Abstract
To explore the expression of leukemia‐related protein 16 (LRP16) in invasive ductal breast carcinoma and analyze its correlation with clinicopathological feature and prognosis, immunohistochemistry was performed on 100 cases of invasive ductal breast carcinoma. Medical records were reviewed and clinicopathological analysis was performed. Leukemia‐related protein 16 expression was detected in 33 of 100 cases (33%) of the invasive ductal breast carcinoma. Expression of LRP16 in carcinoma was obviously higher than that in normal breast tissue. LRP16 protein expression was found in 27.6% (21/76) of carcinoma at stage I and II, and 50.0% (12/24) of carcinoma at stage III and IV. LRP16 expression was found correlative with metastasis in the axillary lymph node (P = 0.001), stage (P = 0.042), estrogen receptor (ER) expression (P = 0.001), fragile histidine triad (FHIT) expression (P = 0.015) and CD133 expression (P = 0.038), but not with grade (P = 0.543), tumor size (P = 0.263), age (P = 0.840), menopause (P = 0.701) and HER‐2 gene amplification (P = 0.463). The difference of the mean disease free survival (DFS) time between cancer patients with LRP16 expression (43.7 months) and those without (77.7 months) was statistically significant (Log rank = 9.989, P = 0.002). The difference of the mean overall survival (OS) time between cancer patients with LRP16 expression (50.0 months) and those without (120.0 months) was statistically significant (Log rank = 9.977, P = 0.002). Our finding suggests that expression of LRP16 protein is correlated with the stage, metastasis, prognosis and expression of ER, progesterone receptor, Ki‐67, CD133 and FHIT in invasive ductal breast carcinoma. (Cancer Sci 2010)
Breast cancer has become the second most frequent cause of death in women, threatening females all over the world. In China, the incidence of breast cancer increases very rapidly and breast cancer has become the most common female malignant tumor. In spite of advances in diagnosis and treatment, almost one‐fourth of women with this neoplasm will die. The major causes of treatment failure and/or death for breast cancer patients are tumor recurrence and metastasis. The use of adjuvant and palliative therapies in patients with breast carcinoma rely primarily on prognostic factors, such as tumor grade and size, axillary nodal status, distant metastasis and candidate biomarkers, such as hormone receptor (nuclear estrogen receptor [nER] and progesterone receptor [PR]) expression, and c‐erbB2/HER‐2/neu amplification/overexpression. Furthermore, expression of hormone receptors and overexpression of c‐erbB2 help in guiding therapeutic strategies and predict response to chemotherapy, endocrine therapy and specific immunotherapy with the antibody, trastuzumab. Therefore, such biomarkers in breast neoplasms provide information regarding the outcome of patients. A study in search of additional biomarkers is necessary for patients with breast cancer.
Leukemia‐related protein 16 (LRP16), which was originally recognized and isolated from human lymphocytes in 1999, was identified as an estrogen‐responsive gene.( 1 ) It localizes on chromosome 11q12.1 and encodes nuclear factor.( 1 , 2 , 3 , 4 , 5 ) It is expressed in testicle, ovaries, mucosa of colon, prostate, small intestine, spleen, thymus( 2 , 3 ) and gastric or colorectal carcinoma.( 6 , 7 ) Leukemia‐related protein 16 is also an estrogen receptor α (ERα) coactivator. Its expression level is strongly dependent on estrogen activities. It is involved in the estrogen signaling pathway and can strengthen the ERα responsive gene activation.
Previously, the mRNA level of LRP16 in breast carcinoma was found to be higher than normal breast tissues by Northern blot (40.9%) and semi‐quantitative reverse transcription–polymerase chain reaction (RT‐PCR) (30.0%).( 8 ) Leukemia‐related protein 16 overexpression is closely correlated to positive rates of ER and PR, Ki‐67 level, tumor diameter and axillary lymph node metastasis of breast cancer, and might be involved in the proliferation and metastasis of human breast cancer. However, no immunohistochemical and clinicopathological studies with follow‐up data of LRP16 protein have been performed in invasive ductal breast carcinoma.
CD133 is a pentaspan transmembrane glycoprotein, with a molecular weight of 120 kDa.( 9 ) Although it was initially considered to be a marker of hematopoietic stem cells, CD133 mRNA transcript could also be found in normal non‐lymphoid hematopoietic tissue.( 9 ) CD133 is overexpressed in various solid tumors,( 10 , 11 , 12 , 13 , 14 ) including colon cancer and glioblastoma.( 15 , 16 ) Recently, detecting expression of CD133 in invasive ductal breast carcinomas has been reported by Liu et al. ( 17 ) and CD133 expression may be of help in more accurately predicting the aggressive properties of breast cancer and determining optimal treatment. However, the authors did not perform the survival analysis by follow up. CD133 expression as a prognostic marker has been found in colorectal cancer( 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 ) and brain tumors,( 26 , 27 , 28 ) but it is still unclear if CD133 can be used as a prognostic marker in pancreatic cancer,( 29 ) ovarian cancer,( 30 ) hepatocellular cancer( 31 , 32 ) and non‐small cell lung carcinoma.( 33 ) There is no report on CD133 expression in the prognosis of invasive ductal breast carcinoma. Like the significance of CD133,( 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 ) we have also recently reported( 7 ) that LRP16 is a prognostic factor in colorectal cancer. It is interesting to explore the correlation between CD133 and LRP16 in invasive ductal breast carcinoma. FHIT is a well‐recognized putative tumor suppressor associated with the prognosis of breast cancer.( 34 , 35 ) and recently the importance of FHIT has been raised as the guardian of the preneoplastic genome.( 36 ) We wonder if there is any association between FHIT and LRP16. Amplification of the HER‐2 gene, as well as expression of ER, PR and Ki‐67 is also a well‐recognized prognostic and molecular targeting marker associated with the prognosis in breast cancer.( 37 , 38 ) There has been no investigation into the correlation between expression of LRP16 and expression of FHIT, CD133, ER, PR, Ki‐67 and amplification of HER‐2 in breast cancer so far. Here, we investigated the expression of LRP16 protein in 100 invasive ductal breast carcinoma specimens by immunohistochemistry and fluorescence in situ hybridization (FISH), and explored the possible correlation between expression of LRP16 protein and expression of FHIT, CD133, ER, PR and Ki‐67, amplification of the HER‐2 gene and clinicopathological features in invasive ductal breast carcinoma.
Materials and Methods
Patients and specimens. One hundred patients who had undergone modified radical mastectomy for treatment of invasive ductal breast carcinomas during 1998–2000 at the Chinese People’s Liberation Army Hospital, Beijing, China were confirmed histologically and were enrolled in this study. Ethical approval for the study was not required by our institution as the experiments carried out did not relate to patient’s privacy, impairment or treatment. Paraffin tissue of tumor specimens were retrieved from the archives of the Department of Pathology. Clinical information, such as tumor size, grade, stage and axillary lymph node status were obtained from medical records and the pathology reports (Table 1).
Table 1.
Correlation between leukemia‐related protein 16 (LRP16) expression and clinicopathological features in 100 patients with invasive ductal breast carcinoma
| Clinicopathological features | LRP16 | P‐value | |
|---|---|---|---|
| Positive (%) | Negative | ||
| Age (years) | |||
| ≥50 | 13 (34.2) | 25 | 0.840 |
| <50 | 20 (32.3) | 42 | |
| Menopause | |||
| Yes | 13 (36.1) | 23 | 0.701 |
| No | 20 (31.2) | 44 | |
| Tumor size | |||
| ≤2 cm | 4 (20.0) | 16 | 0.263 |
| >2∼≤5 cm | 25 (34.7) | 47 | |
| >5 cm | 4 (50.0) | 4 | |
| Axillary lymph node metastasis | |||
| Negative | 8 (16.7) | 40 | 0.004 |
| 1–3 | 11 (47.8) | 12 | |
| ≥4 | 14 (48.3) | 15 | |
| Grade | |||
| I∼II | 26 (35.6) | 47 | 0.474 |
| III | 7 (25.9) | 20 | |
| Stage | |||
| I∼II | 21 (27.6) | 55 | 0.042 |
| III∼IV | 12 (50.0) | 12 | |
| HER‐2 amplification | |||
| Positive | 10 (40.0) | 15 | 0.463 |
| Negative | 23 (30.7) | 52 | |
| ER expression | |||
| Positive | 28 (46.7) | 32 | 0.001 |
| Negative | 5 (12.5) | 35 | |
| PR expression | |||
| Positive | 25 (42.4) | 34 | 0.019 |
| Negative | 8 (19.5) | 33 | |
| CD133 expression | |||
| Positive | 23 (41.8) | 32 | 0.038 |
| Negative | 10 (22.2) | 35 | |
| FHIT expression | |||
| Positive | 19 (26.0) | 54 | 0.015 |
| Negative | 14 (51.9) | 13 | |
| Ki‐67 expression | |||
| Positive | 24 (46.2) | 28 | 0.005 |
| Negative | 9 (18.8) | 39 | |
ER, estrogen receptor; FHIT, fragile histidine triad; HER‐2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Immunohistochemical analysis. Immunohistochemical staining was done on 3–4‐μm slides from formalin‐fixed, paraffin‐embedded tissues. Paraffin slides were then deparaffinized in xylene and rehydrated. Antigen retrieval was performed with slides heated in 0.01 M citrate buffer (pH 6.0) in a microwave oven for 5 min at 100°C. After antigen retrieval, the slides were then cooled in running tap water. The slides were rinsed with PBS and the endogenous peroxidase was inactivated with 3% hydrogen peroxide. After blocking with 10% goat serum, the slides were incubated with primary polyclonal rabbit antibody to human LRP16 (recognized and isolated in 1999 by the Department of Molecular Biology of our hospital) diluted 1:1000, rabbit monoclonal antibody to human ER, PR, Ki‐67 (Zymed Laboratories Inc., South San Francisco, CA, USA) and rabbit polyclonal antibody to human CD133 (Abcam Inc., Cambridge, MA, USA) and to human FHIT (Zymed Laboratories Inc.) diluted 1:100 in blocking solution overnight at 4°C. The sections were rinsed in PBS and incubated for 20 min with polyperoxidase‐anti‐mouse/rabbit IgG (Zymed Laboratories Inc.) and then peroxidase reactivity was visualized using a 3,3′‐Diaminobenzidine (DAB) substrate kit (Zymed Labratories Inc.). Finally, the sections were counterstained with hematoxylin and mounted. Negative control sections were incubated with normal rabbit serum instead of the primary antibody. Positive and negative controls were included in each run.
Evaluation of immunohistochemistry. Antigen expression was evaluated independently by two authors using light microscopy. Both observers were unaware of the clinical outcome. Equivocal cases were re‐assessed on a double‐headed microscope to establish a final score. For each sample, at least five fields (inside the tumor and in the area exhibiting tumor invasion; ×400) and >500 cells were analyzed. Using a semiquantitative scoring system( 39 ) microscopically and referring to each antigen scoring method in other studies,( 40 , 41 ) two observers evaluated the intensity, extent and subcellular distribution of LRP16, CD133 and FHIT. In scoring LRP16 protein expression, both the extent and intensity of immunopositivity in the cell nucleus were considered.( 6 ) Scores were applied as follows: score 0, negative staining in all cells; score 1+, weakly positive or focally positive staining in <10% of the cells; score 2+, moderately positive staining covering 10–50% of the cells; and score 3+, strongly positive staining, including >50% of the cells. For statistical analysis, as well as to reduce intraobserver variability, the immunohistochemical scores were further grouped into two categories: negative (0 and 1+) and positive (2+ and 3+).( 17 )
FISH for HER‐2 gene amplification. Human epidermal growth factor receptor 2 (HER‐2) gene amplification was detected by fluorescence in situ hybridization (FISH) on all tumors. The FISH analysis was performed with the commercially available double‐color FISH probe (PathVysion; Vysis, Abbott Park, IL, USA) consisted of two probes: 17q11.2‐q12 (labeled with Spectrum Orange) covering the whole HER‐2 gene; and the control, centromeric chromosome 17p11.1‐q11.1 (labeled with Spectrum Green) hybridizing the alpha satellite DNA. The FISH‐fixed glass microscope slides with tissue sections were baked overnight at 65°C, deparaffinized in two 10‐min changes of xylene, transferred through two 3‐min changes of 100% ethanol, one 3‐min change of 85% ethanol, one 3‐min change of 70% ethanol and immersed for 15 min in pure water at 90°C. The slides were then incubated for 7–15 min in protease solution at 37°C. The slides were then briefly washed in ×2 sodium saline citrate (×2 SSC; pH 7.2) at room temperature, dehydrated through 70%, 85%, 100% ethanol and acetone, and then allowed to air dry. To denature the DNA, the slides were placed in 78.5°C preheated 70% formamide/×2 SSC for 8 min and then dehydrated in a graded series of concentrations of ethanol that were precooling in −20°C. After drying in the open‐air, 10 μL of probe, which was destructured at 75.5°C for 7 min was applied onto each slide, the cover slip was placed and sealed with rubber cement, and then hybridized overnight at 42.8°C. After 16–18 h of hybridization, the slides were washed in 46°C preheated post‐hybridization buffer (×2 SSC/0.1% sodium dodecyl sulfate) for 5 min and rinsed in 70% ethanol. After air‐drying (out of direct light), the slides were counterstained with 15 μL DAPI/anti‐fade solution and the cover slip was applied.
The FISH analysis was performed by two pathologists who were blinded to the clinical diagnoses at the time of the evaluation. The slides were scanned using an OLYMPUS BX51 fluorescent microscope (OLYMPUS BX51, Tokyo, Japan) equipped with a 100‐watt mercury lamp and single band pass filter set to detect DAPI, Rhodamine (17q11.2‐q12) and FITC (chromosome 17) at ×1000. Thirty randomly selected invasive tumor nuclei in each of two separate, distinct microscopic areas were evaluated. Cases were scored as negative by FISH when the HER‐2 with a HER‐2 to chromosome enumeration probe (CEP) 17 ratio <1.8 by counting at least 30 interphase nuclei, and those cases with a HER‐2 to CEP 17 ratio >2.2 were scored as positive. In addition, more randomly selected invasive tumor nuclei (for example, a total of 100 nuclei) would be evaluated if the HER‐2 with a HER‐2 to CEP 17 ratio was between 1.8 and 2.2.( 42 , 43 )
Statistical analysis. Fisher’s exact test (two‐sided), Pearson Chi‐squared test for trends in proportions, Spearman’s correlation coefficient test, and Kaplan–Meier’s method with log rank test or Cox Regression method for univariate or multivariate overall survival analysis were used to assess the associations between expression of CD133 or FHIT and clinicopathological indices by SPSS 15.0 for Windows (Chicago, IL, USA). A value of P < 0.05 was considered statistically significant.
Results
Clinicopathological characteristics of the patients and tumors. The age of the patients ranged from 28–92 years, with an average of 49 years. Seventeen were at grade 1, 55 at grade 2 and 27 at grade 3, according to the histological grading. Sixteen were at stage I, 60 at stage II, 21 at stage III and 4 at stage IV, according to the clinical staging of TNM, respectively. Lymphatic metastasis in regional nodes at operation was confirmed in 52 cancers in the present study. All 100 women were followed after surgical treatment for a mean period of 45.3 months (range, 8–131 months); 33 cases recurred and 17 cases died. The details of patient characteristics and descriptive statistics for the tumors are shown in Table 1.
Correlation between expression of the LRP16 protein and clinicopathological features. Expression of the LRP16 protein was positive in 33 (33.0%) of 100 invasive ductal breast carcinomas. Leukemia‐related protein 16 was expressed in the nucleus (Fig. 1) or predominantly in the nucleus and minimally in the cytoplasm of the tumor cell. Leukemia‐related protein 16 was also found to be positive in the nucleus of cancer cells at the invasive front (Fig. 2). The relationships between LRP16 expression and the clinicopathological features of the tumors are shown in Table 1. The LRP16 expression level was high in tumors with metastasis in the axillary lymph nodes (48.1%), stage III∼IV (50.0%) and significantly correlated with metastasis of the axillary lymph nodes (P = 0.004) and the clinical stage (P = 0.042). There was no statistically significant association between LRP16 expression and age, menopause, tumor size and grade (Table 1).
Figure 1.
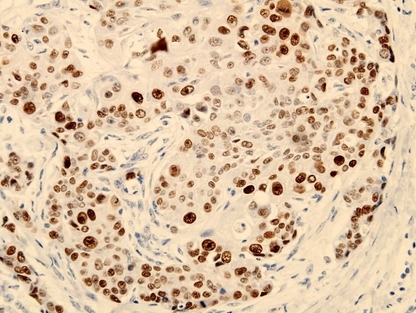
Expression of leukemia‐related protein 16 (LRP16) in invasive ductal breast carcinoma. LRP16 was expressed positive in the nucleus of cancer cells. (LRP16, ×400.)
Figure 2.
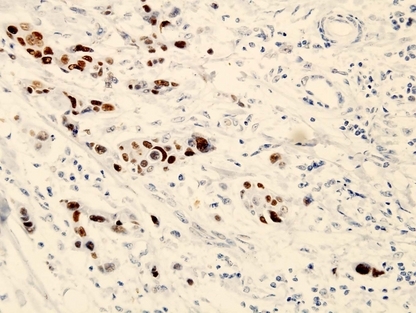
Expression of leukemia‐related protein 16 (LRP16) in invasive ductal breast carcinoma. LRP16 was expressed positive in the nucleus of cancer cells at the invasive front. (LRP16, ×400.)
Correlation between expression of LRP16 and expression of ER, PR, Ki‐67, FHIT, CD133 or amplification of HER‐2. Estrogen receptor expression was positive in 60 (60%) of 100 cases of invasive ductal breast carcinoma and inversely associated with histological grade (P = 0.01), but not with metastasis in the axillary lymph nodes (P = 0.936) and the clinical stage (P = 0.448). By Kaplan–Meier analysis, the overall mean survival time was 118.5 months for ER‐positive patients, but 52.1 months for ER‐negative cases (Log Rank = 6.897, P = 0.009); the disease‐free mean survival time was 78.3 months for ER‐positive patients, but 44.4 months for ER‐negative cases (Log Rank = 6.426, p = 0.011). The expression of LRP16 protein was correlated with the expression of ER (P = 0.001), PR (P = 0.019) and Ki‐67 (P = 0.005) (Table 1). In the ER‐positive cases, six patients died between 18 and 50 months; five of them were LRP16 positive. Whereas in the ER‐nagative cases, 11 patients died between 8–37 months; six of them were LRP16 positive. Forty‐three patients received endocrine therapy in this group of 60 patients with ER‐positive expression. Among them only three (3/43) patients died between 21–37 months and all three cases showed positive expression of LRP16 protein in their carcinomas. Sixty‐five patients received chemotherapy and 12 of them were dead between 13–41 months, in which six cases (50%) were carcinomas with LRP16 positive.
FHIT expression was positive in 73 (73%) of 100 cases of invasive ductal breast carcinomas (Fig. 3) and inversely associated with histological grade (P = 0.026), but not with tumor size (P = 0.569), metastasis in the axillary lymph nodes (P = 0.176) and the clinical stage (P = 0.787). An inverse correlation between LRP16 and FHIT was found (P = 0.015) (Table 1).
Figure 3.
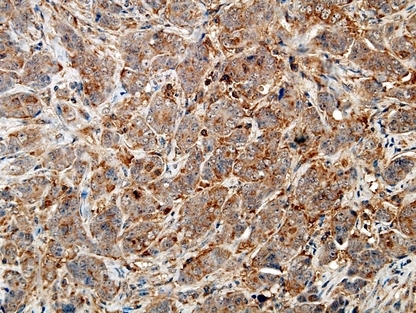
Expression of FHIT protein in invasive ductal breast carcinoma. FHIT was expressed positive in the cytoplasm of cancer cells. (FHIT, ×400.)
CD133 expression was positive in 55 (55%) of 100 invasive ductal breast carcinomas. CD133 was expressed in the tumor cell membrane and cytoplasm (Fig. 4). The CD133 expression level was high in tumors with metastasis in more than or equal to four axillary lymph nodes (72.4%), stage III∼IV (79.2%) and grade III carcinoma (60.7%), and significantly correlated with metastasis of the axillary lymph nodes (P = 0.010) and clinical stage (P = 0.006). There was no statistically significant association between CD133 expression and tumor size (P = 0.575) or grade (P = 0.479). Correlation between LRP16 expression and CD133 expression was found (P = 0.038) (Table 1).
Figure 4.
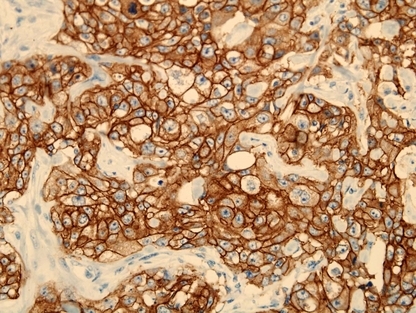
Expression of CD133 protein in invasive ductal breast carcinoma. CD133 was expressed positive in the membrane and cytoplasm of cancer cells. (CD133, ×400.)
The HER‐2 gene was amplified in 25 (25%) of 100 cases of invasive ductal breast carcinomas (Fig. 5) and correlated positively with the clinical stage (P = 0.037) and inversely with FHIT (P = 0.027), but not with tumor size (P = 0.375), histological grade (P = 0.115) or metastasis in the axillary lymph nodes (P = 0.360). Correlation between the expression of LRP16 and amplification of HER‐2 was not found to be significant (P = 0.463) (Table 1).
Figure 5.
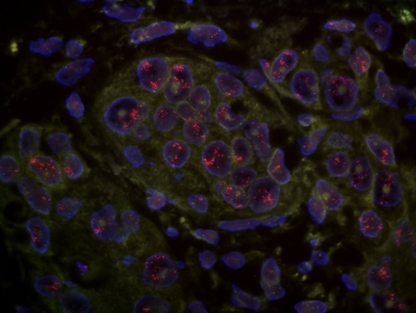
Amplification of the human epidermal growth factor receptor 2 (HER‐2) gene in invasive ductal breast carcinoma. The HER‐2 gene (red) was amplified positive in the nucleus of cancer cells. (HER‐2, ×1000.)
Survival. In 100 cases of invasive ductal breast carcinoma, the prognosis for LRP16‐positive patients was significantly poorer than that for LRP16‐negative patients (P = 0.024; Table 2). Moreover, in the 100 patients with a modified radical mastectomy, the overall survival rate and the disease‐free survival rate for LRP16‐positive patients was significantly poorer than that of LRP16‐negative patients (P = 0.002, P = 0.002, 6, 7). In the 100 invasive ductal breast carcinoma patients, a univariate analysis (Table 2) revealed the overall survival to significantly correlate with expression of LRP16 (P = 0.002), CD133 (P = 0.002), FHIT (P = 0.0001), amplification of HER‐2 (P = 0.001), clinical stage (P = 0.009), lymph node metastasis (P = 0.023) and the number of lymph node metastasis (P = 0.004). The disease‐free survival (Table 2) significantly correlates with expression of LRP16 (P = 0.002), CD133 (P = 0.001), FHIT (P = 0.0001), amplification of HER‐2 (P = 0.010), histological grade (P = 0.020), lymph node metastasis (P = 0.0001) and the number of lymph node metastasis (P = 0.0001). A multivariate analysis revealed expression of LRP16 was an independent prognostic factor (P = 0.040) (Table 3).
Table 2.
Results of univariate analyses of DFS and OS
| Marker | N | Mean DFS (months) | P‐value | Mean OS (months) | P‐value |
|---|---|---|---|---|---|
| HER‐2 | |||||
| + | 25 | 41.72 ± 4.55 | 0.010 | 48.33 ± 4.29 | 0.001 |
| − | 75 | 75.54 ± 4.01 | 117.59 ± 4.62 | ||
| ER | |||||
| + | 60 | 78.34 ± 4.10 | 0.011 | 118.55 ± 4.94 | 0.009 |
| − | 40 | 44.40 ± 3.72 | 52.13 ± 3.11 | ||
| PR | |||||
| + | 57 | 82.16 ± 3.91 | 0.0001 | 122.07 ± 4.40 | 0.0001 |
| − | 43 | 42.15 ± 3.59 | 51.05 ± 3.06 | ||
| CD133 | |||||
| + | 55 | 43.88 ± 3.03 | 0.0001 | 52.56 ± 2.56 | 0.002 |
| − | 45 | 85.60 ± 4.27 | 124.77 ± 4.40 | ||
| FHIT | |||||
| + | 73 | 81.88 ± 3.53 | 0.0001 | 123.68 ± 3.16 | 0.0001 |
| − | 27 | 30.18 ± 3.63 | 44.36 ± 4.16 | ||
| LRP16 | |||||
| + | 33 | 43.66 ± 3.44 | 0.002 | 50.21 ± 3.21 | 0.002 |
| − | 67 | 77.68 ± 4.30 | 120.21 ± 4.24 | ||
| Ki‐67 | |||||
| + | 52 | 44.79 ± 3.10 | 0.011 | 53.39 ± 2.60 | 0.019 |
| − | 48 | 80.41 ± 4.59 | 118.89 ± 5.89 | ||
DFS, disease‐free survival; ER, estrogen receptor; FHIT, fragile histidine triad; HER‐2, human epidermal growth factor receptor 2; OS, overall survival; PR, progesterone receptor.
Figure 6.
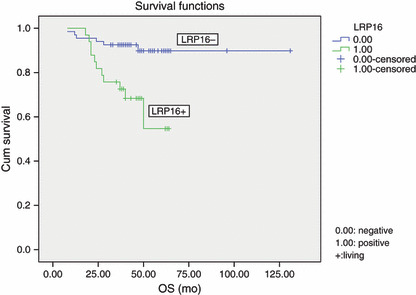
Kaplan–Meier survival analysis by leukemia‐related protein 16 (LRP16) status (n = 100). The y‐axis represents the percentage of patients, and the x‐axis represents their survival in months (“censored” means living). The dotted line represents the LRP16‐positive patients with a trend of worse survival than the solid line representing the LRP16‐negative patients (Log rank = 9.98; P = 0.002). Mean overall survival (OS) time was 50.2 months for the LRP16‐positive group and 120.2 months for the LRP16‐negative group.
Figure 7.
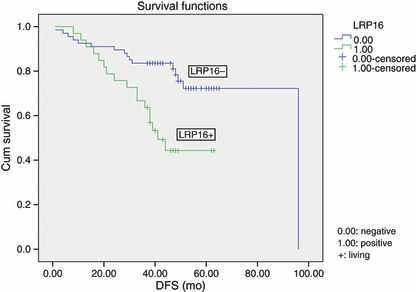
Kaplan–Meier survival analysis by leukemia‐related protein 16 (LRP16) status (n = 100). The y‐axis represents the percentage of patients, and the x‐axis represents their survival in months (“censored” means living). The dotted line represents the LRP16‐positive patients with a trend of worse survival than the solid line representing the LRP16‐negative patients (Log rank = 9.99; P = 0.002). The mean disease‐free survival (DFS) time was 43.7 months for the LRP16‐positive group and 77.7 months for the LRP16‐negative group.
Table 3.
Results of multivariate analyses of overall survival time
| Variables | B | SE | Wald | df | Sig. | Exp(B) |
|---|---|---|---|---|---|---|
| Age | 0.879 | 0.800 | 1.206 | 1 | 0.272 | 2.408 |
| Menopause | −0.154 | 1.017 | 0.023 | 1 | 0.880 | 0.857 |
| Size | 0.210 | 0.587 | 0.128 | 1 | 0.721 | 1.234 |
| Grade | 0.653 | 0.522 | 1.567 | 1 | 0.211 | 1.921 |
| Stage | 0.172 | 0.388 | 0.197 | 1 | 0.657 | 1.188 |
| HER‐2 | 2.562 | 0.636 | 0.780 | 1 | 0.377 | 1.754 |
| ER | −1.380 | 1.086 | 1.567 | 1 | 0.204 | 0.252 |
| PR | −0.855 | 0.973 | 0.773 | 1 | 0.379 | 0.425 |
| CD133 | −0.036 | 0.857 | 0.002 | 1 | 0.967 | 0.965 |
| FHIT | −1.597 | 0.630 | 6.428 | 1 | 0.011 | 0.203 |
| LRP16 | 1.499 | 0.731 | 4.198 | 1 | 0.040 | 4.475 |
| Ki‐67 | 0.540 | 0.798 | 0.458 | 1 | 0.498 | 1.716 |
B, partial regression coefficient; df, degree of freedom; Exp(B), relative risk degree for B; FHIT, fragile histidine triad. HER‐2, human epidermal growth factor receptor 2; SE, standard error; Sig, probability; Wald, statistic for (B/SE)2.
Discussion
Leukemia‐related protein 16 was originally recognized and isolated from human lymphocytes in 1999.( 1 ) It was identified as an estrogen responsive gene.( 1 , 2 , 3 , 4 , 5 ) Expression of LRP16 was found in different tissues in varying degree, including ovary, testicle, prostate, small intestine, spleen, thymus and stomach.( 2 , 3 , 6 ) Furthermore, LRP16 is overexpressed in tumors, compared with their matched normal tissues.( 2 ) Some studies have indicated that LRP16 may play an important role in the carcinogenesis and progression of hormone‐dependent breast cancer.( 5 , 44 ) Overexpression of LRP16 significantly stimulated MCF‐7 cell proliferation by promoting G1/S transition.( 4 ) Suppression of the endogenous LRP16 in ERα‐positive MCF‐7 cells not only inhibits cell growth but also significantly attenuates the cellular estrogen‐responsive proliferation ability and sensitizes tumor cells to radiation.( 44 , 45 , 46 ) However, some authors thought expression of LRP16 in ERα‐negative cells had no effect on proliferation.( 44 ) Expression of the LRP16 gene was dependent on the estrogen activities;( 47 , 48 ) LRP16 was also involved in estrogen signaling and could strengthen the ERα‐responsive gene activation, therefore, it is also considered as an ERα coactivator.( 44 ) A previous study in a smaller sample of breast carcinoma has demonstrated that ERα/PR status, tumor size and axillary lymph node metastasis were closely correlated with LRP16 mRNA overexpression.( 8 ) The present study investigated LRP16 protein in a bigger sample of invasive ductal breast carcinoma with follow‐up data and is first to confirm that LRP16 expression in the protein level was correlative with metastasis in the axillary lymph node (P = 0.004) and clinical stage (P = 0.042). Furthermore, we also found expression of the LRP16 protein was greatly associated with overall survival (P = 0.002) and disease‐free survival (P = 0.002), suggesting LRP16 expression might be a prognostic factor in invasive ductal breast carcinoma. Activation of the ER signaling pathway plays an important role in multi‐tissue development.( 49 , 50 , 51 , 52 ) Therefore, we propose that LRP16, a coactivator of ERα, may display an important function in the carcinogenesis and progression of breast cancer. Besides, with ER, PR and Ki‐67 (all P < 0.05) we also found that LRP16 expression was correlated with FHIT expression (P = 0.015) and CD133 expression (P = 0.038), implying co‐operation of those important gene markers in the mechanism of breast cancer. Although the detailed molecular mechanism involved in this process is unclear, the present study has potential clinical benefits. Leukemia‐related protein 16 expression that could be detected by immunohistochemistry might be a useful molecular marker to predict the prognosis in invasive ductal breast carcinoma patients.
Since endocrine therapy is frequently performed in ER‐positive breast cancer patients, ER‐positive breast carcinoma cases generally show a better prognosis than the ER‐negative patients. It is interesting that LRP16 immunoreactivity was closely associated with poor prognosis, although a great majority of LRP16‐positive breast carcinoma was ER‐positive in our data. In the ER‐positive carcinoma in the present study, there are five of six patients who died with LRP16 positive, suggesting that LRP16 expression may be associated with a poor prognosis in the ER‐positive group. The mechanism is still not clear yet; one explanation may be that LRP16 is closely related to metastasis in the axillary lymph nodes (P = 0.004) and the clinical stage (P = 0.042), but ER is not (P = 0.936 and P = 0.448) in this group of breast carcinoma. Even in 43 cases receiving endocrine therapy, all three cases that died were LRP16 positive, suggesting that LRP16 seems to be a poor prognostic marker in the ER‐positive group by our results. However, it needs further investigation by both basic mechanical research and a larger clinical sample of invasive ductal breast carcinoma. Moreover, LRP16 and ERα could inter‐regulate each other, as LRP16 is an ERα coactivator. This study raises the possibility that anti‐estrogen therapy could be used in patients with high LRP16 expression. This information may help us individualize patient care (e.g. progression and prognosis of patients after operation). In the present study, LRP16 expression was commonly up‐regulated in invasive ductal breast carcinoma and was associated with a shortened survival time of patients in univariate analyses. However, further investigations in larger samples of breast cancer and clinicopathological correlation are necessary to confirm this finding.
Conclusions
Our study has suggested that expression of the LRP16 protein could be correlated with expression of ER, PR, Ki‐67, FHIT and CD133 proteins, and could be considered as a prognostic factor for overall survival and disease‐free survival in invasive ductal breast carcinoma.
Disclosure Statement
The authors declare that they have no competing interests. This research did not receive any specific grant from any funding agency in the public, commercial or not‐for‐profit sector.
Acknowledgments
The authors thank all colleagues in the Department of Molecular Biology, Chinese People’s Liberation Army General Hospital for their help and support with this study. All authors have contributed significantly, and all authors are in agreement with the content of the manuscript.
References
- 1. Yu L, Han WD, Lou FD, Wang QS, Zhao Y, Caligiuri MA. Cloning of leukemia associated gene LRP16 in acute myeloid leukemia. Junyi Jinxiu Xueyuan Xuebao 2000; 21: 81–4. [Google Scholar]
- 2. Han WD, Yu L, Lou FD et al. Cloning and expression characterization of the full length cDNA for a novel leukemia‐associated gene LRP16. Zhongguo Shengwu Huaxue yu Fenzi Shengwu Xuebao 2001; 17: 58–63. [Google Scholar]
- 3. Han WD, Lou FD, Yu L et al. SAGE pattern of LRP16 gene and its expression in normal blood and leukemia cells. Junyi Jinxiu Xueyuan Xuebao 2002; 23: 161–3. [Google Scholar]
- 4. Han WD, Mu YM, Lu XC et al. Estrogen stimulates human breast cancer MCF‐7 cell proliferation by up‐regulation of LRP16 mRNA via activation of estrogen receptor‐α. Zhonghua Neifenmi Daixie Zazhi 2004; 20: 165–8. [Google Scholar]
- 5. Han WD, Mu YM, Lu XC et al. Up‐regulation of LRP16 mRNA by 17beta‐estradiol through activation of estrogen receptor alpha (ERalpha), but not ERbeta, and promotion of human breast cancer MCF‐7 cell proliferation: a preliminary report. Endocr Relat Cancer 2003; 10: 217–24. [DOI] [PubMed] [Google Scholar]
- 6. Li YZ, Zhao P, Han WD. Clinicopathological significance of LRP16 protein in 336 gastric carcinoma patients. World J Gastroenterol 2009; 15: 4833–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xi HQ, Zhao P, Han WD. Clinicopathological significance and prognostic value of LRP16 expression in colorectal carcinoma. World J Gastroenterol 2010; 16: 1644–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao DX, Han WD, Zhao YL et al. Expression and clinical significance of LRP16 gene in human breast cancer. Ai Zheng 2006; 25: 866–70. [PubMed] [Google Scholar]
- 9. Miraglia S, Godfrey W, Yin AH et al. A novel five‐transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 1997; 90: 5013–21. [PubMed] [Google Scholar]
- 10. Hilbe W, Dirnhofer S, Oberwasserlechner F et al. CD133 positive endothelial progenitor cells contribute to the tumour vasculature in non‐small cell lung cancer. J Clin Pathol 2004; 57: 965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh SK, Hawkins C, Clarke ID et al. Identification of human brain tumour initiating cells. Nature 2004; 432: 396–401. [DOI] [PubMed] [Google Scholar]
- 12. Zhou L, Wei X, Cheng L, Tian J, Jiang JJ. CD133, one of the markers of cancer stem cells in Hep‐2 cell line. Laryngoscope 2007; 117: 455–60. [DOI] [PubMed] [Google Scholar]
- 13. Al‐Hajj M, Wicha MS, Benito‐Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100: 3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schrot RJ, Ma JH, Greco CM, Arias AD, Angelastro JM. Organotypic distribution of stem cell markers in formalin‐fixed brain harboring glioblastoma multiforme. J Neurooncol 2007; 85: 149–57. [DOI] [PubMed] [Google Scholar]
- 15. Ricci‐Vitiani L, Ricci‐Vitiani L, Lombardi DG et al. Identification and expansion of human colon‐cancer‐initiating cells. Nature 2007; 445: 111–5. [DOI] [PubMed] [Google Scholar]
- 16. O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007; 445: 106–10. [DOI] [PubMed] [Google Scholar]
- 17. Liu Q, Li J, Zheng X, Jin F, Dong H. Expression of CD133, PAX2, ESA, and GPR30 in invasive ductal breast carcinomas. Chin Med J 2009; 122: 2763–9. [PubMed] [Google Scholar]
- 18. Horst D, Kriegl L, Engel J, Jung A, Kirchner T. CD133 and nuclear beta‐catenin: the marker combination to detect high risk cases of low stage colorectal cancer. Eur J Cancer 2009; 45: 2034–40. [DOI] [PubMed] [Google Scholar]
- 19. Horst D, Kriegl L, Engel J, Kirchner T, Jung A. CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br J Cancer 2008; 99: 1285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kojima M, Ishii G, Atsumi N, Fujii S, Saito N, Ochiai A. Immunohistochemical detection of CD133 expression in colorectal cancer: a clinicopathological study. Cancer Sci 2008; 99: 1578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q, Chen ZG, Du CZ, Wang HW, Yan L, Gu J. Cancer stem cell marker CD133+ tumour cells and clinical outcome in rectal cancer. Histopathology 2009; 55: 284–93. [DOI] [PubMed] [Google Scholar]
- 22. Horst D, Kriegl L, Engel J, Kirchner T, Jung A. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest 2009; 27: 844–50. [DOI] [PubMed] [Google Scholar]
- 23. Li CY, Li BX, Liang Y et al. Higher percentage of CD133+ cells is associated with poor prognosis in colon carcinoma patients with stage IIIB. J Transl Med 2009; 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi D, Lee HW, Hur KY et al. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol 2009; 15: 2258–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horst D, Scheel SK, Liebmann S et al. The cancer stem cell marker CD133 has high prognostic impact but unknown functional relevance for the metastasis of human colon cancer. J Pathol 2009; 219: 427–34. [DOI] [PubMed] [Google Scholar]
- 26. Cheng JX, Liu BL, Zhang X. How powerful is CD133 as a cancer stem cell marker in brain tumors? Cancer Treat Rev 2009; 35: 403–8. [DOI] [PubMed] [Google Scholar]
- 27. Zhang M, Song T, Yang L et al. Nestin and CD133: valuable stem cell‐specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res 2008; 27: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zeppernick F, Ahmadi R, Campos B et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res 2008; 14: 123–9. [DOI] [PubMed] [Google Scholar]
- 29. Maeda S, Shinchi H, Kurahara H et al. CD133 expression is correlated with lymph node metastasis and vascular endothelial growth factor‐C expression in pancreatic cancer. Br J Cancer 2008; 98: 1389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ferrandina G, Martinelli E, Petrillo M et al. CD133 antigen expression in ovarian cancer. BMC Cancer 2009; 9: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song W, Li H, Tao K et al. Expression and clinical significance of the stem cell marker CD133 in hepatocellular carcinoma. Int J Clin Pract 2008; 62: 1212–8. [DOI] [PubMed] [Google Scholar]
- 32. Salnikov AV, Kusumawidjaja G, Rausch V et al. Cancer stem cell marker expression in hepatocellular carcinoma and liver metastases is not sufficient as single prognostic parameter. Cancer Lett 2009; 275: 185–93. [DOI] [PubMed] [Google Scholar]
- 33. Salnikov AV, Gladkich J, Moldenhauer G, Volm M, Mattern J, Herr I. CD133 is indicative for a resistance phenotype but does not represent a prognostic marker for survival of non‐small cell lung cancer patients. Int J Cancer 2010; 126: 950–8. [DOI] [PubMed] [Google Scholar]
- 34. Arun B, Kilic G, Yen C et al. Loss of FHIT expression in breast cancer is correlated with poor prognostic markers. Cancer Epidemiol Biomarkers Prev 2005; 14: 1681–5. [DOI] [PubMed] [Google Scholar]
- 35. Guler G, Uner A, Guler N et al. Concordant loss of fragile gene expression early in breast cancer development. Pathol Int 2005; 55: 471–8. [DOI] [PubMed] [Google Scholar]
- 36. Pichiorri F, Palumbo T, Suh SS et al. Fhit tumor suppressor: guardian of the preneoplastic genome. Future Oncol 2008; 4: 815–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Curigliano G, Viale G, Bagnardi V et al. Clinical relevance of HER2 overexpression/amplification in patients with small tumor size and node‐negative breast cancer. J Clin Oncol 2009; 27: 5693–9. [DOI] [PubMed] [Google Scholar]
- 38. Todorović‐Raković N, Nesković‐Konstantinović Z, Nikolić‐Vukosavljević D. Metastatic breast cancer survival according to HER2 and Topo2a gene status. Dis Markers 2009; 26: 171–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perathoner A, Pirkebner D, Brandacher G et al. 14‐3‐3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin Cancer Res 2005; 11: 3274–9. [DOI] [PubMed] [Google Scholar]
- 40. Immervoll H, Hoem D, Sakariassen PO, Steffensen OJ, Molven A. Expression of the “stem cell marker” CD133 in pancreas and pancreatic ductal adenocarcinomas. BMC cancer 2008; 8: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tong QS, Zheng LD, Tang ST et al. Expression and clinical significance of stem cell marker CD133 in human neuroblastoma. World J Pediatr 2008; 4: 58–62. [DOI] [PubMed] [Google Scholar]
- 42. Bartlett JS, Campbell F, Mallon EA. Determination of HER2 amplification by in situ hybridization: when should chromosome 17 also be determined? Am J Clin Pathol 2008; 130: 920–6. [DOI] [PubMed] [Google Scholar]
- 43. Shah SS, Ketterling RP, Goetz MP et al. Impact of American Society of Clinical Oncology/College of American Pathologists guideline recommendations on HER2 interpretation in breast cancer. Hum Pathol 2010; 41: 103–6. [DOI] [PubMed] [Google Scholar]
- 44. Han WD, Zhao YL, Meng YG et al. Estrogenically regulated LRP16 interacts with estrogen receptor alpha and enhances the receptor’s transcriptional activity. Endocr Relat Cancer 2007; 14: 741–53. [DOI] [PubMed] [Google Scholar]
- 45. Han WD, Zhao YL, Li Q et al. Inhibition of cell proliferation by small interference RNA against LRP16 gene in human breast cancer MCF‐7 cells. Zhongguo Zhongliu Yanjiu Zazhi 2004; 16: 239–45. [Google Scholar]
- 46. Han WD, Yang D, Li Q et al. Improvement of radiation sensitivity by inhibiting expression of the human LRP16 gene in tumor cells. Junyi Jinxiu Xueyuan Xuebao 2005; 26: 183–5. [Google Scholar]
- 47. Zhao YL, Han WD, Li Q et al. Mechanism of transcriptional regulation of LRP16 gene expression by 17‐beta estradiol in MCF‐7 human breast cancer cells. J Mol Endocrinol 2005; 34: 77–89. [DOI] [PubMed] [Google Scholar]
- 48. Lu XC, Lou FD, Han WD et al. Analysis of LRP16 gene promoter activity. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2006; 14: 146–9. [PubMed] [Google Scholar]
- 49. Gerits N, Kostenko S, Moens U. In vivo functions of mitogen‐activated protein kinases: conclusions from knock‐in and knock‐out mice. Transgenic Res 2007; 16: 281–314. [DOI] [PubMed] [Google Scholar]
- 50. Morissette M, Jourdain S, Al Sweidi S, Menniti FS, Ramirez AD, Di Paolo T. Role of estrogen receptors in neuroprotection by estradiol against MPTP toxicity. Neuropharmacology 2007; 52: 1509–20. [DOI] [PubMed] [Google Scholar]
- 51. Zaitsu M, Narita S, Lambert KC et al. Estradiol activates mast cells via a non‐genomic estrogen receptor‐alpha and calcium influx. Mol Immunol 2007; 44: 1977–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morales LB, Loo KK, Liu HB, Peterson C, Tiwari‐Woodruff S, Voskuhl RR. Treatment with an estrogen receptor alpha ligand is neuroprotective in experimental autoimmune encephalomyelitis. J Neurosci 2006; 26: 6823–33. [DOI] [PMC free article] [PubMed] [Google Scholar]


