4X depletion. (a) MDA‐MB‐231 cells were transfected with either siControl (non‐specific siRNA molecules), siRPS4X, empty or YB‐1 expression vectors and treated with the indicated cisplatin concentrations. Experiments were performed in duplicate. Cells were treated with cisplatin for 18 h, after which they were plated (1000 cells per plate) in 100‐mm Petri dishes and cultured for 10 days without drug to allow colony formation. The y‐axis is in logarithmic scale. (b) Tandem affinity purification (TAP) or TAP‐YB‐1 MCF7 cells were transfected with siControl or siRPS4X molecules as indicated and treated with cisplatin for 18 h. The percentage of live cells was counted with a hemocytometer the next day. Experiments were performed in triplicate. Data are the mean ± SEM.
Discussion
Cisplatin is an effective first‐line therapy for many types of cancer. However, a major problem with the cisplatin regimen is the appearance of resistant tumor cells during the course of treatment.( 1 ) Another problem is the lack of knowledge of a common mechanism that confers cisplatin resistance in a large panel of breast cancer types. In recent years, several studies have indicated the importance of YB‐1 overexpression in predicting drug resistance and patient outcome in several cancer types.( 2 , 8 , 9 , 10 , 11 ) This is particularly true for breast cancer.( 2 , 22 , 23 , 24 , 25 ) High YB‐1 levels in breast cancer are associated with doxorubicin, paclitaxel, and cisplatin resistance.( 10 , 14 , 26 , 27 , 28 ) In the present study, we were particularly interested in how YB‐1 confers cisplatin resistance in the MCF7 and MDA‐MB‐231 breast cancer cell lines. Previous results have indicated a very different expression profile for MCF7 and MDA‐MB‐231 cells overexpressing YB‐1.( 29 ) Thus, it was impossible to find a common mechanism underlying cisplatin resistance in both cell lines based on global expression changes conferred by YB‐1 overexpression. To decipher the mechanism by which YB‐1 confers cisplatin resistance, we identified proteins that interacted with a tagged YB‐1 in MCF7 cells. To determine which YB‐1‐interacting proteins were most relevant for the antisense target strategy against cisplatin resistance, we adopted an integrative approach. The antisense technology has been described as a potential anticancer therapy for the treatment of several clinical cancers and there are currently several molecules in phase I and II trials.( 19 ) Following proteomics analyses of YB‐1 interactors, we focused on genes localized to chromosome rearrangements that are overexpressed in breast cancers based on information available in public databases. We then performed an siRNA screen of the YB‐1‐interacting proteins that consistently exhibited altered cisplatin cytotoxicity in both MCF7 and MDA‐MB‐231 breast cancer cell lines, followed by western validations of the knock down. Using this integrative approach, we found that the RPS4X protein was the only protein that could be a potential general target in breast cancers for the control of cisplatin resistance. The interaction of RPS4X with YB‐1 is direct because RNAses were used in the extraction buffers before mass spectrometry analyses. The exact mechanism by which the RPS4X/YB‐1 complex confers cisplatin resistance is not known. One interesting hypothesis from our results is that YB‐1 confers cisplatin resistance by inhibiting the activity of the RPS4X ribosomal protein during translation. Interestingly, although total protein levels of RPS4X did not change in TAP‐YB‐1‐expressing MCF7 cells compared with TAP‐expressing cells, nuclear RPS4X levels were increased in the former (Fig. 1c). Thus, YB‐1 could slow down translation by inducing a ribosomal stress that could, in turn, lead to cell growth arrest. Indeed, it has been known for a long time that overexpression of YB‐1 in cells results in the inhibition of translation.( 30 ) Despite this effect on translation and cell growth, recent studies have indicated that YB‐1 reduces the proliferation of breast cancer cells, but enhances the invasive potential of these same cells in tissues.( 31 , 32 ) In fact, YB‐1 binds only to a subset of mRNAs that have an impact on cell growth and transformation/metastatic phenotypes.( 31 , 32 ) It has been suggested that the reduced growth rate could constitute a significant event in the survival of cancer cells following a major stress like cisplatin treatment.( 33 , 34 ) Reduction or inhibition of RPS4X activity as a result of YB‐1 interaction would reduce cell proliferation and allow cancer cells to better resist cisplatin treatment over time. Of note, RPS4X also affects cell cycle progression in temperature‐sensitive RPS4X mutated baby hamster kidney cells (BHK21‐13) because they are blocked in the G1 and G2 phases of the cell cycle after they are shifted to non permissive temperatures.( 35 ) Our knockdown results are in accord with these findings. Depletion of RPS4X decreased BrdU incorporation in both MCF7 cells and MDA‐MB‐231 cells. The same result was observed with the overexpression of YB‐1 in these cell lines, suggesting that both proteins affect a common molecular mechanism important for cisplatin resistance. In addition, we did not observe a further increase in resistance following transfection of YB‐1 into RPS4X‐depleted breast cancer cell lines, suggesting that the pathway involving YB‐1 is not independent of that involving RPS4X. Although the microarray data from the NCBI indicated increased expression in several breast cancer tissues, the use of PrognoScan (http://gibk21.bse.kyutech.ac.jp/PrognoScan/index.html, accessed Jan 5, 2011) indicated a decreased expression of RPS4X with overall survival of several breast cancer patients. Epidemiological studies, including immunohistochemistry with antibodies against both YB‐1 and RPS4X in breast tumors from drug‐resistant and ‐sensitive patients, are warranted.
Disclosure statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Cisplatin resistance of TAP and TAP‐YB‐1 clones.
Fig. S2. Depletion of PABPC1, MRPS7, and RB1CC1 with specific siRNA molecules in MCF7 cells.
Fig. S3. Response of transfected MCF7 cells to cisplatin.
Fig. S4. Response of transfected MDA‐MB‐231 cells to cisplatin.
Fig. S5. Examples of FACS cell cycle analysis of MDA‐MB‐231 cells transfected with an empty vector (left) and a YB‐1 expression vector (right).
Table S1. List of proteins potentially interacting with YB‐1, their chromosomal localization, and expression levels in breast cancer.
Table S2. Sequence of each siRNA used to establish dose–response curves.
Data S1.Materials and methods.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Acknowledgments
The authors are grateful to Nancy Roberge (Centre de Recherche en Cancérologie, Quebec City, QC, Canada) for FACS analyses and to Meraj Aziz (Translational Genomics Research Institute) for analysis of the dose–response curves. This work was supported, in part, by the Cancer Research Society, Inc. and the Canadian Institutes of Health Research (to ML). ML is a senior scholar from the Fonds de la Recherche en Santé du Québec (FRSQ). SPT is a scholar of the Quebec‐Clinical Research Organization in Cancer Consortium, financed by the Pfizer‐FRSQ Innovation Award.
References
- 1. Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res 2001; 478: 23–43. [DOI] [PubMed] [Google Scholar]
- 2. Janz M, Harbeck N, Dettmar P et al. Y‐box factor YB‐1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI‐1. Int J Cancer 2002; 97: 278–82. [DOI] [PubMed] [Google Scholar]
- 3. Swamynathan SK, Nambiar A, Guntaka RV. Role of single‐stranded DNA regions and Y‐box proteins in transcriptional regulation of viral and cellular genes. FASEB J 1998; 12: 515–22. [DOI] [PubMed] [Google Scholar]
- 4. Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB‐1 binds A/C‐rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J 2001; 20: 3821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ashizuka M, Fukuda T, Nakamura T et al. Novel translational control through an iron‐responsive element by interaction of multifunctional protein YB‐1 and IRP2. Mol Cell Biol 2002; 22: 6375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evdokimova V, Ruzanov P, Anglesio MS et al. Akt‐mediated YB‐1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol 2006; 26: 277–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohga T, Uchiumi T, Makino Y et al. Direct involvement of the Y‐box binding protein YB‐1 in genotoxic stress‐induced activation of the human multidrug resistance 1 gene. J Biol Chem 1998; 273: 5997–6000. [DOI] [PubMed] [Google Scholar]
- 8. Bargou RC, Jurchott K, Wagener C et al. Nuclear localization and increased levels of transcription factor YB‐1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med 1997; 3: 447–50. [DOI] [PubMed] [Google Scholar]
- 9. Rubinstein DB, Stortchevoi A, Boosalis M, Ashfaq R, Guillaume T. Overexpression of DNA‐binding protein B gene product in breast cancer as detected by in vitro‐generated combinatorial human immunoglobulin libraries. Cancer Res 2002; 62: 4985–91. [PubMed] [Google Scholar]
- 10. Yahata H, Kobayashi H, Kamura T et al. Increased nuclear localization of transcription factor YB‐1 in acquired cisplatin‐resistant ovarian cancer. J Cancer Res Clin Oncol 2002; 128: 621–6. [DOI] [PubMed] [Google Scholar]
- 11. Oda Y, Ohishi Y, Saito T et al. Nuclear expression of Y‐box‐binding protein‐1 correlates with P‐glycoprotein and topoisomerase II alpha expression, and with poor prognosis in synovial sarcoma. J Pathol 2003; 199: 251–8. [DOI] [PubMed] [Google Scholar]
- 12. Koike K, Uchiumi T, Ohga T et al. Nuclear translocation of the Y‐box binding protein by ultraviolet irradiation. FEBS Lett 1997; 417: 390–4. [DOI] [PubMed] [Google Scholar]
- 13. Hayakawa H, Uchiumi T, Fukuda T et al. Binding capacity of human YB‐1 protein for RNA containing 8‐oxoguanine. Biochemistry 2002; 41: 12. [DOI] [PubMed] [Google Scholar]
- 14. Ise T, Nagatani G, Imamura T et al. Transcription factor Y‐box binding protein 1 binds preferentially to cisplatin‐modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res 1999; 59: 342–6. [PubMed] [Google Scholar]
- 15. Gaudreault I, Guay D, Lebel M. YB‐1 promotes strand separation of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities, and binds several DNA repair proteins. Nucleic Acids Res 2004; 32: 316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izumi H, Imamura T, Nagatani G et al. Y box‐binding protein‐1 binds preferentially to single‐stranded nucleic acids and exhibits 3′→5′ exonuclease activity. Nucleic Acids Res 2001; 29: 1200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marenstein DR, Ocampo MT, Chan MK et al. Stimulation of human endonuclease III by Y box‐binding protein 1 (DNA‐binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J Biol Chem 2001; 276: 21. [DOI] [PubMed] [Google Scholar]
- 18. Guay D, Garand C, Reddy S, Schmutte C, Lebel M. The human NTH1 enzyme is a relevant target to potentiate cisplatin cytotoxicity in YB‐1 overexpressing tumor cells. Cancer Sci 2008; 99: 762–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Di Cresce C, Koropatnick J. Antisense treatment in human prostate cancer and melanoma. Curr Cancer Drug Targets 2010; 10: 555–65. [DOI] [PubMed] [Google Scholar]
- 20. de Souza‐Pinto NC, Mason PA, Hashiguchi K et al. Novel DNA mismatch‐repair activity involving YB‐1 in human mitochondria. DNA Repair 2009; 8: 704–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rhodes DR, Yu J, Shanker K et al. ONCOMINE: a cancer microarray database and integrated data‐mining platform. Neoplasia 2004; 6: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang J, Tan PH, Li KB, Matsumoto K, Tsujimoto M, Bay BH. Y‐box binding protein, YB‐1, as a marker of tumor aggressiveness and response to adjuvant chemotherapy in breast cancer. Int J Oncol 2005; 26: 607–13. [PubMed] [Google Scholar]
- 23. Habibi G, Leung S, Law JH et al. Redefining prognostic factors for breast cancer: YB‐1 is a stronger predictor of relapse and disease‐specific survival than estrogen receptor or HER‐2 across all tumor subtypes. Breast Cancer Res 2008; 10: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dahl E, En‐Nia A, Wiesmann F et al. Nuclear detection of Y‐box protein‐1 (YB‐1) closely associates with progesterone receptor negativity and is a strong adverse survival factor in human breast cancer. BMC Cancer 2009; 9: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gluz O, Mengele K, Schmitt M et al. Y‐box‐binding protein YB‐1 identifies high‐risk patients with primary breast cancer benefiting from rapidly cycled tandem high‐dose adjuvant chemotherapy. J Clin Oncol 2009; 27: 6144–51. [DOI] [PubMed] [Google Scholar]
- 26. Fujita T, Ito K, Izumi H et al. Increased nuclear localization of transcription factor Y‐box binding protein 1 accompanied by up‐regulation of P‐glycoprotein in breast cancer pretreated with paclitaxel. Clin Cancer Res 2005; 11: 8837–44. [DOI] [PubMed] [Google Scholar]
- 27. To K, Fotovati A, Reipas KM et al. Y‐box binding protein‐1 induces the expression of CD44 and CD49f leading to enhanced self‐renewal, mammosphere growth, and drug resistance. Cancer Res 2010; 70: 2840–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang JY, Ha SA, Yang YS, Kim JW. p‐Glycoprotein ABCB5 and YB‐1 expression plays a role in increased heterogeneity of breast cancer cells: correlations with cell fusion and doxorubicin resistance. BMC Cancer 2010; 10: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guay D, Evoy AA, Paquet E et al. The strand separation and nuclease activities associated with YB‐1 are dispensable for cisplatin resistance but overexpression of YB‐1 in MCF7 and MDA‐MB‐231 breast tumor cells generates several chemoresistance signatures. Int J Biochem Cell Biol 2008; 40: 2492–507. [DOI] [PubMed] [Google Scholar]
- 30. Davydova EK, Evdokimova VM, Ovchinnikov LP, Hershey JW. Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res 1997; 25: 2911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evdokimova V, Tognon C, Ng T et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB‐1 promotes an epithelial–mesenchymal transition. Cancer Cell 2009; 15: 402–15. [DOI] [PubMed] [Google Scholar]
- 32. Evdokimova V, Tognon C, Ng T, Sorensen PH. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB‐1. Cell Cycle 2009; 8: 2901–6. [DOI] [PubMed] [Google Scholar]
- 33. LaRue KE, Khalil M, Freyer JP. Microenvironmental regulation of proliferation in multicellular spheroids is mediated through differential expression of cyclin‐dependent kinase inhibitors. Cancer Res 2004; 64: 1621–31. [DOI] [PubMed] [Google Scholar]
- 34. Xing H, Wang S, Hu K et al. Effect of the cyclin‐dependent kinases inhibitor p27 on resistance of ovarian cancer multicellular spheroids to anticancer chemotherapy. J Cancer Res Clin Oncol 2005; 131: 511–9. [DOI] [PubMed] [Google Scholar]
- 35. Watanabe M, Furuno N, Goebl M et al. Molecular cloning of the human gene, CCG2, that complements the BHK‐derived temperature‐sensitive cell cycle mutant tsBN63: identity of CCG2 with the human X chromosomal SCAR/RPS4X gene. J Cell Sci 1991; 100: 35–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Cisplatin resistance of TAP and TAP‐YB‐1 clones.
Fig. S2. Depletion of PABPC1, MRPS7, and RB1CC1 with specific siRNA molecules in MCF7 cells.
Fig. S3. Response of transfected MCF7 cells to cisplatin.
Fig. S4. Response of transfected MDA‐MB‐231 cells to cisplatin.
Fig. S5. Examples of FACS cell cycle analysis of MDA‐MB‐231 cells transfected with an empty vector (left) and a YB‐1 expression vector (right).
Table S1. List of proteins potentially interacting with YB‐1, their chromosomal localization, and expression levels in breast cancer.
Table S2. Sequence of each siRNA used to establish dose–response curves.
Data S1.Materials and methods.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


