Abstract
To identify differentially regulated molecules related to early and late stages of tumor promotion in a rat two‐stage thyroid carcinogenesis model by an antithyroid agent, sulfadimethoxine, microarray‐based microdissected lesion‐specific gene expression profiling was carried out. Proliferative lesions for profiling were divided into two categories: (i) focal follicular cell hyperplasias (FFCH) and adenomas (Ad) as early lesions; and (ii) carcinomas (Ca) as more advanced. In both cases, gene expression was compared with that in surrounding non‐tumor follicular cells. Characteristically, upregulation of cell cycle‐related genes in FFCH + Ad, downregulation of genes related to tumor suppression and transcription inhibitors of inhibitor of DNA binding (Id) family proteins in Ca, and upregulation of genes related to cell proliferation and tumor progression in common in FFCH + Ad and Ca, were detected. The immunohistochemical distributions of molecules included in the altered expression profiles were further examined. In parallel with microarray data, increased localization of ceruloplasmin, cyclin B1, and cell division cycle 2 homolog A, and decreased localization of poliovirus receptor‐related 3 and Id3 were observed in all types of lesion. Although inconsistent with the microarray data, thyroglobulin immunoreactivity appeared to reduce in Ca. The results thus suggest cell cycling facilitation by induction of M‐phase‐promoting factor consisting of cyclin B1 and cell division cycle 2 homolog A and generation of oxidative responses as evidenced by ceruloplasmin accumulation from an early stage, as well as suppression of cell adhesion involving poliovirus receptor‐related 3 and inhibition of cellular differentiation regulated by Id3. Decrease of thyroglobulin in Ca may reflect dedifferentiation with progression. (Cancer Sci 2009; 100: 617–625)
Abbreviations:
- Ad
adenoma
- Ca
carcinoma
- Ccnb1
cyclin B1
- Cdc2
cell division cycle 2
- DHPN
N‐bis(2‐hydroxypropyl)nitrosamine
- FFCH
focal follicular cell hyperplasia
- Id
inhibitor of DNA binding
- IGSF
immunoglobulin superfamily
- NTF
non‐tumor follicles
- PCR
polymerase chain reaction
- Pvrl3
poliovirus receptor‐related 3
- RT
reverse transcription
- SDM
sulfadimethoxine
- TSH
thyroid‐stimulating hormone
Although clinically recognized thyroid Ca constitute less than 1% of all human malignant tumors, it is the most common endocrine cancer (90% of cases) and is responsible for more deaths than all other endocrine cancers combined.( 1 ) Ca of the thyroid is usually of follicular cell origin, but the medullary carcinoma arises from parafollicular or C cells. In humans, causative factors for thyroid Ca are not well understood except for secondary occurrence after radiotherapy.( 2 ) In rats, on the other hand, thyroid follicular cell tumors can be produced by administration of antithyroid agents, such as by propylthiouracil,( 3 ) methimazole,( 4 ) and 3‐amino‐1,2,4‐triazole,( 5 ) in an initiation‐promotion model.
Many chemicals that can induce thyroid tumors in rodents cause disruption of the thyroid–pituitary axis through induction of hypothyroidism.( 6 ) The putative mechanism for this carcinogenesis is believed to be non‐genotoxic, decrease in the serum levels of triiodothyronine and thyroxine causing suppression of negative feedback through the pituitary and an increase in serum TSH. TSH then stimulates thyroid functions, including growth and proliferation of follicular cells.( 7 , 8 ) However, detailed molecular mechanisms remain to be resolved.( 6 )
SDM is a broad‐spectrum antimicrobial sulfonamide that has been shown to effectively induce thyroid follicular cell tumors in a rat two‐stage thyroid carcinogenesis model after initiation with DHPN.( 9 ) The anti‐thyroidal effects of this drug are mediated through inhibition of iodination reactions catalyzed by thyroid peroxidase, resulting in reduction of thyroid hormone synthesis and increased levels of TSH in the bloodstream.( 10 )
Histological lesion‐specific gene expression profiling provides valuable information on the mechanisms underlying lesion development. We have established molecular analysis methods for DNA, RNA, and proteins in paraffin‐embedded small‐tissue specimens utilizing an organic solvent‐based fixative, methacarn,( 11 , 12 , 13 ) and applied them for analyses of microdissected lesions.( 14 , 15 ) With regard to mRNA expression analysis, expression fidelity in the methacarn‐fixed paraffin‐embedded tissues was found to be very close to that in the unfixed frozen tissues in both the real‐time RT‐PCR and oligonucleotide microarray systems, suggesting a great advantage of methacarn in analyses of microdissected lesions after paraffin embedding.( 14 , 15 )
In the present study, to identify differentially regulated molecules related to thyroid carcinogenesis through hypothyroidism, we carried out global gene expression profiling of early and late‐stage proliferative lesions obtained after promotion with SDM in a rat two‐stage carcinogenesis model. Localization of representative molecules showing altered expression was further analyzed immunohistochemically.
Materials and Methods
Chemicals and animals. DHPN (CAS no. 53609‐64‐6) and SDM (CAS no. 122‐11‐2) were purchased from Nacalai Tesque (Kyoto, Japan) and Sigma (St Louis, MO, USA), respectively. Male 5‐week‐old F344 rats were purchased from Japan SLC (Hamamatsu, Japan) and housed four to five rats per polycarbonate cage with sterilized softwood chips as bedding in a barrier‐sustained animal room conditioned at 24 ± 1°C and 55 ± 5% humidity, with a 12:12 h L : D cycle. They received CRF‐1 (Oriental Yeast Co., Tokyo, Japan) as a basal diet and water ad libitum throughout the experimental period, including the 1 week of acclimation.
Experimental design. At 6 weeks of age, 30 rats were injected subcutaneously with 2800 mg/kg body weight DHPN. Another group of nine animals was injected with the vehicle saline as non‐treated controls. One week later, 25 DHPN‐initiated animals were administered SDM at 1000 p.p.m. in the drinking water ad libitum for up to 15 weeks. The other five DHPN‐initiated animals were maintained on tap water for 10 weeks as a DHPN‐alone group. At week 10 after SDM treatment, 10 animals were killed for microdissection of FFCH + Ad as well as NTF in each animal. The other SDM‐promoted animals were further maintained until week 15, when 12 rats were killed for microdissection of Ca. Five untreated controls and the DHPN‐alone group were killed at week 10 of SDM promotion, and the four remaining untreated controls at week 15. All animals were killed by exsanguination from the abdominal aorta under deep anesthesia with ether. The animal protocol was reviewed and approved by the Animal Care and Use Committee of the National Institute of Health Sciences, Japan.
Preparation of tissue specimens and microdissection. Caudal halves of the bilateral thyroid tissues of SDM‐promoted animals were immersed in methacarn solution for 2 h at 4°C.( 11 ) Tissue samples were then dehydrated, immersed in xylene, and embedded in paraffin as described previously.( 15 ) Embedded tissue blocks were stored at 4°C until microdissection.( 16 )
For microarray analysis, 4 µm‐thick sections between 10 16 µm‐thick serial sections were prepared. The 16 µm‐thick sections were mounted onto PEN‐foil film (Leica Microsystems, Welzlar, Germany) overlaid on glass slides, dried in an incubator overnight at 37°C, and then stained using an LCM staining kit (Ambion, Austin, TX, USA). All microdissections were carried out within 2 h of tissue staining. The histopathological identity of each FFCH, Ad, and Ca, as well as NTF, was determined under microscopic observation of the adjacent 4 µm‐thick sections stained with hematoxylin–eosin according to published criteria (Fig. 1).( 17 ) In the present carcinogenesis model with SDM promotion, capsular invasive carcinomas are generated in addition to less‐frequent parenchymal Ca.( 18 ) In the present study, parenchymal proliferative lesions including the latter were subjected to laser microbeam microdissection (Leica Microsystems). Approximately 20 sections of bilateral thyroids were subjected to microdissection in one animal, and the microdissected samples (NTF, FFCH + Ad, and Ca) were collected and stored in separate 1.5‐mL sample tubes at –80°C until extraction of total RNA.
Figure 1.
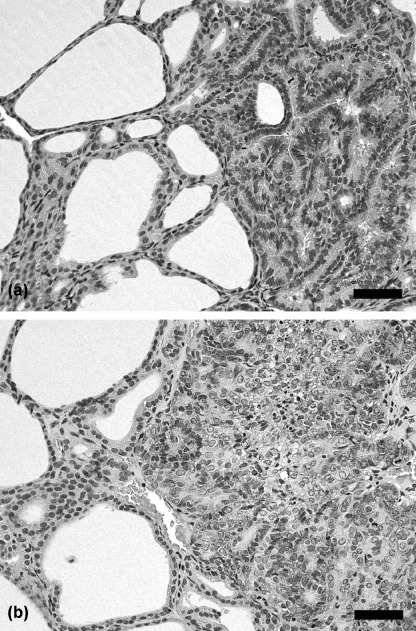
Representative proliferative lesions developing after promotion with sulfadimethoxine (SDM) for (a) 10 or (b) 15 weeks in a rat two‐stage thyroid carcinogenesis model. (a) Adenoma with follicular features, showing expansive growth with minor atypia. (b) Carcinoma showing obvious cellular atypia, consisting of follicular and solid growth elements with structural irregularity. Note focal necrosis and fibrosis. Hematoxylin–eosin staining. Scale bars = 50 µm.
RNA isolation, amplification, and microarray analysis. Total RNA extraction from each histological sample and quantitation of the RNA yield were carried out according to methods described previously.( 15 )
For microarray analysis, equal amounts of extracted total RNA samples from two animals were mixed (100 ng/sample) and subjected to amplification, consisting of RT and subsequent two‐step in vitro transcription, using a MessageAmp II aRNA Kit (Ambion).
Second‐round‐amplified biotin‐labeled antisense RNA was subjected to hybidization with a GeneChip Rat Genome 230 2.0 Array (Affymetrix Inc., Santa Clara, CA, USA). RNA samples collected from two animals were subjected to analysis with individual microarrays (n = 5/histological preparation).
Selection of genes and normalization of expression data were carried out using GeneSpring software (ver7.2; Silicon Genetics, Redwood City, CA, USA). To normalize chip‐wide variation in intensity, per chip normalization was performed by dividing the signal strength for each gene with the level of the 50th percentile of the measurement in the chip, and dividing the value by the average intensity in the samples of NTF. Genes showing signals judged to be ‘absent’ in all 10 samples of NTF and each proliferative lesion group (FFCH + Ad or Ca) for comparison were excluded. Then, genes showing expression change with differences at least 2‐fold in magnitude in the proliferative lesion groups from the NTF, as well as the ‘presence’ signal in more than four of five samples in the histological lesion group (NTF or each proliferative lesion group) showing higher expression values in comparison, were selected. Genes showing altered expression in common in both FFCH + Ad and Ca were also selected.
Real‐time RT‐PCR. Quantitative real‐time RT‐PCR was carried out for confirmation of expression values obtained with microarrays using an ABI Prism 7900HT (Applied Biosystems, Foster City, CA, USA). The following 11 genes (eight upregulated and three downregulated in proliferative lesions) were selected as targets: chitinase 3‐like 1, ceruloplasmin, solute carrier family 2 (facilitated glucose transporter) member 3, solute carrier family 16 (monocarboxylic acid transporters) member 6, glucagon, prolactin receptor, phosphatidylinositol 4‐kinase type 2α, and actinin α1 as upregulated examples; and Pvrl3, retinoic acid induced 3, and glucosaminyl (N‐acetyl) transferase 1 core 2 as downregulated examples. RT was carried out using first‐round antisense RNA prepared for microarray analysis. Real‐time PCR analysis of ceruloplasmin, glucagon, and glucosaminyl (N‐acetyl) transferase 1 core 2 was carried out using ABI Assays‐on‐Demand TaqMan probe and primer sets from Applied Biosystems (available at https://products.appliedbiosystems.com/ab/en/US/adirect/ab?cmd=catNavigate2&catID=601267) (n = 5/histological preparation). For measurement of transcript levels of chitinase 3‐like 1, solute carrier family 2 (facilitated glucose transporter) member 3, solute carrier family 16 (monocarboxylic acid transporters) member 6, prolactin receptor, phosphatidylinositol 4‐kinase type 2α, actinin α1, Pvrl3, and retinoic acid induced 3, primer sets were designed using Primer Express software (Version 2.0; Applied Biosystems), and the corresponding primer sequences are shown in Table 1. Amplified transcript levels were measured with the SYBR Green detection system (n = 5/histological preparation). For quantification of expression data, a standard curve method was applied using the first‐round antisense RNA prepared for microarray analysis from NTF as a standard sample. Expression values were normalized to two housekeeping genes, glyceraldehyde 3‐phosphate dehydrogenase and hypoxanthine‐guanine phosphoribosyltransferase, as described previously.( 14 )
Table 1.
Sequences of primers used for real‐time reverse transcription–polymerase chain reaction with the SYBR Green detection system
| Gene | Accession no. | Sense/Antisense | Sequence |
|---|---|---|---|
| Chi3l1 | AA945643 | Sense | 5′‐TCGTTAACAGGGATGACCTGTATCT‐3′ |
| Antisense | 5′‐GGGTAGGACGGTGGGATTGT‐3′ | ||
| Slc2a3 | AA901341 | Sense | 5′‐AAGCTGGCCATTGGCAAAT‐3′ |
| Antisense | 5′‐CTAGCCTCTTGTGCTCCTCCAT‐3′ | ||
| Slc16a6 | AA859652 | Sense | 5′‐AAAGGTGTTTCGACTGCATTCTC‐3′ |
| Antisense | 5′‐CCCCATGTACCAAGCACTGTT‐3′ | ||
| Prlr | AW142962 | Sense | 5′‐TGTCGCATAAGGTCCCCTCTT‐3′ |
| Antisense | 5′‐GCTTGGCAATTTGTAGGGAAAG‐3′ | ||
| Pi4KII | BE097981 | Sense | 5′‐CCCCTTTCTCTTTCCTTCTGGTA‐3′ |
| Antisense | 5′‐ACAGCAAGTTCCAGGACAGTCA‐3′ | ||
| Actn1 | BE119221 | Sense | 5′‐AAGAAGGCGGTGTCTGTAAGCT‐3′ |
| Antisense | 5′‐CCGTCCCTTGGCTTTGAA‐3′ | ||
| Pvrl3 | AW525315 | Sense | 5′‐GGCAAGACACTGGTTCTACACAAT‐3′ |
| Antisense | 5′‐AAGGCCCGAAGAATGTTTTTC‐3′ | ||
| Rai3 | BI276110 | Sense | 5′‐GGAGCAAGTGCCAGGAATTTAT‐3′ |
| Antisense | 5′‐CAGTTTTTTCCAGCCAGGAGAA‐3′ |
Actn1, actinin α1; Chi3l1, chitinase 3‐like 1; Pi4KII, phosphatidylinositol 4‐kinase type 2α; Prlr, prolactin receptor; Pvrl3, poliovirus receptor‐related 3; Rai3, retinoic acid induced 3; Slc2a3, solute carrier family 2 (facilitated glucose transporter) member 3; Slc16a6, solute carrier family 16 (monocarboxylic acid transporters) member 6.
Immunohistochemistry. The cranial halves of the bilateral thyroids of SDM‐promoted animals were subjected to fixation in 10% phosphate‐buffered formalin (pH 7.4) solution for 2 days at room temperature, and prepared for histopathological examination. In untreated controls and DHPN‐alone cases, whole thyroid tissue was fixed in buffered formalin and prepared similarly.
Immunohistochemistry was carried out with antibodies against ceruloplasmin (clone 8, mouse IgG1, 1:50; BD Transduction Laboratories, San Jose, CA, USA), Ccnb1 (clone V152, mouse IgG1, 1:100; Thermo Fisher Scientific Inc., Fremont, CA, USA), Cdc2 (clone A17, mouse IgG2a, 1:200; GeneTex, San Antonio, TX, USA), thyroglobulin (clone SPM221, mouse IgG1, 1:250; Spring Bioscience, Fremont, CA, USA), Pvrl3 (goat IgG, 1:100; R&D Systems, Minneapolis, MN, USA), and Id3 (rabbit IgG, 1:100; ProteinTech Group, Chicago, IL, USA). For confirmation of positive immunoreactivity of antigens examined, normal rat tissues, such as the liver for ceruloplasmin,( 19 ) duodenal mucosa for Ccnb1 and Cdc2, and thyroid for thyroglobulin,( 20 ) Pvrl3, and Id3, were used. For each antigen, subcellular or extracellular localization was examined and compared to the cases previously reported.( 21 , 22 , 23 , 24 , 25 , 26 ) Optimal conditions for antigen retrieval were also determined using positive control tissues. For antigen retrieval, deparaffinized sections were heated in 10 mM citrate buffer (pH 6.0) by autoclaving for 10 min before incubation with the Ccnb1 and Pvrl3 antibodies, or for 20 min before incubation for ceruloplasmin, Cdc2, and Id3. No antigen retrieval treatment was carried out for thyroglobulin. Immunodetection was carried out utilizing a Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA, USA), with 3,3′‐diaminobenzidine/H2O2 as the chromogen, as described previously.( 15 ) As negative controls for immunoreactivity, normal serum from mouse, goat, or rabbit was applied to rat positive control tissues with appropriate dilutions instead of the primary antibodies. Sections were counterstained with hematoxylin.
Analysis of immunoreactivity. For all antigens examined in the present study, immunoreactivity was essentially unaltered in the thyroids between the groups of untreated controls and DHPN alone, and these animals did not develop any proliferative lesion. Therefore, immunoreactivity scores in these groups were counted together and represented as ‘normal follicular cells’ with three randomly selected microscopic areas at 200‐fold magnification in one animal. Animals examined for ‘normal follicular cells’ were nine untreated controls and five treated with DHPN. In the SDM‐promoted cases, immunoreactivity scores were counted in the histological categories of NTF, FFCH + Ad, and Ca. With regard to NTF, three randomly selected microscopic areas at 200‐fold magnification were subjected to evaluation for each animal. With Ca, both capsular invasive and parenchymal Ca were analyzed. Immunoreactivity in each histological type was not essentially different between the cases after 10 and 15 weeks of promotion, and therefore immunolocalization scores were counted together for these two time points. The numbers of animals examined for proliferative lesions and surrounding NTF were 10 and 11 after 10 and 15 weeks of promotion, respectively.
For ceruloplasmin, Ccnb1, Cdc2, Id3, and thyroglobulin, immunolocalization was scored as 0 (negative), 1 (slight), 2 (moderate), or 3 (prominent). In the case of Pvrl3, scores were 0 (negative), 1 (partially positive), or 2 (positive). Detailed criteria for the immunoreactivity for each molecule given in Table 2 were determined after due consultation of two independent pathologists. Immunoreactivity score in each lesion was double‐checked by one pathologist and then cross‐checked by another pathologist.
Table 2.
Scoring criteria for immunohistochemical localization
| Molecule | Immunolocalization for evaluation | Score of immunoreactivity | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| Ceruloplasmin | Luminal cellular surface | — | Weakly positive, a few follicles | Strongly positive, focal follicular populations | Strongly positive, majority of follicles |
| Ccnb1 | Cytoplasm | — | Weakly positive, a few cells | Weakly positive, majority of cells | Strongly positive, majority of cells |
| Cdc2 | Cytoplasm and nucleus | — | 1–10 cells/400× field | 10–30 cells/400x field | > 30 cells/400× field |
| Thyroglobulin | Cytoplasm | — | < 20% cells | 20–70% cells | > 70% cells |
| Pvrl3 | Intercellular membrane | — | Focally positive | Entirely positive | Not applied |
| Id3 | Nucleus | — | Weakly positive, a few cells | Weakly positive, majority of cells | Strongly positive, majority of cells |
Ccnb1, Cyclin B1; Cdc2, cell division cycle 2; Id3, inhibitor of DNA binding 3; Pvrl3, poliovirus receptor‐related 3.
Data analysis. Expression values from the real‐time RT‐PCR were analyzed by Student's or Welch's t‐test following a test for equal variance. Scores for immunoreactivity were assessed with Mann–Whitney's U‐test, comparing NTF and normal follicles, FFCH + Ad or Ca. For the microarray data, the statistical analysis was carried out with GeneSpring software, and the significance of gene expression changes was analyzed by Student's t‐test or ANOVA between NTF and normal follicles, FFCH + Ad or Ca.
Results
Microarray analysis. As genes showing altered expression specifically in FFCH + Ad, 40 examples were upregulated and 20 examples were downregulated, as compared with surrounding NTF (Table 3; Supporting Tables S1, S2). In the Ca cases, the numbers of genes specifically upregulated and downregulated were 69 and 142, respectively. Representative genes with known annotations associated with carcinogenesis are listed in Table 3. Interestingly, a cluster of cell cycle‐related genes were found to be upregulated specifically in FFCH + Ad, such as ubiquitin‐like with PHD and RING finger domains 1, kinesin family member 23, cyclin A2, M‐phase phosphoprotein, topoisomerase (DNA) 2α, Cdc2 homolog A, Ccnb1, and cyclin‐dependent kinase inhibitor 3. Extracellular matrix proteins, laminin γ2, and fibronectin 1 also showed upregulation in FFCH + Ad. No particular functional cluster was observed for downregulated genes in FFCH + Ad cases. Among the upregulated genes in Ca, examples with functions in transport (ceruloplasmin) and biosynthesis (thyroglobulin) were found, whereas downregulated genes typically involved functions in tumor suppression, such as: decorin, reversion‐inducing‐cysteine‐rich protein with kazal motifs, creatine kinase mitochondrial 1 ubiquitous, retinoblastoma 1, lysyl oxidase, and NAD(P)H dehydrogenase quinine 1. Transcript levels for genes encoding signal transduction molecules and transcription factors were also downregulated in Ca. All isoforms of Id were found to be reduced.
Table 3.
List of representative genes with known functional annotations associated with carcinogenesis showing altered expression specifically in thyroid proliferative lesions of each category induced in rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold) †
| Gene function | Accession no. | Gene title | Symbol | FFCH + Ad | Ca |
|---|---|---|---|---|---|
| FFCH + Ad | |||||
| Upregulated genes (of 40 genes in total) | |||||
| Cell cycle | BE098732 | Ubiquitin‐like, containing PHD and RING finger domains, 1 | Uhrf1 | 2.43 | 1.81 |
| Cell cycle | BE113443 | Kinesin family member 23 ‡ | Kif23 | 2.20 | 1.09 |
| Cell cycle | AA998516 | Cyclin A2 | Ccna2 | 2.13 | 1.29 |
| Cell cycle | BM385445 | Topoisomerase (DNA) 2α | Top2a | 2.12 | 1.37 |
| Cell cycle | BE110723 | M‐phase phosphoprotein 1 ‡ | Mphosph1 | 2.09 | 1.35 |
| Cell cycle | NM_019296 | Cell division cycle 2 homolog A (S. pombe) | Cdc2a | 2.04 | 1.45 |
| Cell cycle | X64589 | Cyclin B1 | Ccnb1 | 2.01 | 1.40 |
| Cell cycle | BE113362 | Cyclin‐dependent kinase inhibitor 3 ‡ | Cdkn3 | 1.97 | 1.18 |
| Metastasis | BM385282 | Laminin, γ2 | Lamc2 | 2.90 | 1.25 |
| Metastasis | AA893484 | Fibronectin 1 | Fn1 | 2.16 | 1.40 |
| Downregulated genes (of 20 genes in total) | |||||
| Metastasis | BE117767 | Immunoglobulin superfamily, member 4A ‡ | Igsf4a | 0.46 | 0.73 |
| Cell differentiation | BG666709 | N‐myc downstream regulated 4 | Ndr4 | 0.50 | 0.51 |
| Ca | |||||
| Upregulated genes (of 69 genes in total) | |||||
| Biosynthesis | AI500952 | Thyroglobulin | Tg | 1.72 | 2.65 |
| Transport | AF202115 | Ceruloplasmin | Cp | 1.99 | 2.45 |
| Transport | BE106526 | Solute carrier family 6 (neurotransmitter transporter, GABA), member 11 | Slc6a11 | 1.44 | 1.98 |
| Cell growth | M57668 | Prolactin receptor | Prlr | 1.78 | 2.42 |
| Cell cycle | NM_133578 | Dual specificity phosphatase 5 | Dusp5 | 1.74 | 2.02 |
| Proto‐oncogene | NM_012874 | v‐ros UR2 sarcoma virus oncogene homolog 1 (avian) | Ros1 | 1.59 | 1.98 |
| Metastatic regulator | AI175048 | Sine oculis homeobox homolog 1 (Drosophila) | Six1 | 1.62 | 1.96 |
| Glycolysis | BI294137 | Hexokinase 2 | Hk2 | 1.20 | 1.87 |
| Downregulated genes (of 142 genes in total) | |||||
| Tumor suppressor | BM390253 | Decorin | Dcn | 0.76 | 0.27 |
| Tumor suppressor | AW523759 | Reversion‐inducing‐cysteine‐rich protein with kazal motifs ‡ | Reck | 0.65 | 0.32 |
| Tumor suppressor | BI301453 | Creatine kinase, mitochondrial 1, ubiquitous | Ckmt1 | 0.49 | 0.35 |
| Tumor suppressor | AI178012 | Retinoblastoma 1 | Rb1 | 0.75 | 0.38 |
| Tumor suppressor | NM_017061 | Lysyl oxidase | Lox | 0.81 | 0.39 |
| (BI304009) | (0.91) | (0.49) | |||
| Tumor suppressor | J02679 | NAD(P)H dehydrogenase, quinone 1 | Nqo1 | 0.57 | 0.42 |
| Signal transduction | U78517 | cAMP‐regulated guanine nucleotide exchange factor II | Rapgef4 | 0.65 | 0.32 |
| Signal transduction | AA945708 | Calcitonin receptor‐like | Calcrl | 0.59 | 0.34 |
| Signal transduction | BI295477 | G protein‐coupled receptor 116 | Gpr116 | 0.61 | 0.45 |
| Signal transduction | NM_030829 | G protein‐coupled receptor kinase 5 | Gprk5 | 0.66 | 0.48 |
| Cell adhesion | NM_031050 | Lumican | Lum | 0.99 | 0.33 |
| Transcription | AF000942 | Inhibitor of DNA binding 3, dominant negative helix‐loop‐helix protein | Id3 | 0.57 | 0.33 |
| Transcription | BE116009 | Inhibitor of DNA binding 4 | Idb4 | 0.56 | 0.40 |
| Transcription | NM_053713 | Kruppel‐like factor 4 (gut) | Klf4 | 0.67 | 0.40 |
| Transcription | M86708 | Inhibitor of DNA binding 1, helix‐loop‐helix protein (splice variation) | Id1 | 0.64 | 0.44 |
| Transcription | AI008792 | Inhibitor of DNA binding 2, dominant negative helix‐loop‐helix protein | Id2 | 0.68 | 0.48 |
| Metastasis suppressor | AI578087 | transmembrane 4 superfamily member 1 ‡ | Tm4sf1 | 0.64 | 0.36 |
| (AW435343) | (0.74) | (0.48) | |||
| Apoptosis | AA892770 | Glutamate‐cysteine ligase, catalytic subunit | Gclc | 0.61 | 0.41 |
| Tumor metastasis | NM_133526 | Transmembrane 4 superfamily member 3 | Tm4sf3 | 0.70 | 0.41 |
Ad, adenoma; Ca, carcinoma; FFCH, focal follicular cell hyperplasias.
Proliferative lesions were divided into two categories, i.e. FFCH + Ad and Ca.
Predicted gene identity.
With regard to genes showing altered expression in common in FFCH + Ad and Ca, totals of 93 and 53 were upregulated and downregulated, respectively (Table 4; Supporting Table S3). Upregulated genes included examples linked to transport, cell proliferation, and tumor progression. In particular, multiple gene probes in the array showed increased signals for ceruloplasmin and solute carrier family 16 (monocarboxylic acid transporters) member 6. Among the genes that showed downregulation, no particular functional clusters were apparent. Two gene probes for Pvrl3 in the array demonstrated downregulation. Real‐time RT‐PCR for validation of microarray data was carried out for 11 genes showing commonly altered expression with FFCH + Ad and Ca, eight upregulated and three downregulated, the results being summarized in Table 5. In both FFCH + Ad and Ca, many expression changes were similar with the two analysis systems, despite a lower magnitude of alteration observed with PCR data for the upregulated genes when the values were normalized to hypoxanthine‐guanine phosphoribosyltransferase levels. With regard to downregulated genes, variability of PCR data in the NTF after normalization to the hypoxanthine‐guanine phosphoribosyltransferase level was slightly higher than with glyceraldehyde 3‐phosphate dehydrogenase (data not shown), and therefore statistical significance was not attained for FFCH + Ad.
Table 4.
List of representative genes with known functional annotations associated with carcinogenesis showing altered expression in common with all types of thyroid proliferative lesions induced in rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold) †
| Gene function | Accession no. | Gene title | Symbol | FFCH + Ad | Ca |
|---|---|---|---|---|---|
| Upregulated genes (of 93 genes in total) | |||||
| Adhesion | AA945643 | Chitinase 3‐like 1 | Chi3l1 | 7.46 | 8.55 |
| Adhesion | AI169104 | Platelet factor 4 | Pf4 | 2.52 | 3.71 |
| Angiogenesis | NM_021751 | Prominin 1 | Prom1 | 4.26 | 5.53 |
| Transport | NM_012532 | Ceruloplasmin | Cp | 3.32 | 4.12 |
| (AF202115) | (2.43) | (2.79) | |||
| Transport | AA901341 | Solute carrier family 2 (facilitated glucose transporter), member 3 | Slc2a3 | 2.68 | 2.53 |
| Transport | AA859652 | Solute carrier family 16 (monocarboxylic acid transporters), member 6 | Slc16a6 | 2.28 | 2.33 |
| (BG372184) | (2.25) | (2.24) | |||
| Cell proliferation | AF411318 | Metallothionein | Mt1a | 2.11 | 3.77 |
| Cell proliferation | AA819913 | Carbohydrate (keratan sulfate Gal‐6) sulfotransferase 1 ‡ | Chst1 | 2.77 | 3.75 |
| Cell proliferation | NM_022278 | Glutaredoxin 1 (thioltransferase) | Glrx1 | 3.63 | 3.70 |
| Cell proliferation | NM_013122 | Insulin‐like growth factor binding protein 2 | Igfbp2 | 2.51 | 2.78 |
| Cell proliferation | AI101583 | Transient receptor potential cation channel, subfamily V, member 6 | Trpv6 | 2.01 | 2.25 |
| Cell proliferation | BI290527 | T‐box 2 ‡ | Tbx2 | 2.66 | 2.19 |
| Biosynthesis | BE097981 | Phosphatidylinositol 4‐kinase type 2α | Pi4KII | 2.66 | 3.36 |
| Signal transduction | NM_012707 | Glucagon | Gcg | 3.07 | 2.79 |
| Cell growth | AW142962 | Prolactin receptor | Prlr | 2.22 | 2.46 |
| Tumor progression | BE102969 | Ets variant gene 4 (E1A enhancer binding protein, E1AF) ‡ | Etv4 | 2.62 | 2.25 |
| Tumor progression | BG379319 | Transforming growth factor, beta induced | Tgfbi | 3.41 | 2.41 |
| Tumor progression | BE120425 | Calcium/calmodulin‐dependent protein kinase II gamma | Camk2g | 2.17 | 1.90 |
| Cell cycle | NM_133309 | Calpain 8 | Capn8 | 2.06 | 2.55 |
| Cytoskeleton | BE119221 | Actinin, α1 | Actn1 | 2.11 | 2.09 |
| Downregulated genes (of 53 genes in total) | |||||
| Adhesion | AW525315 | Poliovirus receptor‐related 3 ‡ | Pvrl3 | 0.46 | 0.28 |
| (AI103913) | (0.44) | (0.31) | |||
| Transcription | NM_013060 | Inhibitor of DNA binding 2, dominant negative helix‐loop‐helix protein | Id2 | 0.47 | 0.28 |
| Biosynthesis | NM_022276 | Glucosaminyl (N‐acetyl) transferase 1, core 2 | Gcnt1 | 0.44 | 0.30 |
| Signal transduction | BI276110 | Retinoic acid induced 3 ‡ | Rai3 | 0.48 | 0.33 |
| Apoptosis | AI227742 | Bcl‐2‐related ovarian killer protein | Bok | 0.44 | 0.36 |
Ad, adenomas; Ca, carcinomas; FFCH, focal follicular cell hyperplasias.
Proliferative lesions were divided into two categories, i.e. FFCH + Ad and Ca.
Predicted gene identity.
Table 5.
Validation of microarray data by real‐time reverse transcription‐polymerase chain reaction (PCR)
| Gene | FFCH + Ad | Ca | ||||
|---|---|---|---|---|---|---|
| Microarray | Real‐time PCR normalized by | Microarray | Real‐time PCR normalized by | |||
| Hprt | Gapdh | Hprt | Gapdh | |||
| Chi3l1 | 7.40 ± 1.06* | 7.35 ± 2.85*** | 10.73 ± 4.13 | 8.67 ± 2.67* | 8.14 ± 3.49*** | 14.09 ± 4.61*** |
| Cp | 3.29 ± 0.27** | 3.18 ± 0.89**** | 4.69 ± 1.19**** | 4.12 ± 0.62** | 3.50 ± 0.84**** | 6.15 ± 0.83**** |
| Slc2a3 | 2.66 ± 0.25** | 2.47 ± 0.66**** | 3.55 ± 0.93 | 2.52 ± 0.31** | 2.41 ± 0.41**** | 4.15 ± 0.28*** |
| Scl16a6 | 2.29 ± 0.53** | 2.20 ± 0.73*** | 3.26 ± 1.12**** | 2.23 ± 0.26** | 2.19 ± 0.63*** | 3.88 ± 0.89**** |
| Gcg | 3.07 ± 0.60* | 2.37 ± 0.72*** | 3.46 ± 0.88 | 2.96 ± 1.21* | 2.55 ± 1.17*** | 4.40 ± 1.69*** |
| Prlr | 2.22 ± 0.61* | 1.53 ± 0.36 | 2.21 ± 0.51**** | 2.51 ± 0.80** | 1.87 ± 0.36*** | 3.23 ± 0.43**** |
| Pi4KII | 2.67 ± 0.89* | 2.01 ± 0.74 | 2.96 ± 1.12**** | 3.30 ± 0.95** | 2.17 ± 0.59*** | 3.84 ± 0.83**** |
| Actn1 | 2.13 ± 0.47* | 1.61 ± 0.48 | 2.35 ± 0.73**** | 2.17 ± 0.82* | 1.52 ± 0.38 | 2.67 ± 0.56**** |
| Pvrl3 | 0.44 ± 0.07* | 0.40 ± 0.08 | 0.59 ± 0.10**** | 0.31 ± 0.05* | 0.35 ± 0.05*** | 0.62 ± 0.12**** |
| Rai3 | 0.48 ± 0.06* | 0.42 ± 0.06 | 0.63 ± 0.09**** | 0.34 ± 0.08* | 0.32 ± 0.06*** | 0.58 ± 0.17**** |
| Gcnt1 | 0.44 ± 0.08* | 0.63 ± 0.14 | 0.95 ± 0.17 | 0.31 ± 0.09* | 0.37 ± 0.05*** | 0.67 ± 0.08**** |
Gapdh, glyceraldehyde 3‐phosphate dehydrogenase; Hprt, hypoxanthine‐guanine phosphoribosyltransferase.
Values are mean ± SD (n = 5) when the expression level in non‐tumor follicles was calculated as 1. A single RNA sample for measurement was an equal mixture of total RNA from the same category tissue preparations from two animals.
: Significantly different from non‐tumor follicles at P < 0.05 and P < 0.01, respectively (Student's t‐test calculated by GeneSpring).
: Significantly different from non‐tumor follicles at P < 0.05 and P < 0.01, respectively (Student's t‐test).
Immunolocalization in the thyroid in relation to proliferative lesions. Ceruloplasmin was immunolocalized mainly at the luminal surfaces of cell membranes of follicular cells, almost specific to the proliferative lesions (Fig. 2a). Diffuse or granular immunoreactivity was also observed in the follicular lumina of lesions showing cell surface immunoreactivity. In parallel with the upregulation of transcript levels both in microarray and real‐time RT‐PCR analyses, immunolocalization of ceruloplasmin was specifically observed in all types of proliferative lesions, with statistical significance in the scores as compared with NTF. Although the intensity was weak, increased cytoplasmic staining was also observed in some Ca.
Figure 2.
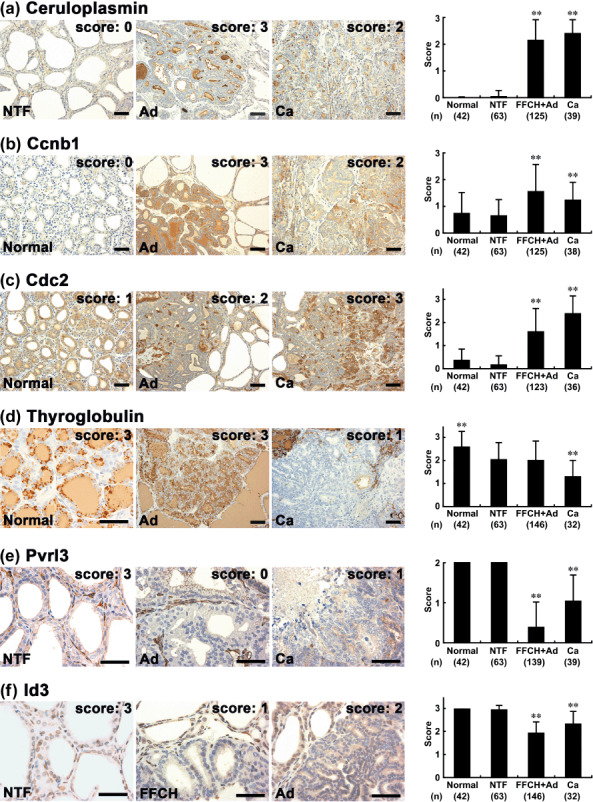
Immunohistochemical distributions of ceruloplasmin, cyclin B1 (Ccnb1), cell division cycle 2 (Cdc2), thyroglobulin, poliovirus receptor‐related 3 (Pvrl3), and inhibitor of DNA binding 3 (Id3) in normal follicular cells of untreated or N‐bis(2‐hydroxypropyl)nitrosamine (DHPN)‐treated animals and in proliferative lesions (focal follicular cell hyperplasia [FFCH], adenoma [Ad], carcinoma [Ca]) or surrounding non‐tumor follicles (NTF) after promotion with sulfadimethoxine (SDM) for 10 or 15 weeks. (a) Ceruloplasmin in the NTF, Ad, and Ca. Left: NTF nearly lacking ceruloplasmin expression. Middle and right: Ad and Ca showing ceruloplasmin immunoreactivity in the luminal surfaces of cellular membranes of follicular cells. Diffuse or granular immunoreactivity is also evident in the follicular lumina. (b) Ccnb1 in normal follicles, Ad, and Ca. Note fine granular immunoreactivity of Ccnb1 in the cytoplasm of follicular cells. Left: Weak and sparse Ccnb1 localization in normal follicular cells. Middle: Strong and diffuse immunoreactivity in an Ad. Right: Ca demonstrating Ccnb1 immunoreactivity with variable intensity. (c) Cdc2 in normal follicles, Ad, and Ca. Strong Cdc2 immunoreactivity both in the cytoplasm and nucleus of follicular cells. Left: Rather few immunoreactive cells in normal follicles. Middle: An Ad showing sparse Cdc2 immunoreactivity. Right: Note increased numbers of immunoreactive cells in a Ca. (d) Thyroglobulin in normal follicles, Ad, and Ca. Left: Strong and granular thyroglobulin immunoreactivity in the cytoplasm of normal follicular cells as well as diffuse immunoreactivity in the follicular lumina. Middle: Ad showing strong and diffuse thyroglobulin immunoreactivity. Right: A Ca lacking thyroglobulin immunoreactivity in the neoplastic cells. (e) Pvrl3 in the NTF, Ad, and Ca. Left: Diffuse intercellular membrane localization of Pvrl3 in cells comprising NTF. Middle: Ad entirely lacking Pvrl3 expression. Right: Ca showing an irregular and weak intercellular expression pattern. (f) Id3 in NTF, FFCH, and Ad. Left: Diffuse nuclear immunoexpression of Id3 in NTF. Middle: FFCH showing sparse nuclear Id3 expression. Right: Ad showing immunoreactivity in moderate numbers of neoplastic cells. The graphs show scores (mean + SD) for immunohistochemical findings. **P < 0.01 versus NTF (Mann–Whitney's U‐test). (a–f) Scale bar = 50 µm.
Ccnb1 was immunolocalized in the cytoplasm of follicular cells with fine granular immunoreactivity (Fig. 2b). In the normal follicles and NTF, weak and sparse immunolocalization was typical. In the proliferative lesions, the expression pattern of this molecule was either sparse or diffuse, and staining was weak with the former and either weak or strong with the latter. In parallel with upregulation of transcript levels in microarray analysis, a significant increase in the immunolocalization scores was observed in FFCH + Ad as compared with surrounding NTF. Although statistically significant elevation was still evident, immunoreactivity was less intense in Ca than in FFCH + Ad.
Immunoreactivity of Cdc2 was strong and localized both to the cytoplasm and nucleus of tumor follicular cells (Fig. 2c). In the normal follicles and NTF, immunoreactive cells were rather few. Although microarray data showed upregulation only in FFCH + Ad, both FFCH + Ad and Ca showed statistically significant increases in the immunolocalization scores as compared with NTF. Ca demonstrated the highest scores.
Thyroglobulin showed strong and granular immunolocalization in the cytoplasm of normal and non‐tumor follicular cells as well as diffuse immunoreactivity in the follicular lumina (Fig. 2d). Although microarray data showed upregulation of the transcript levels in Ca, follicular proliferative lesions showed large variability in the immunoreactivity, and Ca showed a significant decrease in immunolocalization scores as compared with NTF.
Pvrl3 showed intercellular membrane immunolocalization in the normal and non‐tumor follicular cells (Fig. 2e). With both FFCH + Ad and Ca, in parallel with the downregulation of the transcript levels common to microarray and real‐time RT‐PCR analyses, statistically significant decreased immunolocalization scores were observed as compared with NTF. However, the magnitude of the decrease in FFCH + Ad was stronger than in the Ca case, contrasting with the transcript data.
Inhibitor of DNA binding 3 showed nuclear immunoexpression in normal and non‐tumor follicular cells (Fig. 2f). Although microarray analysis showed statistically significant downregulation of transcript levels only in Ca, statistically significant decreases in the nuclear immunolocalization scores were also observed in FFCH + Ad.
Discussion
With the present microdissected lesion‐specific gene expression profiling, alteration was found for 60 genes specifically in FFCH + Ad, 211 genes specifically in Ca, and 146 genes in common in both, as compared with surrounding NTF. On selection of these with known annotations associated with carcinogenesis, we found upregulation of cell cycle‐related genes specifically in the early proliferative lesions, represented by FFCH and Ad. In the advanced Ca lesions, downregulation of genes related to tumor suppression and those encoding transcriptional inhibitors of Id family proteins appeared specific. These genes may play stage‐dependent roles during carcinogenesis. In particular, selective activation of cell‐cycle molecules in the early stages is considered to be essential for lesions to undergo efficient replication in response to TSH‐stimulation, and loss of tumor‐suppressor functions may be necessary for acquisition of a malignant phenotype during the progression stage. As genes upregulated in common in all types of proliferative lesions, we also found examples related to cell proliferation, suggesting roles for these molecules consistently throughout carcinogenic processes.
Ceruloplasmin, a copper‐containing plasma protein mainly synthesized in the liver, is known to act as a ferroxidase preventing production of toxic Fe and controlling membrane lipid oxidation, while also functioning in angiogenesis and blood coagulation.( 19 ) High levels of antioxidant production likely result from high amounts of reactive oxygen species, which have been implicated in mitogenic signaling and angiogenesis.( 27 , 28 ) As with high ceruloplasmin levels in the sera of cancer patients, overexpression of ceruloplasmin has been reported in many human malignancies, such as those in the lungs, kidneys, and ovaries.( 29 , 30 , 31 ) With regard to ceruloplasmin in thyroid tumors, it has been demonstrated in follicular cell carcinomas as well as papillary carcinomas, but follicular Ad lack expression.( 32 , 33 , 34 ) In the present study, ceruloplasmin could be immunohistochemically demonstrated in all types of proliferative lesion, in line with transcriptional upregulation. In contrast to the generally benign nature of human Ad, FFCH and Ad in SDM‐promoted cases show high cell‐proliferation activity similarly to Ca,( 9 ) and this biological behavior may be linked to the increased ceruloplasmin immunoreactivity found in our early proliferative lesions. As discussed by Kondi‐Pafiti et al., strong cytoplasmic localization of ceruloplasmin is mainly observed in Ca of human cases, and immunolocalization in luminal secretions (as present in our rat cases) is rare, suggesting a defective catabolism of ceruloplasmin in human Ca.( 34 )
Cdc2 exerts protein kinase activity by forming complexes with cyclin A2, Ccnb1, and p13suc1 and acts as an active subunit of the M‐phase‐promoting factor and the M‐phase‐specific histone H1 kinase.( 35 ) In the present study, increased expression of Ccnb1 and Cdc2, as with other cyclin‐related molecules such as cyclin A2, was detected specifically in early proliferative lesions by microarray analysis. Immunohistochemically, we also observed increased expression of Ccnb1 and Cdc2 in both FFCH + Ad and Ca. Chen et al. reported coordinated and increased expression of Cdc2 and Ccnb1 in parallel with the pathological grade of human gliomas.( 36 ) They also showed that increased expression of Cdc2 and Ccnb1 contributes to chromosomal instability in tumor cells through alteration of the spindle checkpoint. Thus, the coordinated upregulation of Cdc2 and Ccnb1 observed in the present study might be important as a driving force for both promotion and progression. In another thyroid carcinogenesis study we recently carried out using propylthiouracil as a promoter, a concordant increase of Cdc2 and a cell proliferation marker Ki‐67 was found in the proliferative lesions (K. Ago and M. Shibutani, 2008, unpublished data).
Thyroglobulin, a scaffold protein for thyroid hormonogenesis and a storage element for thyroid hormones and iodide, is specifically expressed in the thyroid in response to TSH stimulation.( 20 ) In human malignancies, the presence of thyroglobulin in cancer cells indicates a thyroidal origin.( 20 ) In the present study, although the reason remains unclear, some discrepancy was evident between the mRNA and immunolocalization levels in Ca, the increase in expression on microarray analysis contrasting with the decrease in protein finding. In human thyroid Ca, an inverse relationship between loss of differentiation and thyroglobulin immunoreactivity has been observed, positive cases being less anaplastic,( 37 ) suggesting that the decrease in thyroglobulin in our Ca might have been linked with dedifferentiation, leading to loss of TSH control with malignancy.
Cell adhesion molecules contribute cell‐to‐cell or cell‐to‐substratum interactions by homophilic or heterophilic processes.( 38 ) Pvrl molecules, also known as nectins, are adhesion receptors belonging to the IGSF that are involved in cell‐to‐cell spreading of viruses.( 39 ) Although Pvrl3 protein has not been extensively investigated, it may be a new adhesion molecule expressed on lymphatic endothelial cells.( 40 ) Recent studies have revealed that nectins and nectin‐like molecules, in cooperation with integrin and the platelet‐derived growth factor receptor, are crucial for mechanisms underlying contact inhibition of cell movement and proliferation.( 41 ) Reduced Pvrl3 expression in all types of proliferative lesions in the present study may reflect acquisition of growth advantage. Interestingly, Pvrl has been shown to be the heterophilic binding partner of another IGSF‐type adhesion molecule, tumor suppressor in lung cancer 1 (TSLC1)/IGSF4, a recently identified tumor‐suppressor gene.( 42 , 43 )
Inhibitor of DNA binding proteins, composed of four members of the helix‐loop‐helix transcription factors, are known to act as dominant‐negative regulators of basic helix‐loop‐helix transcription factors, and function to inhibit differentiation and enhance cell proliferation.( 44 , 45 ) In many human malignancies, upregulation of Id has been reported.( 46 ) However, in the present study, all Id isoforms showed downregulation in Ca by microarray analysis, and immunohistochemically, Id3‐immunoreactive cells were reduced in all types of proliferative lesions. Similar findings have been reported for human ovarian tumors, in which downregulation of Id3 was noted in 70% of 38 cases.( 47 ) Also, the expression of Id1, Id3, and Id4 was downregulated in microdissected human thyroid Ca compared with surrounding tissues by microarray analysis in one recent study.( 48 ) Although the reason for the inconsistency in the expression alterations between tumor types is not clear, it is possible that gene control mechanisms of Id proteins may differ with the cell type of origin.
In conclusion, we here found differentially regulated genes that may play key roles in early and late stages of thyroid carcinogenesis by microarray analysis of microdissected proliferative lesions developing after promotion with SDM in a two‐stage model. Immunohistochemical analysis of representative proliferative lesions indicated facilitation of the cell cycle in early lesions by forming an M‐phase promoting factor, as evidenced by the synchronized localization of Ccnb1 and Cdc2, and generation of oxidative stress responses by ceruloplasmin accumulation, as well as reduction of cellular adhesion involving Pvrl3 and cellular differentiation related to transcriptional control by Id3. Decreased expression of thyroglobulin in Ca may reflect dedifferentiation. Although further studies should address particular roles in the processes of thyroid carcinogenesis, the molecules identified in the present study provide pointers to understanding the mechanism of non‐genotoxic carcinogenesis and should help in efforts to secure human health.
Supporting information
Table S1. List of genes showing altered expression in microdissected focal follicular cell hyperplasias (FFCH) + adeomas (Ad) induced in the thyroids of rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold).
Table S2. List of genes showing altered expression in microdissected carinomas (Ca) induced in the thyroid of rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold).
Table S3. List of genes showing altered expression in common in all types of thyroid proliferative lesion induced in rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgments
We thank Miss Tomomi Morikawa and Ayako Kaneko for their technical assistance in conducting the animal study. This work was supported by Health and Labour Sciences Research Grants (Research on Food Safety) from the Ministry of Health, Labour, and Welfare of Japan. We all authors disclose here that there are no conflicts of interest that could inappropriately influence the outcome of the present study.
References
- 1. Thyroid Carcinoma Task Force . AACE/AAES medical/surgical guidelines for clinical practice: management of thyroid carcinoma. American Association of Clinical Endocrinologists. American College of Endocrinology. Endocr Pract 2001; 7: 202–20. [PubMed] [Google Scholar]
- 2. Schneider AB, Sarne DH. Long‐term risks for thyroid cancer and other neoplasms after exposure to radiation. Nat Clin Pract Endocrinol Metab 2005; 1: 82–91. [DOI] [PubMed] [Google Scholar]
- 3. Kitahori Y, Hiasa Y, Konishi N, Enoki N, Shimoyama T, Miyashiro A. Effect of propylthiouracil on the thyroid tumorigenesis induced by N‐bis (2‐hydroxypropyl) nitrosamine in rats. Carcinogenesis 1984; 5: 657–60. [DOI] [PubMed] [Google Scholar]
- 4. Jemec B. Studies of the goitrogenic and tumorigenic effect of two goitrogens in combination with hypophysectomy or thyroid hormone treatment. Cancer 1980; 45: 2138–48. [DOI] [PubMed] [Google Scholar]
- 5. Hiasa Y, Ohshima M, Kitahori Y, Yuasa T, Fujita T, Iwata C. Promoting effects of 3‐amino‐1,2,4‐triazole on the development of thyroid tumors in rats treated with N‐bis (2‐hydroxypropyl) nitrosamine. Carcinogenesis 1982; 3: 381–4. [DOI] [PubMed] [Google Scholar]
- 6. Hard GC. Recent developments in the investigation of thyroid regulation and thyroid carcinogenesis. Environ Health Perspect 1998; 106: 427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith P, Williams ED, Wynford‐Thomas D. In vitro demonstration of a TSH‐specific growth desensitising mechanism in rat thyroid epithelium. Mol Cell Endocrinol 1987; 51: 51–8. [DOI] [PubMed] [Google Scholar]
- 8. Brewer C, Yeager N, Di Cristofano A. Thyroid‐stimulating hormone initiated proliferative signals converge in vivo on the mTOR kinase without activating AKT. Cancer Res 2007; 67: 8002–6. [DOI] [PubMed] [Google Scholar]
- 9. Mitsumori K, Onodera H, Takahashi M et al . Effect of thyroid stimulating hormone on the development and progression of rat thyroid follicular cell tumors. Cancer Lett 1995; 92: 193–202. [DOI] [PubMed] [Google Scholar]
- 10. Roy G, Mugesh G. Bioinorganic chemistry in thyroid gland: effect of antithyroid drugs on peroxidase‐catalyzed oxidation and iodination reactions. Bioinorg Chem Appl 2006; 2006: 23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shibutani M, Uneyama C, Miyazaki K, Toyoda K, Hirose M. Methacarn fixation: a novel tool for analysis of gene expressions in paraffin‐embedded tissue specimens. Lab Invest 2000; 80: 199–208. [DOI] [PubMed] [Google Scholar]
- 12. Uneyama C, Shibutani M, Masutomi N, Takagi H, Hirose M. Methacarn fixation for genomic DNA analysis in microdissected, paraffin‐embedded tissue specimens. J Histochem Cytochem 2002; 50: 1237–45. [DOI] [PubMed] [Google Scholar]
- 13. Takagi H, Shibutani M, Kato N et al . Microdissected region‐specific gene expression analysis with methacarn‐fixed, paraffin‐embedded tissues by real‐time RT‐PCR. J Histochem Cytochem 2004; 52: 903–13. [DOI] [PubMed] [Google Scholar]
- 14. Takagi H, Shibutani M, Lee KY et al . Impact of maternal dietary exposure to endocrine‐acting chemicals on progesterone receptor expression in microdissected hypothalamic medial preoptic areas of rat offspring. Toxicol Appl Pharmacol 2005; 208: 127–36. [DOI] [PubMed] [Google Scholar]
- 15. Shibutani M, Lee KY, Igarashi K et al . Hypothalamus region‐specific global gene expression profiling in early stages of central endocrine disruption in rat neonates injected with estradiol benzoate or flutamide. Dev Neurobiol 2007; 67: 253–69. [DOI] [PubMed] [Google Scholar]
- 16. Lee KY, Shibutani M, Inoue K, Kuroiwa KUM, Woo GH, Hirose M. Methacarn fixation – effects of tissue processing and storage conditions on detection of mRNAs and proteins in paraffin‐embedded tissues. Anal Biochem 2006; 351: 36–43. [DOI] [PubMed] [Google Scholar]
- 17. Hardisty JF, Boorman GA. Thyroid gland. In: Boorman GA, Eustis SL, Elwell MR, Montgomery CA Jr, MacKenzie WF, eds. Pathology of the Fischer Rat, Reference and Atlas. San Diego: Academic Press, 1990: 519–36. [Google Scholar]
- 18. Imai T, Onose J, Hasumura M, Ueda M, Takizawa T, Hirose M. Sequential analysis of development of invasive thyroid follicular cell carcinomas in inflamed capsular regions of rats treated with sulfadimethoxine after N‐bis (2‐hydroxypropyl) nitrosamine‐initiation. Toxicol Pathol 2004; 32: 229–36. [DOI] [PubMed] [Google Scholar]
- 19. Goldstein IM, Kaplan HB, Edelson HS, Weissmann G. Ceruloplasmin. A scavenger of superoxide anion radicals. J Biol Chem 1979; 254: 4040–5. [PubMed] [Google Scholar]
- 20. Lin JD. Thyroglobulin and human thyroid cancer. Clin Chim Acta 2008; 388: 15–21. [DOI] [PubMed] [Google Scholar]
- 21. Ziak M, Meier M, Novak‐Hofer I, Roth J. Ceruloplasmin carries the anionic glycan oligo/poly α2,8 deaminoneuraminic acid. Biochem Biophys Res Commun 2002; 295: 597–602. [DOI] [PubMed] [Google Scholar]
- 22. Scott IS, Morris LS, Bird K et al . A novel immunohistochemical method to estimate cell‐cycle phase distribution in archival tissue: implications for the prediction of outcome in colorectal cancer. J Pathol 2003; 201: 187–97. [DOI] [PubMed] [Google Scholar]
- 23. Yamamoto H, Monden T, Ikeda K et al . Coexpression of cdk2/cdc2 and retinoblastoma gene products in colorectal cancer. Br J Cancer 1995; 71: 1231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faggiano A, Caillou B, Lacroix L et al . Functional characterization of human thyroid tissue with immunohistochemistry. Thyroid 2007; 17: 203–11. [DOI] [PubMed] [Google Scholar]
- 25. Kuramitsu K, Ikeda W, Inoue N, Tamaru Y, Takai Y. Novel role of nectin: implication in the co‐localization of JAM‐A and claudin‐1 at the same cell–cell adhesion membrane domain. Genes Cells 2008; 13: 797–805. [DOI] [PubMed] [Google Scholar]
- 26. Carroll M, Hamzeh M, Robaire B. Expression, localization, and regulation of inhibitor of DNA binding (Id) proteins in the rat epididymis. J Androl 2006; 27: 212–24. [DOI] [PubMed] [Google Scholar]
- 27. Lelkes PI, Hahn KL, Sukovich DA, Karmiol S, Schmidt DH. On the possible role of reactive oxygen species in angiogenesis. Adv Exp Med Biol 1998; 454: 295–310. [DOI] [PubMed] [Google Scholar]
- 28. Suh YA, Arnold RS, Lassegue B et al . Cell transformation by the superoxide‐generating oxidase Mox1. Nature 1999; 401: 79–82. [DOI] [PubMed] [Google Scholar]
- 29. Stassar MJ, Devitt G, Brosius M et al . Identification of human renal cell carcinoma associated genes by suppression subtractive hybridization. Br J Cancer 2001; 85: 1372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hough CD, Cho KR, Zonderman AB, Schwartz DR, Morin PJ. Coordinately up‐regulated genes in ovarian cancer. Cancer Res 2001; 61: 3869–76. [PubMed] [Google Scholar]
- 31. Wang KK, Liu N, Radulovich N et al . Novel candidate tumor marker genes for lung adenocarcinoma. Oncogene 2002; 21: 7598–604. [DOI] [PubMed] [Google Scholar]
- 32. Tuccari G, Barresi G. Immunohistochemical demonstration of ceruloplasmin in follicular adenomas and thyroid carcinomas. Histopathology 1987; 11: 723–31. [DOI] [PubMed] [Google Scholar]
- 33. Song B. Immunohistochemical demonstration of epidermal growth factor receptor and ceruloplasmin in thyroid diseases. Acta Pathol Jpn 1991; 41: 336–43. [DOI] [PubMed] [Google Scholar]
- 34. Kondi‐Pafiti A, Smyrniotis V, Frangou M, Papayanopoulou A, Englezou M, Deligeorgi H. Immunohistochemical study of ceruloplasmin, lactoferrin and secretory component expression in neoplastic and non‐neoplastic thyroid gland diseases. Acta Oncol 2000; 39: 753–6. [DOI] [PubMed] [Google Scholar]
- 35. Draetta G, Luca F, Westendorf J, Brizuela L, Ruderman J, Beach D. cdc2 protein kinase is complexed with both cyclin A and B. Evidence for proteolytic inactivation of MPF. Cell 1989; 56: 829–38. [DOI] [PubMed] [Google Scholar]
- 36. Chen H, Huang Q, Dong J, Zhai DZ, Wang AD, Lan Q. Overexpression of CDC2/CyclinB1 in gliomas, and CDC2 depletion inhibits proliferation of human glioma cells in vitro and in vivo . BMC Cancer 2008; 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wiseman SM, Griffith OL, Deen S et al . Identification of molecular markers altered during transformation of differentiated into anaplastic thyroid carcinoma. Arch Surg 2007; 142: 717–29. [DOI] [PubMed] [Google Scholar]
- 38. Simmons DL. Dissecting the modes of interactions amongst cell adhesion molecules. Dev Suppl 1993; ???: 193–203. [PubMed] [Google Scholar]
- 39. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor‐related protein 1 and poliovirus receptor. Science 1998; 280: 1618–20. [DOI] [PubMed] [Google Scholar]
- 40. Nelson GM, Padera TP, Garkavtsev I, Shioda T, Jain RK. Differential gene expression of primary cultured lymphatic and blood vascular endothelial cells. Neoplasia 2007; 9: 1038–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin‐like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol 2008; 9: 603–15. [DOI] [PubMed] [Google Scholar]
- 42. Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci 2005; 96: 543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wakayama T, Sai Y, Ito A et al . Heterophilic binding of the adhesion molecules poliovirus receptor and immunoglobulin superfamily 4A in the interaction between mouse spermatogenic and Sertoli cells. Biol Reprod 2007; 76: 1081–90. [DOI] [PubMed] [Google Scholar]
- 44. Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id. A negative regulator of helix‐loop‐helix DNA binding proteins. Cell 1990; 61: 49–59. [DOI] [PubMed] [Google Scholar]
- 45. Ellis HM, Spann DR, Posakony JW. Extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix‐loop‐helix proteins. Cell 1990; 61: 27–38. [DOI] [PubMed] [Google Scholar]
- 46. Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell 2003; 3: 525–30. [DOI] [PubMed] [Google Scholar]
- 47. Arnold JM, Mok SC, Purdie D, Chenevix‐Trench G. Decreased expression of the Id3 gene at 1p36.1 in ovarian adenocarcinomas. Br J Cancer 2001; 84: 352–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nikolova DN, Zembutsu H, Sechanov T et al . Genome‐wide gene expression profiles of thyroid carcinoma: Identification of molecular targets for treatment of thyroid carcinoma. Oncol Rep 2008; 20: 105–21. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of genes showing altered expression in microdissected focal follicular cell hyperplasias (FFCH) + adeomas (Ad) induced in the thyroids of rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold).
Table S2. List of genes showing altered expression in microdissected carinomas (Ca) induced in the thyroid of rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold).
Table S3. List of genes showing altered expression in common in all types of thyroid proliferative lesion induced in rats using a two‐stage thyroid carcinogenesis model (≥2‐fold, ≤0.5‐fold).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


