Abstract
To comprehensively screen for genetic events underlying colorectal cancer, we performed suppression subtraction hybridization analysis on an advanced colon cancer. Because Dickkopf‐4, a member of the Dickkopf family acting as a Wnt‐signaling modulator, was identified as one of the upregulated genes in this specimen, we investigated expression profiles of all the Dickkopf family members in 55 colorectal tumors (21 cancers and 34 adenomas). We also investigated mechanisms regulating the expression of Dickkopf‐4 in these cancers in vitro and in vivo. Compared with normal adjacent mucosae, Dickkopf‐4 (median 27.4, P < 0.01) and ‐2 (median 51.4, P < 0.01) were strongly expressed in colorectal cancers. The level of Dickkopf‐4 was positively correlated with fibroblast growth factor‐20 (rs = 0.61, P = 0.00017), a representative β‐catenin transcriptional target gene, and with the degree of nuclear accumulation of β‐catenin in colorectal tumors. Dickkopf‐4 was induced by activated β‐catenin in vitro. Reciprocally, recombinant Dickkopf‐4 significantly inhibited T‐cell factor/lymphocyte enhancer factor reporter activity stimulated by recombinant Wnt3a in human embryonic kidney 293 cells. We conclude that Dickkopf‐4 and ‐2 are significantly upregulated in most colorectal tumors, and that Dickkopf‐4 upregulation reflects activation of the Wnt/canonical pathway. (Cancer Sci 2009; 100: 1923–1930)
The Wnt signaling pathway plays an important role in the carcinogenesis of colorectal cancer. Wnt ligands bind to Frizzled receptors and low‐density lipoprotein receptor‐related protein 5/6 (LRP5/6) coreceptors located at the plasma membrane and activate both canonical and non‐canonical pathways. Activation of the Wnt/canonical pathway leads to the cytosolic stabilization and accumulation of β‐catenin, which translocates to the nucleus and forms a complex with the transcription factor T‐cell factor/lymphocyte enhancer factor (TCF/LEF), thus regulating target gene expression. Constitutive activation of the Wnt/canonical pathway due to mutation of the adenomatous polyposis coli gene (APC), axis inhibitor protein (AXIN), or β‐catenin has been observed in various human cancers, and most frequently in colorectal cancer.( 1 )
The Dickkopf (DKK) family was identified as a group of secreted Wnt modulators; vertebrates express four DKK proteins (DKK1, ‐2, ‐3, and ‐4). DKK1 acts as an inhibitor of the Wnt/canonical pathway through binding to LRP5/6 and Kremen, thus inducing LRP endocytosis and preventing signaling to β‐catenin. DKK4 also acts as an inhibitor of the Wnt/canonical pathway. Interestingly, DKK2 can either activate or inhibit this pathway depending on the cellular context. Thus far, the role of DKK3 is unknown; it does not bind LRP5/6, nor alter the signal.( 2 , 3 )
In several clinical cancers, altered expression of human DKKs has been reported including DKK1 upregulation in multiple myeloma,( 4 ) Wilm's tumor,( 5 ) hepatocellular carcinoma,( 6 ) and breast cancer;( 7 ) DKK1 downregulation in choriocarcinoma( 8 ) and malignant melanoma;( 9 ) DKK2 downregulation in malignant melanoma;( 9 ) and DKK3 downregulation in hepatocellular carcinoma,( 10 ) cholangiocarcinoma,( 10 ) HeLa cervical carcinoma,( 10 ) renal clear cell carcinoma,( 11 ) and malignant melanoma;( 9 ) as well as DKK4 upregulation in gastric cancer.( 12 ) Although their role in these tumors has not yet been established, it is not surprising that DKKs are involved in carcinogenesis considering their actions as Wnt modulators in normal settings. In clinical colorectal cancers, frequent methylation of DKK1, ‐2, ‐3, and ‐4 has been reported.( 13 , 14 ) On the other hand, the expression of these DKKs in clinical specimens has not been investigated in detail.
In the present study, to identify genetic events playing important roles in colorectal cancer, we first comprehensively screened for genes differentially expressed between normal colonic mucosa and colorectal cancer by the suppression subtraction hybridization (SSH) technique using a clinical sample of advanced colon cancer. We found that DKK4 was one of the upregulated genes, and therefore, we next focused on expression of all the DKK family members using tissue samples of 55 clinical colorectal tumors (21 cancers and 34 adenomas). Finally, we explored mechanisms regulating the expression of DKK4 in the Wnt/canonical pathway in colorectal cancers in vitro and in vivo.
Materials and Methods
Tissue samples, RNA extraction. Tissue samples of advanced, well‐differentiated colon cancer and adjacent normal colonic mucosa for use in SSH analysis were obtained from a 60‐year‐old male by endoscopic biopsy. Additional paired tissue samples of 21 primary colorectal cancers and 34 colorectal adenomas were obtained from other patients by endoscopic biopsy. Tissue samples for RNA extraction were immediately placed in RNAlater (Ambion, Austin, TX, USA) and stored at 4°C until use. Total RNA was extracted by the modified acid–guanidium–chloroform method( 15 ) using Isogen (Nippon Gene, Toyama, Japan) according to the manufacturer's protocol. Written informed consent was obtained from patients before collection of the tissue samples, and this study was approved by the ethical committee of Yamanashi University Hospital in accordance with the Helsinki Declaration.
SMART (switching mechanism at 5′ end of RNA template) cDNA synthesis for SSH analysis. Full‐length cDNAs were generated from the total RNA using the SMART–RT‐PCR cDNA synthesis kit (Clontech, Palo Alto, CA, USA)( 16 ) following the manufacturer's instructions. These were then used for SSH analysis. Quantitative analysis of specific genes was performed on cDNAs generated from 1 µg of total RNA in 10‐µL mixture with 200 U of Superscript reverse transcriptase (Gibco, Grand Island, NY, USA) using random hexamer primers.
Suppression SSH, sequencing, and quantitative analysis of identified genes. Subtractive hybridization was performed using a PCR‐Select cDNA subtraction kit (Clontech, Tokyo, Japan) according to the manufacturer's instructions. A total of 10 ng of PCR products were cloned into the pGEM‐T Easy Vector plasmid (Stratagene, Cedar Creek, TX, USA) and transformed to Escherichia coli XL2‐blue Ultracompetent cells (Gibco, Madison, WI, USA). In all, 100 colonies were randomly picked and sequenced using the PRISM dye termination kit (Applied Biosystems, Foster City, CA, USA). BLAST Search 2.0 (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) was used to analyze sequence homologies in the gene database.
Overexpression of the obtained genes was confirmed by semi‐quantitative RT‐PCR, comparing the amount of PCR products by agarose gel electrophoresis at the PCR cycle number in the exponential phase of amplification. The primers used in the quantitative PCR are shown in Supporting Information Table 1. G3PDH was used as an internal control.
Table 1.
Clinicopathologicaldata of study subjects
| Colorectal cancer patients (n = 21) | ||
|---|---|---|
| Sex | % | |
| Male | 13 | 61.9 |
| Female | 8 | 38.1 |
| Age | ||
| Range | 55–87 years | |
| Median | 69 years | |
| Location | ||
| Cecum | 0 | 0.0 |
| Ascending | 3 | 14.3 |
| Transverse | 3 | 14.3 |
| Descending | 1 | 4.8 |
| Sigmoid | 7 | 33.3 |
| Rectum | 7 | 33.3 |
| Dukes | ||
| A | 11 | 52.4 |
| B | 2 | 9.5 |
| C | 8 | 38.1 |
| TNM stage | ||
| T stage | ||
| Tis | 6 | 28.6 |
| T1 | 1 | 4.8 |
| T2 | 5 | 23.8 |
| T3 | 9 | 42.9 |
| T4 | 0 | 0.0 |
| N stage | ||
| N0 | 13 | 61.9 |
| N1 | 7 | 33.3 |
| N2 | 1 | 4.8 |
| M stage | ||
| M0 | 18 | 85.7 |
| M1 | 3 | 14.3 |
| Degree in differentiation | ||
| Well | 15 | 71.4 |
| Moderately | 6 | 28.6 |
| Poorly | 0 | 0.0 |
| Colorectal adenoma patients (n = 34) | ||
|---|---|---|
| Sex | % | |
| Male | 29 | 85.3 |
| Female | 5 | 14.7 |
| Age | ||
| Range | 38–81 years | |
| Median | 65 years | |
| Location | ||
| Cecum | 2 | 5.9 |
| Ascending | 4 | 11.8 |
| Transverse | 6 | 17.6 |
| Descending | 3 | 8.8 |
| Sigmoid | 16 | 47.1 |
| Rectum | 3 | 8.8 |
| Degree of dysplasia | ||
| Low | 23 | 67.6 |
| High | 11 | 32.4 |
Real‐time RT‐PCR analysis. We focused on DKK family members in colorectal cancer, and quantified their expression by real‐time RT‐PCR in clinical samples from 21 colorectal cancers and 34 colorectal adenomas, compared to their corresponding adjacent normal colonic mucosae. The patients’ characteristics are shown in Table 1.
Fibroblast growth factor (FGF)‐20 is a transcriptional target of β‐catenin, and is highly induced by a dominant‐active mutant form of β‐catenin in human embryonic kidney (HEK)‐293T cells.( 17 ) Therefore, FGF20 could serve as a suitable marker reflecting activation of the Wnt/canonical pathway. We compared the expression level of each DKK family member and FGF20 in each colorectal tumor.
Real‐time RT‐PCR was carried out using TaqMan Gene Expression Assays (Applied Biosystems) with a 7500 Real‐Time PCR System (Applied Biosystems) according to the manufacturer's instructions. SDS2.1 software (Applied Biosystems) was used to perform comparative delta Ct analysis. 18srRNA was used as an internal control.
Immunohistochemical staining. To investigate the relationship between DKK4 expression and Wnt/canonical pathway activation, immunohistochemical staining for β‐catenin was performed in all primary colorectal cancer specimens (n = 18).
Deparaffinized sections of formalin‐fixed tissue at 2‐µm thicknesses were stained with primary antibodies specific for β‐catenin (1:100 dilutions; cat. 610153; BD Transduction Laboratories, Palo Alto, CA, USA), DKK4 (N‐term) (1:250 dilution; AP1524a; Abgent, San Diego, CA, USA). Antigen retrieval was achieved by boiling the tissue sections in citrate buffer (0.01 M, pH 6.0) in an autoclave unit for 20 min at 120°C, before incubation with anti‐β‐catenin. Envision kit/HRP (AEC) (cat. K1491; Dako, Carpinteria, CA, USA) was used as the secondary antibody, and diaminobenzidine was used as the chromogen.
Cell line. A human embryonic kidney cell line, HEK293, was maintained in Dulbecco's modified Eagle's medium (Sigma‐Aldrich, Tokyo, Japan) supplemented with 10% fetal bovine serum (Equitech‐Bio, Kerrville, TX, USA) and 1% antibiotic‐antimycotic (Gibco, Madison, WI, USA) in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Transient transfection of HEK293 cells with dominat‐active mutant form of β‐catenin. To test whether DKK4 is induced by activation of the Wnt/canonical pathway in vitro, HEK293 cells were transiently transfected with pFLAG‐cmv4‐β‐catenin ΔN134 (plasmid harboring cDNA‐encoding dominant‐active mutant form of β‐catenin, from which 134 amino acids had been deleted from the NH2‐terminus).
A total of 5 × 105 cells were seeded onto six‐well plates 1 day before transfection. Transfections with pcDNA3.1neo, pFLAG‐cmv4‐β‐catenin wild type (plasmid harboring full‐length human β‐catenin cDNA), and pFLAG‐cmv4‐β‐cateninΔN134 were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. pFLAG‐cmv4‐β‐catenin wild type, and pFLAG‐cmv4‐β‐cateninΔN134 were kindly provided by Dr Miki Shitashige and Dr Tesshi Yamada (National Cancer Center Research Institute, Tokyo, Japan).( 18 ) Cells were harvested 24 h after transfection and the amount of DKK4 expressed was determined by western blotting.
Western blot analysis. Equal amounts of cellular protein (5 µg total protein) were separated by electrophoresis in NuPAGE 4–12% Bis‐Tris Gel (Invitrogen). Primary antibodies specific for β‐catenin (1:500 dilution; BD Transduction Laboratories), DKK4 (N‐term) (1:250 dilution; Abgent), and β‐actin (1:4000 dilution; A1978; Sigma, St. Louis, MO, USA) were used. Secondary antibodies specific for mouse IgG (1:2000 dilution; NA931; Amersham Bioscience, Piscataway, NJ, USA) and rabbit IgG (1:2000 dilution; 7074; Cell Signaling, Danvers, MA, USA) were also used. Staining was performed using ECL western blotting detection reagents (Amersham Bioscience). All western blot experiments were repeated at least three times.
Luciferase reporter assay. In order to investigate the involvement of DKK4 in activation of the Wnt/canonical pathway, TCF/LEF reporter luciferase activity was measured in HEK293 cells stimulated with recombinant mouse Wnt3a protein (rmWnt3a) in the presence or absence of recombinant human DKK4 protein (rhDKK4). As a positive control inhibiting the Wnt/canonical pathway, TCF/LEF reporter luciferase activity stimulated with rmWnt3a in the presence of rhDKK1 was also measured.
To evaluate TCF/LEF transcriptional activity, we used a pair of luciferase reporter constructs, super 8 × TOP‐FLASH and super 8 × FOP‐FLASH (Addgene, Cambridge, MA, USA). Cells were transiently transfected in triplicate with one of these luciferase reporters and pRL‐TK (Promega, Madison, WI, USA) as an internal control using FuGENE 6 transfection reagent (Roche Diagnostics, Mannheim, Germany). Five hours after transfection, the medium was changed to fresh medium with 50 ng/mL or 500 ng/mL of recombinant human DKK1 or ‐4 (R&D Systems, Minneapolis, MN, USA). Twelve hours after addition of rhDKKs, 100 ng/mL of recombinant mouse Wnt3a (R&D Systems) was added. A further 12 hs thereafter, luciferase activity was measured with the dual‐luciferase reporter assay system (Promega), with renilla luciferase activity as an internal control.
Results
Suppression subtraction hybridization (SSH) and confirmation of differentially expressed genes by semiquantitative RT‐PCR. Tissue samples from colon cancer and adjacent normal colonic mucosa were obtained by endoscopic biopsy from a 60‐year‐old man with advanced disease (type 1) in the sigmoid colon. The cDNA generated by SMART–RT‐PCR exhibited a smear pattern representing amplification of all the mRNA species on agarose gel electrophoresis (Fig. 1a, left panel). Several discrete bands derived from highly expressed genes in colon cancer appeared after SSH (Fig. 1a, right panel). After subcloning the SSH‐PCR products into pGEM‐T Easy vectors, nucleotide sequencing was performed on 100 independent clones. Eight known genes, DKK4, tumor overexpressed gene (TOG), budding uninhibited by benzimidazole 1 (BUB1), cobalamin synthetase W‐domain containing (CBWD), gamma‐aminobutyric acid‐A (GABA‐A) receptor, carcinoembryonic antigen (CEA), non‐specific cross‐reacting antigen (NCA), and mammary serine protease inhibitor (Maspin), were represented more than once in colorectal cancer samples (Table 2). The differential expression of these eight genes was confirmed by semiquantitative RT‐PCR using the original colon cancer tissue and the adjacent normal colonic mucosa. All these genes were more strongly expressed in the colon cancer tissue than in the normal colonic mucosa (Fig. 1b). We confirmed that the size of each PCR product was correct, as predicted.
Figure 1.
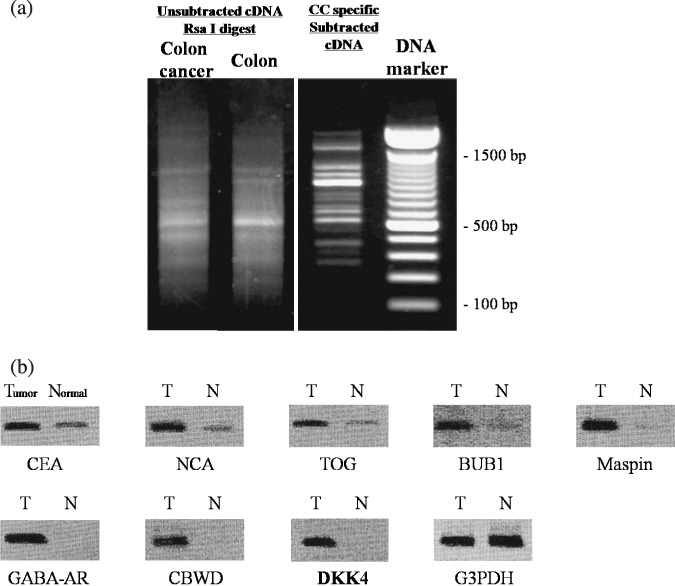
Electrophoretic band patterns of suppression subtraction hybridization (SSH) and gene overexpression in original tester tissue. (a) The cDNAs amplified by SMART–RT‐PCR and cDNA after subtraction by SSH were electrophoresed on 2.0% agarose. The amplified cDNA derived from colon cancer and adjacent colonic mucosa appeared as a smear before SSH (left panel). After SSH, it exhibited several distinct bands (right panel). (b) Semiquantitative RT‐PCR using gene‐specific primer sets for each identified gene. PCR products were analyzed at the PCR cycle number given in the exponential phase of amplification (carcinoembryonic antigen [CEA], 18 cycles; non‐specific cross‐reacting antigen [NCA], 18 cycles; tumor overexpressed gene [TOG], 24 cycles; benzimidazole 1 [BUB1], 24 cycles; mammary serine protease inhibitor [Maspin], 30 cycles; gamma‐aminobutyric acid‐A receptor [GABA‐AR], 30 cycles; cobalamin synthetase W‐domain containing [CBWD], 24 cycles; Dickkopf (DKK)‐4, 35 cycles; G3PDH, 21 cycles). The expression level of housekeeping gene G3PDH remained constant, but genes identified by SSH were clearly more abundant in colon cancer tissues compared to their corresponding adjacent mucosa. T and N indicate colorectal tumor and adjacent normal mucosa, respectively.
Table 2.
Sequence analyses of 100 clones obtained from subtracted colon cancer cDNA
| Name of the clones | No. |
|---|---|
| DKK4 | 19 |
| TOG | 10 |
| BUB1 | 9 |
| CBWD | 7 |
| GABA‐A receptor | 4 |
| CEA | 4 |
| NCA | 4 |
| Maspin | 3 |
| Miscellaneous | 40 |
| Total | 100 |
BUB1, budding uninhibited by benzimidazole 1; CBWD, cobalamin synthetase W‐domain containing; CEA, carcinoembryonic antigen; DKK4, Dickkopf‐4; GABA‐A receptor, gamma‐aminobutyric acid‐A receptor; Maspin, mammary serine protease inhibitor; NCA, non‐specific cross‐reacting antigen; TOG, tumor overexpressed gene.
Quantifying expression of Dickkopf family members in colorectal tumors by real‐time RT‐PCR. Figure 2a shows the expression level of each DKK family member (DKK1, ‐2, ‐3, and ‐4) given as the tumor/normal (T/N) ratio for each patient. In colorectal cancer, the median T/N ratio was 3.34 (range, 0.080–932; P < 0.01) for DKK1, 51.4 (range, 0.60–2901; P < 0.01) for DKK2, 2.81 (range, 0.63–26; P < 0.01) for DKK3, and 27.4 (range, 0.34–558; P < 0.01) for DKK4. In colorectal adenoma, these values were 3.25 (range, 0.094–123; P < 0.05), 6.78 (range, 0.052–104; P < 0.01), 1.01 (range, 5.57 × 10−8–8.18; n.s.), and 3.80 (range, 0.045–471; P < 0.01), respectively. Thus in colorectal cancer, DKK2 and ‐4 were strongly upregulated, whereas DKK1 and ‐3 were weakly upregulated relative to adjacent colorectal mucosa. On the other hand, in colorectal adenoma, DKK1, ‐2 and ‐4 but not DKK3 were weakly upregulated relative to adjacent colorectal mucosa.
Figure 2.
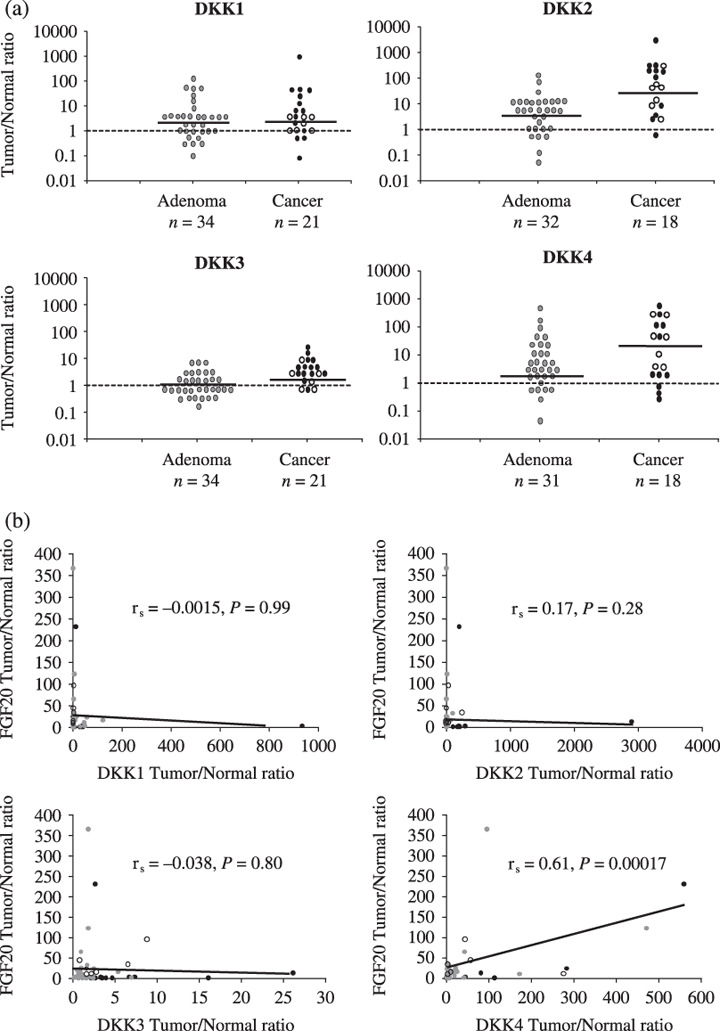
Expression profiles of Dickkopf (DKK) genes in colorectal tumors. (a) Expression level of each DKK family member in colorectal tumors measured by real‐time RT‐PCR.  , advanced cancer;
, advanced cancer;  , early cancer,
, early cancer,  , adenoma. The black horizontal line indicates the median in each group of subjects. The black dashed line indicates equal expression of DKK between tumor and normal mucosa. Data were analyzed with the Wilcoxon signed‐rank test. (b) The relationship between expression levels of DKK family members and fibroblast growth factor (FGF)‐20 in colorectal tumors. Data were analyzed for Spearman's correlation coefficient by rank testing (rs). A significant correlation between the levels of DKK4 and FGF20 was found. There was no such relationship between levels of DKK1, ‐2, ‐3, and FGF20.
, adenoma. The black horizontal line indicates the median in each group of subjects. The black dashed line indicates equal expression of DKK between tumor and normal mucosa. Data were analyzed with the Wilcoxon signed‐rank test. (b) The relationship between expression levels of DKK family members and fibroblast growth factor (FGF)‐20 in colorectal tumors. Data were analyzed for Spearman's correlation coefficient by rank testing (rs). A significant correlation between the levels of DKK4 and FGF20 was found. There was no such relationship between levels of DKK1, ‐2, ‐3, and FGF20.  , advanced cancer;
, advanced cancer;  , early cancer;
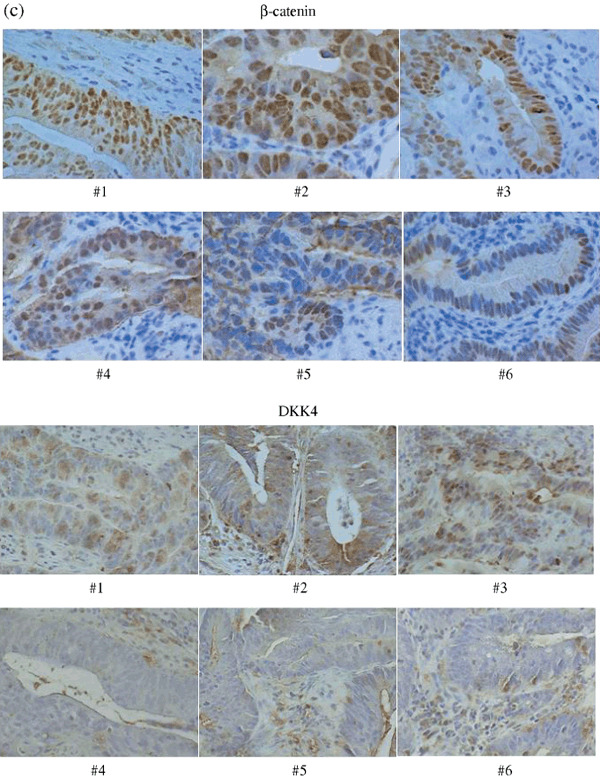
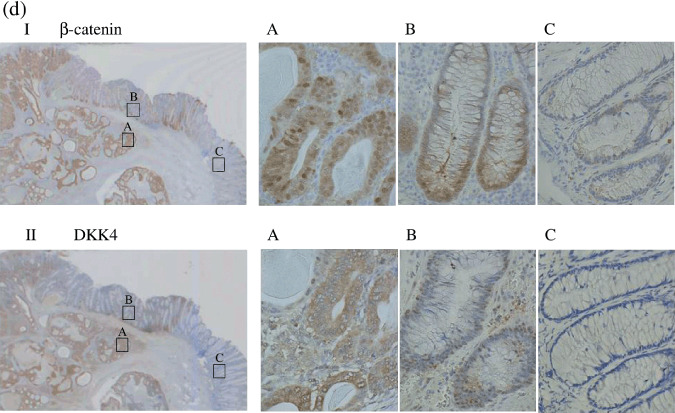
, early cancer;  , adenoma. (c) Immunohistochemical staining with β‐catenin and DKK4 antibody in primary colorectal cancers. All specimens were evaluated under a light microscope at ×400 magnification. There is a strong correlation between nuclear β‐catenin accumulation and expression of DKK4. #, sample number. DKK4 Tumor/Normal ratio: #1 559; #2 283; #3 81.2; #4 2.39; #5 2.21; #6 0.61. (d) Immunohistochemical staining with β‐catenin (upper panel) and DKK4 (lower panel) antibody in the specimen which was obtained by endoscopic mucosal resection showing: I, II, whole image; (a) invasive cancer with intense staining; (b) adenoma with weak staining; (c) normal mucosa with almost negative staining. Magnification a–c, ×400.
, adenoma. (c) Immunohistochemical staining with β‐catenin and DKK4 antibody in primary colorectal cancers. All specimens were evaluated under a light microscope at ×400 magnification. There is a strong correlation between nuclear β‐catenin accumulation and expression of DKK4. #, sample number. DKK4 Tumor/Normal ratio: #1 559; #2 283; #3 81.2; #4 2.39; #5 2.21; #6 0.61. (d) Immunohistochemical staining with β‐catenin (upper panel) and DKK4 (lower panel) antibody in the specimen which was obtained by endoscopic mucosal resection showing: I, II, whole image; (a) invasive cancer with intense staining; (b) adenoma with weak staining; (c) normal mucosa with almost negative staining. Magnification a–c, ×400.
Next, we compared mRNA levels of each DKK in colorectal cancer and adenoma. Receiver–operator curve (ROC) analysis showed that the best T/N ratio cut‐off values to distinguish between colorectal cancer and adenoma were 7, 30, 2.5, and 40 for DKK1, ‐2, ‐3, and ‐4, respectively (Table 3). When these cut‐offs were applied, DKK1–4 were more highly expressed in colorectal cancer than in adenoma (by Fisher's exact probability test, Table 3).
Table 3.
Comparison of Dickkopf (DKK) mRNA level between adenoma and cancer
| DKK1 | P = 0.028 | ≤7 | Total |
|---|---|---|---|
| T/N | <7 | ||
| Carcinoma | 14 | 7 | 21 |
| Adenoma | 31 | 3 | 34 |
| Total | 45 | 10 | 55 |
| DKK2 | P = 0.000046 | ≤30 | Total |
|---|---|---|---|
| T/N | <30 | ||
| Carcinoma | 7 | 11 | 18 |
| Adenoma | 30 | 2 | 32 |
| Total | 37 | 13 | 50 |
| DKK3 | P = 0.00015 | ≤2.5 | Total |
|---|---|---|---|
| T/N | <2.5 | ||
| Carcinoma | 8 | 13 | 21 |
| Adenoma | 30 | 4 | 34 |
| Total | 38 | 17 | 55 |
| DKK4 | P = 0.014 | ≤40 | Total |
|---|---|---|---|
| T/N | <40 | ||
| Carcinoma | 9 | 9 | 18 |
| Adenoma | 26 | 5 | 31 |
| Total | 35 | 14 | 49 |
Correlation between expression of DKKs and FGF20 in colorectal tumors. As shown in Figure 2(b), there was a positive correlation between DKK4 and FGF20 (rs = 0.61, P = 0.00017) in both colorectal cancers and colorectal adenomas. In contrast, no relationship was found between FGF20 and the other three DKKs: rs = –0.0015, P = 0.99 for DKK1; rs = 0.17, P = 0.28 for DKK2; and rs = –0.038, P = 0.80 for DKK3.
Correlation between nuclear β‐catenin accumulation and expression of DKK4. Figure 2(c) (upper panel) shows results from six representative samples. Specimens with high nuclear β‐catenin accumulation (#1, #2, and #3) also strongly expressed DKK4 (T/N ratios of 559, 283, and 81.2, respectively). On the other hand, specimens with little nuclear β‐catenin accumulation (#4, #5, and #6), only weakly expressed DKK4 (T/N ratios of 2.39, 2.21, and 0.61, respectively). The other 12 cancer samples were also stained for β‐catenin, confirming the correlation between nuclear β‐catenin accumulation and DKK4 expression (data not shown).
Immunohistochemical staining for DKK4 was also done in these samples, and was intense in samples #1, #2, and #3, which showed high nuclear β‐catenin accumulation, but was weak or negative in #4, #5, and #6, which showed little nuclear β‐catenin accumulation (Fig. 2c, lower panel).
Immunohistochemical staining for β‐catenin and DKK4 was also performed in a sample obtained by endoscopic mucosal resection, which included normal mucosa, adenoma, and invasive cancer in the same specimen. Figure 2d shows that both nuclear β‐catenin and DKK4 were most intensely stained in invasive cancer, less so in adenoma, and weakly in normal mucosa, demonstrating that the expression of nuclear β‐catenin perfectly correlates with DKK4.
In vitro analysis of the DKK4 gene in the Wnt/canonical pathway. As shown in Figure 3(a), western blot analysis demonstrated that the DKK4 protein was upregulated only by the dominant‐active mutant form of β‐catenin, but not by wild‐type β‐catenin. This indicates that DKK4 is upregulated by activation of the Wnt/canonical pathway in vitro.
Figure 3.
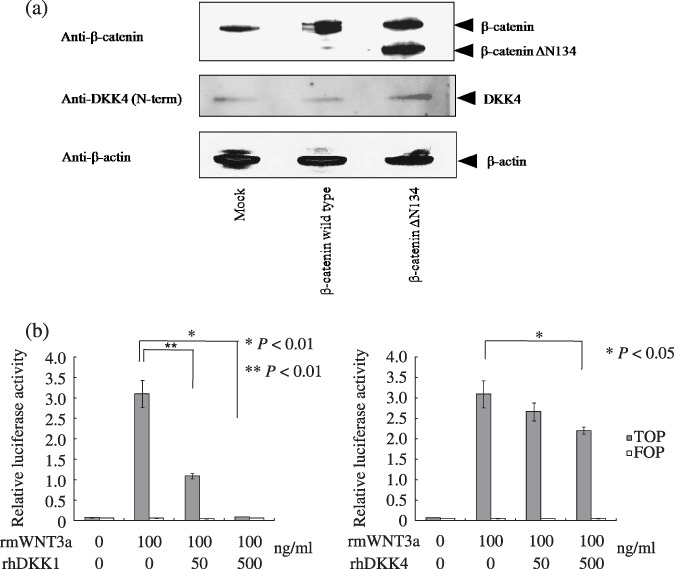
In vitro analysis of Dickkopf (DKK)‐4 gene in the Wnt/canonical pathway using human embryonic kidney (HEK)‐293 cells. (a) Whole cell lysate from HEK293 cells transfected with empty pcDNA3.1neo (Mock), pFLAG‐β‐catenin wild type (β‐catenin wild type), or pFLAG‐β‐catenin ΔN134 (β‐catenin ΔN134), analyzed by immunoblotting with anti‐β‐catenin, anti‐DKK4 (N‐term), and anti‐β‐actin antibodies. (b) Functional involvement of DKK1 and ‐4 in activation of the Wnt‐canonical pathway. HEK293 cells were cotransfected with canonical (super 8 × TOP‐FLASH, solid columns) or mutant (super 8 × FOP‐FLASH, open columns) transcription factor T‐cell factor/lymphocyte enhancer factor (TCF/LEF) luciferase reporters, and pRL‐TK. Five hours after transfection, medium was changed and rhDKK1 and ‐4 was added. rmWnt3a was added 12 h later and luciferase activity was measured 12 h after that. Bars, SD.
In Figure 3(b), addition of rmWnt3a significantly increased the luciferase activity of super 8 × TOP‐FLASH over the control (8 × FOP‐FLASH). However, addition of 500 ng/mL of rhDKK1 together with 100 ng/mL rmWnt3a to the culture medium resulted in significantly decreased luciferase activity (Fig. 3b, left panel). Although the effect was less marked than that of rhDKK1, rhDKK4 also significantly decreased luciferase activity (Fig. 3b, right panel), demonstrating that DKK4 acts as an inhibitor of the Wnt/canonical signaling pathway in non‐tumor cells (Student's t‐test, P < 0.05).
Discussion
In the present study, we investigated expression of DKK family genes in colorectal tumors using biopsy samples. We found that all four DKKs, but especially DKK2 and ‐4, were significantly upregulated in primary colorectal cancers relative to adjacent mucosae. We also found that upregulation of DKKs except DKK3 was already observed in adenomas. Moreover, we demonstrated a significant positive relationship in primary colorectal tumors at the gene expression level between DKK4 and FGF20, and between DKK4 and β‐catenin nuclear accumulation, suggesting that DKK4 is a target gene of the Wnt/canonical pathway. Subsequent in vitro analysis showed DKK4 was indeed induced by the activation of the Wnt/canonical pathway.
Prior to the screening for DKKs, an SSH analysis performed for a single patient with advanced colon cancer had identified eight known genes (DKK4, TOG, BUB1, CBWD, GABA‐A receptor, CEA, NCA, and Maspin) as those isolated more than once from 100 independent clones overexpressed in a colon cancer tissue relative to adjacent mucosa. In addition to previous data on DKK4, other studies had reported upregulation and/or possible contributions of these seven genes to colorectal carcinogenesis (TOG,( 19 ) BUB1,( 20 , 21 ) CBWD,( 22 , 23 ) CEA,( 24 ) NCA,( 24 ) GABA‐A receptor,( 25 ) and Maspin( 26 )). This agreement with previous results suggests that the SSH analysis performed here was reliable and appropriate even though the data were obtained from a single patient.
Secreted modulator molecules in the Wnt pathway such as Wnt inhibitory factor‐1, the secreted Frizzled‐related protein (sFRP) family, and the DKK family have recently been focused on regarding their role in carcinogenesis of different cancers. Because these molecules modulate activation of the Wnt signaling pathway, and that pathway is considered to promote cancer progression, they are themselves believed to function as modulators of tumor progression. Importantly, it was reported that ectopic expression of sFRP1, ‐2 and ‐5, which is known to be downregulated in colon cancer cells, attenuates Wnt/canonical signaling even in the presence of downstream mutations of APC or β‐catenin.( 27 ) Regarding the DKKs, there are several studies to date investigating their role in human colorectal cancers. For example, it was reported that CpG islands of DKKs are frequently methylated in colorectal cancers,( 13 , 14 ) suggesting downregulation of the expression of DKKs in cancer. It was also reported that the DKK4 gene harbors only a small number of relevant CpG islands in its regulatory region to be silenced by CpG methylation.( 14 , 28 ) Downregulation of DKK1( 29 ) and upregulation of DKK3 in colorectal cancer has also been reported.( 30 ) Here, we quantified the expression of all the DKK family members simultaneously and found that DKK4 and ‐2 were both significantly upregulated in primary colorectal cancers and even in adenomas, although to a lesser extent.
Recently, DKK4 was reported to harbor seven TCF1/LEF1 consensus binding sites, and to be induced by activation of the Wnt/canonical pathway in mice and HEK293T cells.( 31 ) A dominant‐active mutant form of β‐catenin upregulates DKK4 in a murine granulosa cell tumor model.( 32 ) On the other hand, the DKK4 gene seems to lack typical CpG islands( 14 , 28 ) as alluded to above. Considering those previous studies, we propose that significant upregulation of DKK4 in colorectal cancer, in which the Wnt/canonical pathway is frequently activated, might have directly reflected the activity of Wnt signaling. Therefore, we examined how DKK4 expression and the Wnt/canonical pathway might be related, and showed a positive correlation between FGF20 and DKK4 expression in primary colorectal cancers and adenomas by real‐time RT‐PCR analysis. We also demonstrated that cancers with high DKK4 expression showed significant nuclear β‐catenin accumulation by immunohistochemistry. In addition, we confirmed that DKK4 protein was strongly expressed in the colorectal cancer epithelium, and furthermore, was perfectly co‐localized with β‐catenin. Finally, we confirmed the induction of DKK4 by a dominant‐active mutant form of β‐catenin in HEK293 cells in vitro. All these results demonstrated that DKK4 is a downstream target of the Wnt/canonical pathway, and its upregulation in primary colorectal cancer reflects the activation of that pathway.
During the preparation of this report, DKK4 dysregulation in colorectal cancer was also reported on by two other groups.( 28 , 33 ) Pendas‐Franco et al. reported upregulation of DKK4 mRNA in 20 of 29 cancers relative to normal mucosae by real‐time RT‐PCR analysis,( 33 ) consistent with our results. However, these investigators showed upregulation of DKK4 mRNA only in colorectal cancers, whereas we document here that DKK4 upregulation is not limited to cancer, but is already present also in adenomas. Moreover, we confirmed upregulation of DKK4 at the protein level by immunohistochemical analysis in colorectal cancers. In contrast, Baehs et al. reported downregulation of DKK4 in 12 of 21 cancers using laser‐captured micro‐dissected samples,( 28 ) contrary to our results. Although the reason for this discrepancy is unclear, in addition to demonstration of DKK4 upregulation by mRNA quantification, we also clearly showed that DKK4 was strongly stained in colorectal cancer epithelium in situ compared to normal mucosa by immunohistochemical analysis, indicating that DKK4 was truly upregulated in colorectal cancer.
For DKK2, to our knowledge, the present study is the first to demonstrate strong expression in clinical samples of colorectal cancers and adenomas. However, the methylation status of DKK2 was already reported to be high (65–70%), suggesting that DKK2 is a target of epigenetic silencing in colorectal tumors.( 13 , 14 ) Although the reason for these potentially discrepant results is also not clear, they are not necessarily mutually exclusive, since tumor tissue consists of heterogeneous cell populations. We have investigated the methylation status of DKK2 in several cancer tissues of our series by methylation‐specific PCR, and found that both methylated and unmethylated bands were observed in most of them (data not shown). This suggests that tumor cells with unmethylated DKK2 overexpress this gene. However, if this were true, what up‐regulates DKK2 in tumor cells? We found six TCF/LEF consensus binding motifs within 2kbp upstream of the transcriptional start site in DKK2 using Genomatix software (http://www.genomatix.de/products/index.html), indicating that DKK2 may also be a downstream target of the Wnt/canonical pathway. However, our finding that DKK2 expression did not parallel the expression of FGF20 implied that some unknown regulatory mechanism other than the activation of Wnt/canonical signaling might exist. This requires investigation in further studies.
Regarding DKK1 and ‐3, we demonstrated that both genes were also upregulated in colorectal tumors, although to a lesser degree than the other two DKKs. Previous studies reported that DKK1 is downregulated in colorectal cancer, contrary to our results.( 29 ) However, a recent report by Maehata et al. showed that DKK1 tended to be upregulated, consistent with our results.( 13 ) For DKK3, the result of a previous study that this was upregulated in colorectal cancer also coincides with our result.( 30 )
What is the role of these DKKs in the carcinogenesis of colorectal cancer? In the present study, we confirmed that DKK4 inhibited Wnt‐induced activation of the Wnt/canonical pathway in HEK293 cells (Fig. 3b). However, we did not observe inhibition of this pathway when rhDKK4 protein was added to the culture medium of colon cancer cell lines SW480 and LoVo (data not shown). Considering that the Wnt/canonical pathway is activated in most human colorectal cancers as a result of APC loss and mutation or β‐catenin mutation,( 34 ) it is possible that DKK4 overexpression is a side effect of Wnt/canonical signal activation. However, it was recently reported that ectopic DKK4 expression increased the migratory and invasive properties of colon cancer cells, and conditioned media from DKK4‐expressing cells enhanced the ability of human primary microvascular endothelial cells to migrate and form capillary‐like tubules.( 33 ) This suggests that DKK4 also has effects in addition to inhibition of the Wnt/canonical signal. For DKK2, we still do not have any data regarding its function. However, as it was reported to act as either an inhibitor or an activator of the Wnt/canonical signal in the normal setting, it is possible that DKK2 functions as a tumor suppressor or tumor activator depending on the circumstance. Further studies are clearly needed.
In conclusion, the present study demonstrated upregulation of DKK4 and ‐2 in human colorectal cancers and that upregulation of DKK4 reflected activation of the Wnt/canonical pathway. Clarifying the mechanisms responsible for this upregulation would further contribute to our understanding of the molecular pathogenesis of colorectal cancer.
Disclosure Statement
We (all authors) do not have a commercial or other association that might pose a conflict of interest.
Supporting information
Table S1. PCR primer sequences for upregulatedgenes detected by suppression subtraction hybridization (SSH) analysis
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgments
We are grateful to Dr Tesshi Yamada and Dr Miki Shitashige (NCCRI, Tokyo, Japan) for their generous donation of the plasmids pFLAG‐cmv4‐beta‐catenin WT and pFLAG‐cmv4‐beta‐catenin ΔN134. This study was supported by a Grant‐Aid for Scientific Research (no. 30397301) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1. Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev 2005; 19: 877–90. [DOI] [PubMed] [Google Scholar]
- 2. Mao B, Wu W, Davidson G et al . Kremen proteins are Dickkopf receptors that regulate Wnt/beta‐catenin signalling. Nature 2002; 417: 664–7. [DOI] [PubMed] [Google Scholar]
- 3. Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 2006; 25: 7469–81. [DOI] [PubMed] [Google Scholar]
- 4. Politou MC, Heath DJ, Rahemtulla A et al . Serum concentrations of Dickkopf‐1 protein are increased in patients with multiple myeloma and reduced after autologous stem cell transplantation. Int J Cancer 2006; 119: 1728–31. [DOI] [PubMed] [Google Scholar]
- 5. Wirths O, Waha A, Weggen S et al . Overexpression of human Dickkopf‐1, an antagonist of wingless/WNT signaling, in human hepatoblastomas and Wilms’ tumors. Lab Invest 2003; 83: 429–34. [DOI] [PubMed] [Google Scholar]
- 6. Patil MA, Chua MS, Pan KH et al . An integrated data analysis approach to characterize genes highly expressed in hepatocellular carcinoma. Oncogene 2005; 24: 3737–47. [DOI] [PubMed] [Google Scholar]
- 7. Voorzanger‐Rousselot N, Goehrig D, Journe F et al . Increased Dickkopf‐1 expression in breast cancer bone metastases. Br J Cancer 2007; 97: 964–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng S, Miao C, Li J, Fan X, Cao Y, Duan E. Dickkopf‐1 induced apoptosis in human placental choriocarcinoma is independent of canonical Wnt signaling. Biochem Biophys Res Commun 2006; 350: 641–7. [DOI] [PubMed] [Google Scholar]
- 9. Kuphal S, Lodermeyer S, Bataille F, Schuierer M, Hoang BH, Bosserhoff AK. Expression of Dickkopf genes is strongly reduced in malignant melanoma. Oncogene 2006; 25: 5027–36. [DOI] [PubMed] [Google Scholar]
- 10. Tsuji T, Miyazaki M, Sakaguchi M, Inoue Y, Namba M. A REIC gene shows down‐regulation in human immortalized cells and human tumor‐derived cell lines. Biochem Biophys Res Commun 2000; 268: 20–4. [DOI] [PubMed] [Google Scholar]
- 11. Kurose K, Sakaguchi M, Nasu Y et al . Decreased expression of REIC/Dkk‐3 in human renal clear cell carcinoma. J Urol 2004; 171: 1314–8. [DOI] [PubMed] [Google Scholar]
- 12. Aung PP, Oue N, Mitani Y et al . Systematic search for gastric cancer‐specific genes based on SAGE data: melanoma inhibitory activity and matrix metalloproteinase‐10 are novel prognostic factors in patients with gastric cancer. Oncogene 2006; 25: 2546–57. [DOI] [PubMed] [Google Scholar]
- 13. Maehata T, Taniguchi H, Yamamoto H et al . Transcriptional silencing of Dickkopf gene family by CpG island hypermethylation in human gastrointestinal cancer. World J Gastroenterol 2008; 14: 2702–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sato H, Suzuki H, Toyota M et al . Frequent epigenetic inactivation of DICKKOPF family genes in human gastrointestinal tumors. Carcinogenesis 2007; 28: 2459–66. [DOI] [PubMed] [Google Scholar]
- 15. Chomczynski P, Sacchi N. Single‐step method of RNA isolation by acid guanidinium thiocyanate‐phenol‐chloroform extraction. Anal Biochem 1987; 162: 156–9. [DOI] [PubMed] [Google Scholar]
- 16. Matz M, Shagin D, Bogdanova E et al . Amplification of cDNA ends based on template‐switching effect and step‐out PCR. Nucleic Acids Res 1999; 27: 1558–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. FGF‐20 and DKK1 are transcriptional targets of beta‐catenin and FGF‐20 is implicated in cancer and development. Embo J 2005; 24: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shitashige M, Naishiro Y, Idogawa M et al . Involvement of splicing factor‐1 in beta‐catenin/T‐cell factor‐4‐mediated gene transactivation and pre‐mRNA splicing. Gastroenterology 2007; 132: 1039–54. [DOI] [PubMed] [Google Scholar]
- 19. Charrasse S, Mazel M, Taviaux S, Berta P, Chow T, Larroque C. Characterization of the cDNA and pattern of expression of a new gene over‐expressed in human hepatomas and colonic tumors. Eur J Biochem 1995; 234: 406–13. [DOI] [PubMed] [Google Scholar]
- 20. Cahill DP, Lengauer C, Yu J et al . Mutations of mitotic checkpoint genes in human cancers. Nature 1998; 392: 300–3. [DOI] [PubMed] [Google Scholar]
- 21. Abal M, Obrador‐Hevia A, Janssen KP et al . APC inactivation associates with abnormal mitosis completion and concomitant BUB1B/MAD2L1 up‐regulation. Gastroenterology 2007; 132: 2448–58. [DOI] [PubMed] [Google Scholar]
- 22. Wade J, Tang YP, Peabody C, Tempelman RJ. Enhanced gene expression in the forebrain of hatchling and juvenile male zebra finches. J Neurobiol 2005; 64: 224–38. [DOI] [PubMed] [Google Scholar]
- 23. Kune G, Watson L. Colorectal cancer protective effects and the dietary micronutrients folate, methionine, vitamins B6, B12, C, E, selenium, and lycopene. Nutr Cancer 2006; 56: 11–21. [DOI] [PubMed] [Google Scholar]
- 24. Kodera Y, Isobe K, Yamauchi M et al . Expression of carcinoembryonic antigen (CEA) and nonspecific crossreacting antigen (NCA) in gastrointestinal cancer; the correlation with degree of differentiation. Br J Cancer 1993; 68: 130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szczaurska K, Mazurkiewicz M, Opolski A. [The role of GABA‐ergic system in carcinogenesis]. Postepy Hig Med Dosw 2003; 57: 485–500. (In Polish.) [PubMed] [Google Scholar]
- 26. Dietmaier W, Bettstetter M, Wild PJ et al . Nuclear Maspin expression is associated with response to adjuvant 5‐fluorouracil based chemotherapy in patients with stage III colon cancer. Int J Cancer 2006; 118: 2247–54. [DOI] [PubMed] [Google Scholar]
- 27. Suzuki H, Watkins DN, Jair KW et al . Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004; 36: 417–22. [DOI] [PubMed] [Google Scholar]
- 28. Baehs S, Herbst A, Thieme SE et al . Dickkopf‐4 is frequently down‐regulated and inhibits growth of colorectal cancer cells. Cancer Lett 2009; 276: 152–9. [DOI] [PubMed] [Google Scholar]
- 29. Gonzalez‐Sancho JM, Aguilera O, Garcia JM et al . The Wnt antagonist DICKKOPF‐1 gene is a downstream target of beta‐catenin/TCF and is downregulated in human colon cancer. Oncogene 2005; 24: 1098–103. [DOI] [PubMed] [Google Scholar]
- 30. Zitt M, Untergasser G, Amberger A et al . Dickkopf‐3 as a new potential marker for neoangiogenesis in colorectal cancer: expression in cancer tissue and adjacent non‐cancerous tissue. Dis Markers 2008; 24: 101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CA, Christiano AM. The Wnt inhibitor, Dickkopf 4, is induced by canonical Wnt signaling during ectodermal appendage morphogenesis. Dev Biol 2007; 305: 498–507. [DOI] [PubMed] [Google Scholar]
- 32. Boerboom D, White LD, Dalle S, Courty J, Richards JS. Dominant‐stable beta‐catenin expression causes cell fate alterations and Wnt signaling antagonist expression in a murine granulosa cell tumor model. Cancer Res 2006; 66: 1964–73. [DOI] [PubMed] [Google Scholar]
- 33. Pendas‐Franco N, Garcia JM, Pena C et al . DICKKOPF‐4 is induced by TCF/beta‐catenin and upregulated in human colon cancer, promotes tumour cell invasion and angiogenesis and is repressed by 1alpha,25‐dihydroxyvitamin D3. Oncogene 2008; 27: 4467–77. [DOI] [PubMed] [Google Scholar]
- 34. Giles RH, Van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta 2003; 1653: 1–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. PCR primer sequences for upregulatedgenes detected by suppression subtraction hybridization (SSH) analysis
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


